Synthesis, Characterization and Assessment of Antioxidant and Melanogenic Inhibitory Properties of Edaravone Derivatives
Abstract
:1. Introduction
2. Experimental
2.1. Materials and Methods
2.2. Synthesis of Edaravone Derivatives
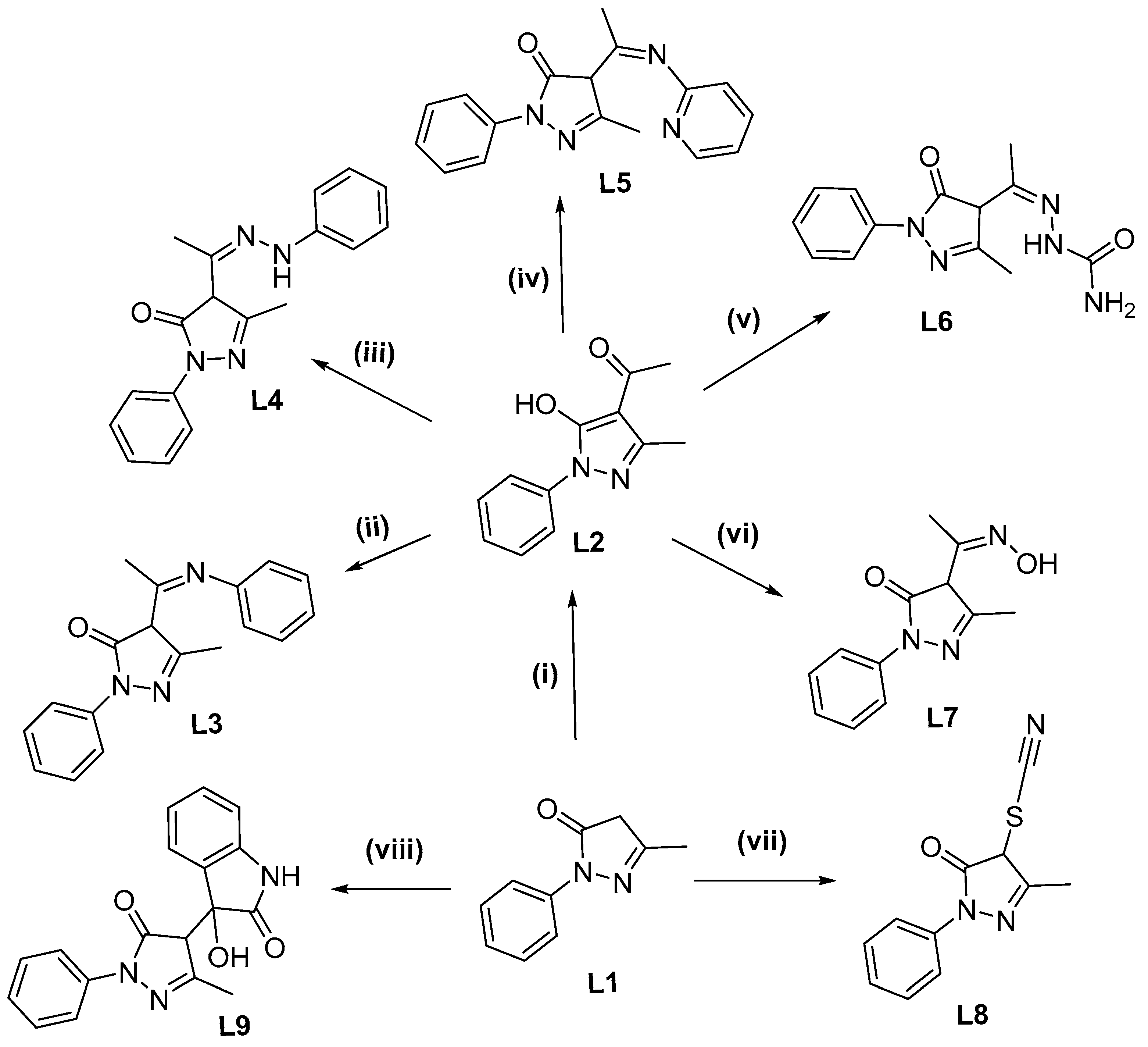
2.2.1. 4-Acetyl-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (L2)
2.2.2. 5-Methyl-2-phenyl-4-(1-(phenylimino)ethyl)-2,4-dihydro-3H-pyrazol-3-one (L3)
2.2.3. 5-Methyl-2-phenyl-4-(1-(2-phenylhydrazineylidene)ethyl)-2,4-dihydro-3H-pyrazol-3-one (L4)
2.2.4. 5-Methyl-2-phenyl-4-(1-(pyridin-2-ylimino)ethyl)-2,4-dihydro-3H-pyrazol-3-one (L5)
2.2.5. 2-(1-(3-Methyl-5-oxo-1-phenyl-4,5-dihydro-1H-pyrazol-4-yl)ethylidene)hydrazine-1-carboxamide (L6)
2.2.6. 4-(1-(Hydroxyimino)ethyl)-5-methyl-2-phenyl-2,4-dihydro-3H-pyrazol-3-one (L7)
2.2.7. 5-Methyl-2-phenyl-4-thiocyanato-2,4-dihydro-3H-pyrazol-3-one (L8)
2.2.8. 3-Hydroxy-3-(3-methyl-5-oxo-1-phenyl-4,5-dihydro-1H-pyrazol-4-yl)indolin-2-one (L9)
2.3. Synthesis of Complexes
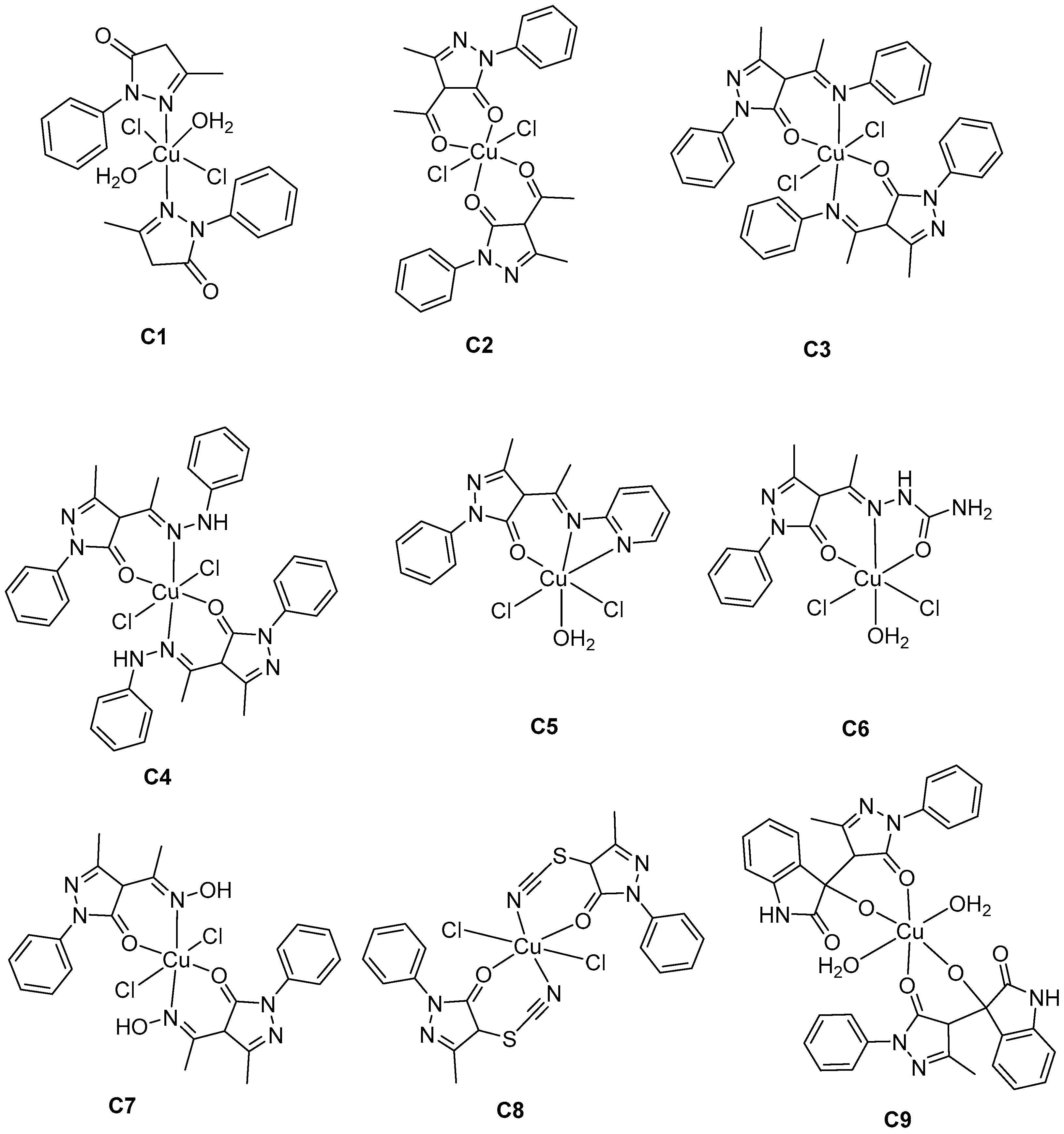
2.3.1. Complex C1
2.3.2. Complex C2
2.3.3. Complex C3
2.3.4. Complex C4
2.3.5. Complex C5
2.3.6. Complex C6
2.3.7. Complex C7
2.3.8. Complex C8
2.3.9. Complex C9
2.4. Evaluation of Antioxidant Properties
2.4.1. Hydrogen Peroxide Scavenging Activity (HPSA) Assay Protocol
2.4.2. Free Radical Scavenging Assay
2.4.3. ABTS (2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) assay
2.4.4. Statistical Analysis
2.5. Computational Studies: Methadology
3. Results and Discussion
3.1. Chemistry
3.1.1. Synthesis
3.1.2. Characterization
Elemental Analysis
Infrared Spectral Analysis
UV–Visible Spectral Analysis
NMR Analysis
Mass Analysis
3.2. Biochemistry: Evaluation of Antioxidant Properties
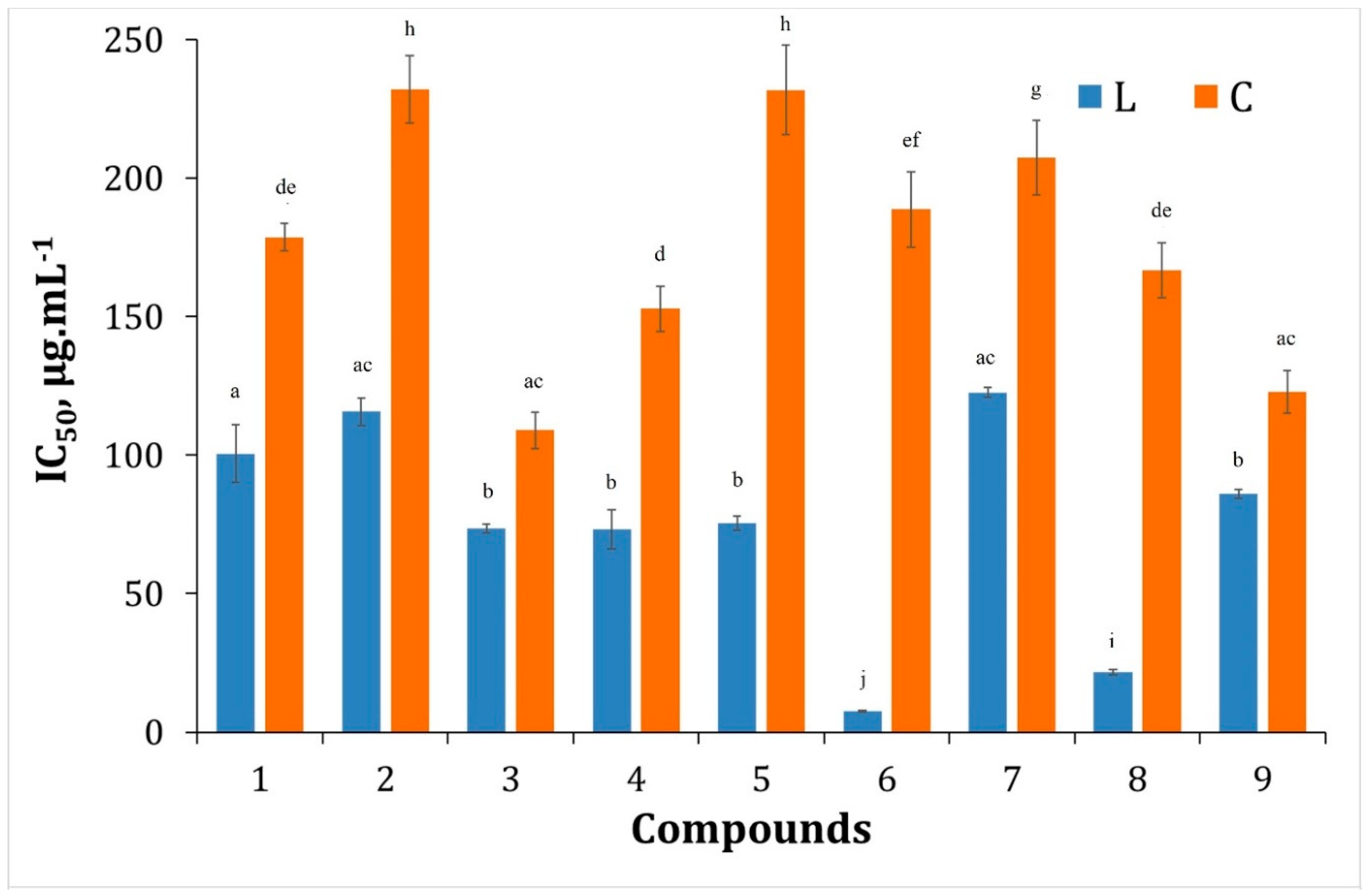
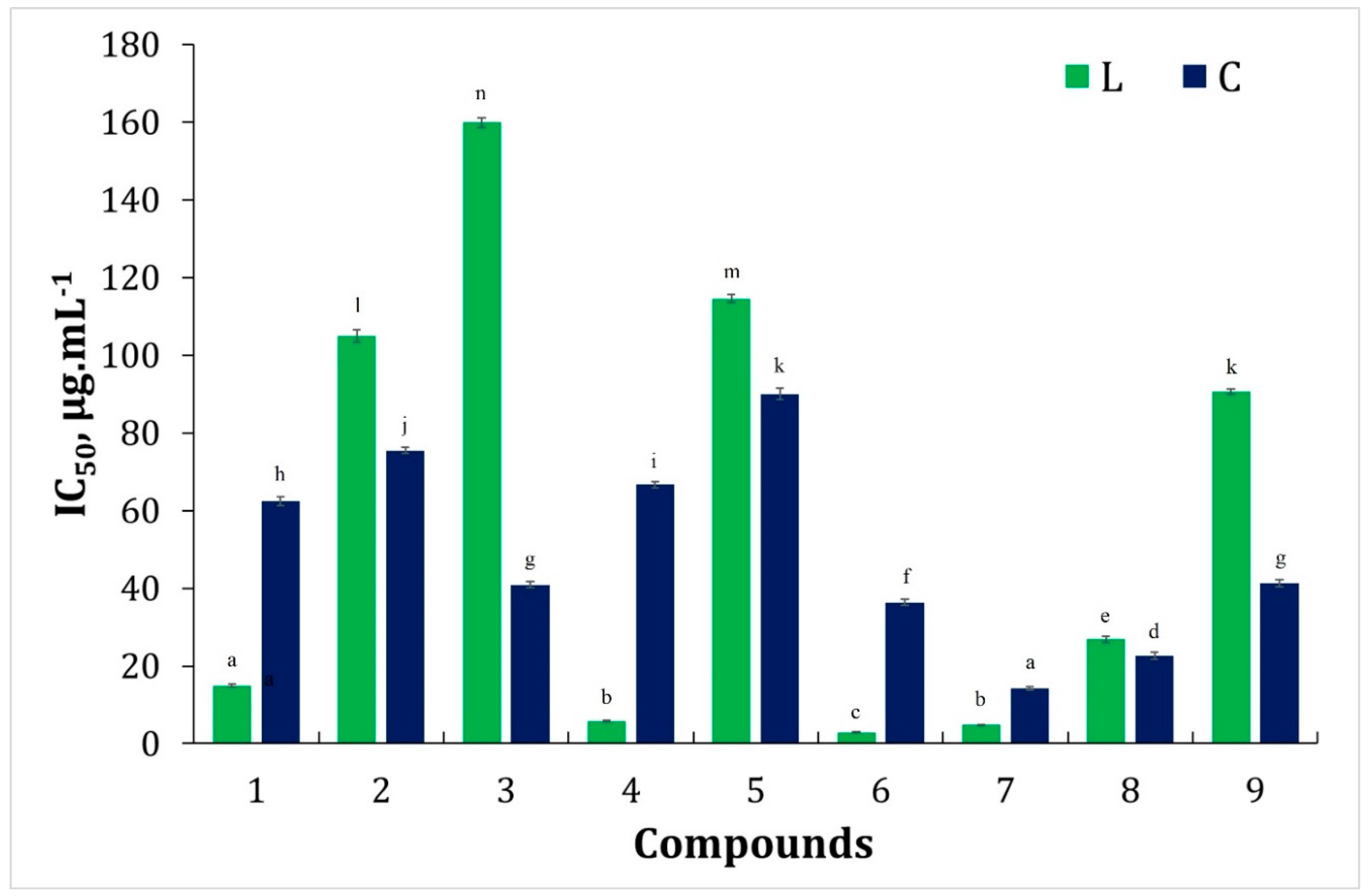
3.2.1. Hydrogen Peroxide Scavenging Activity (HPSA) Assay
3.2.2. DPPH Free Radical Scavenging Activity Assay
3.2.3. ABTS Assay
3.3. Computational Studies
3.3.1. Electronic Properties
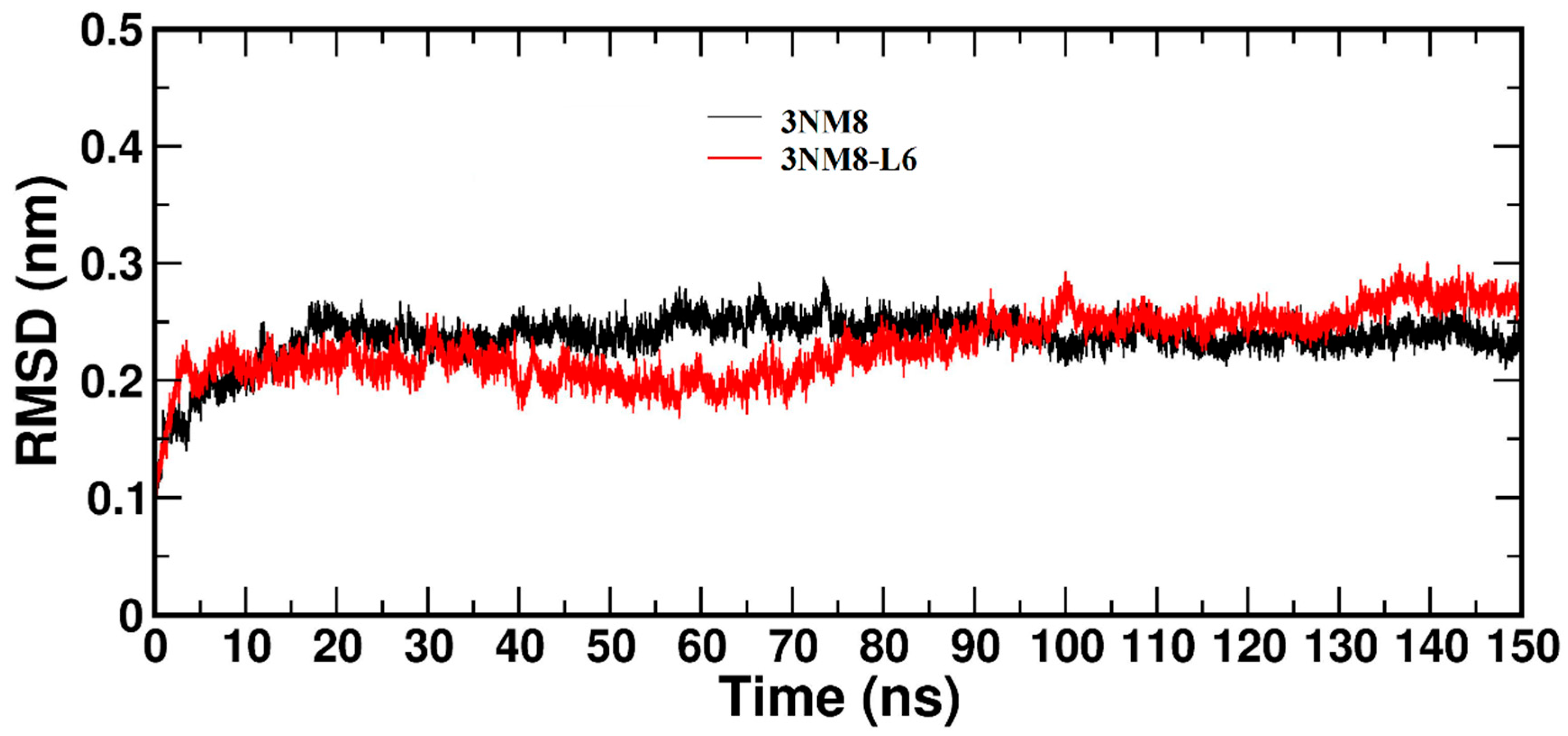
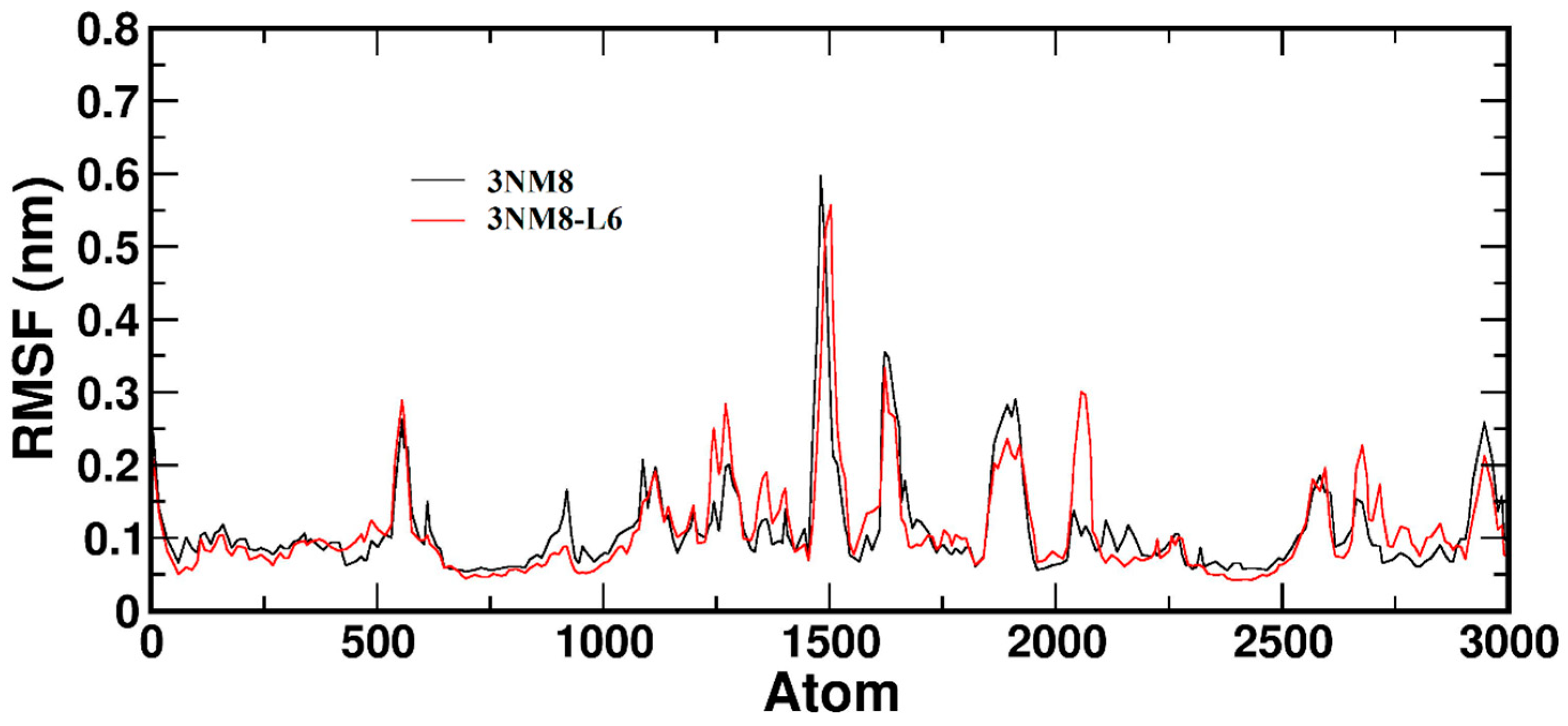
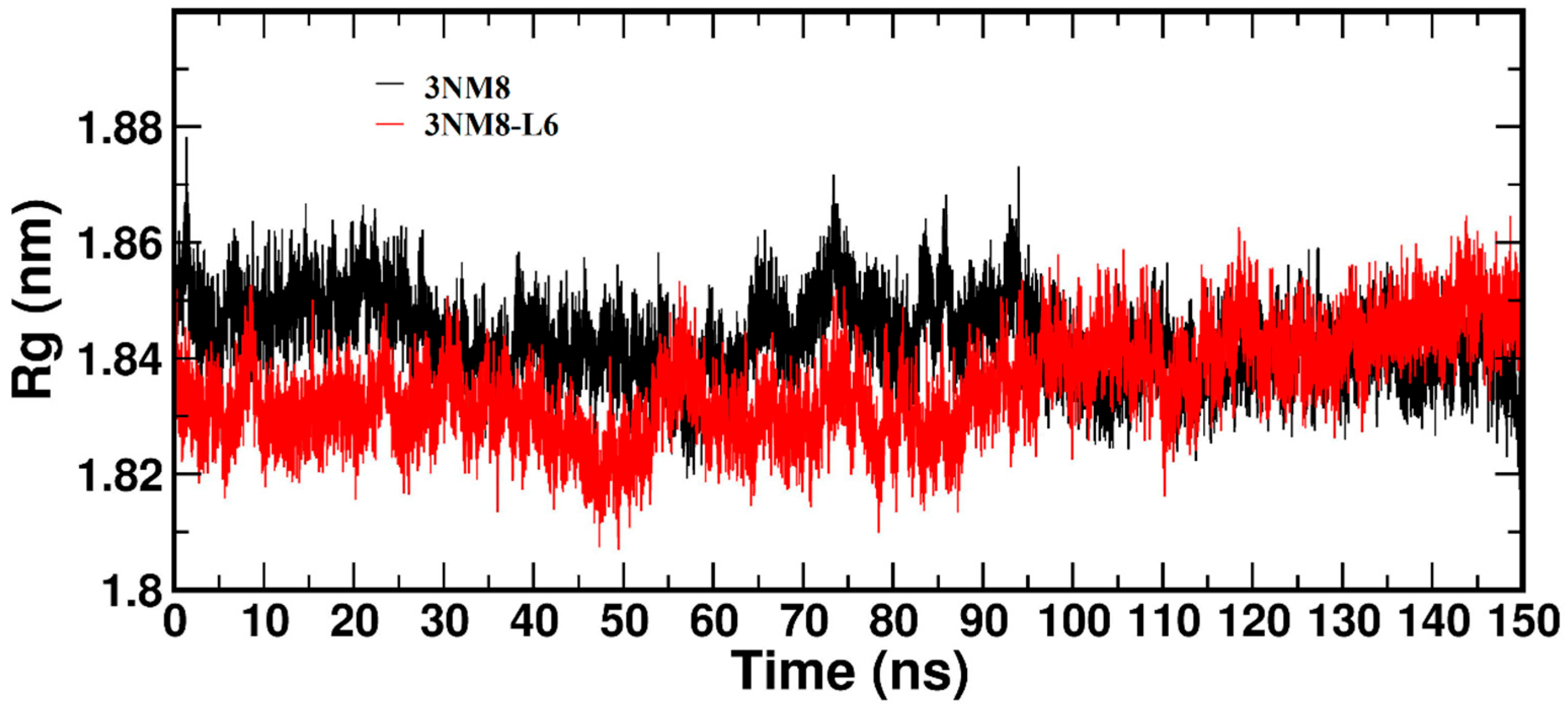
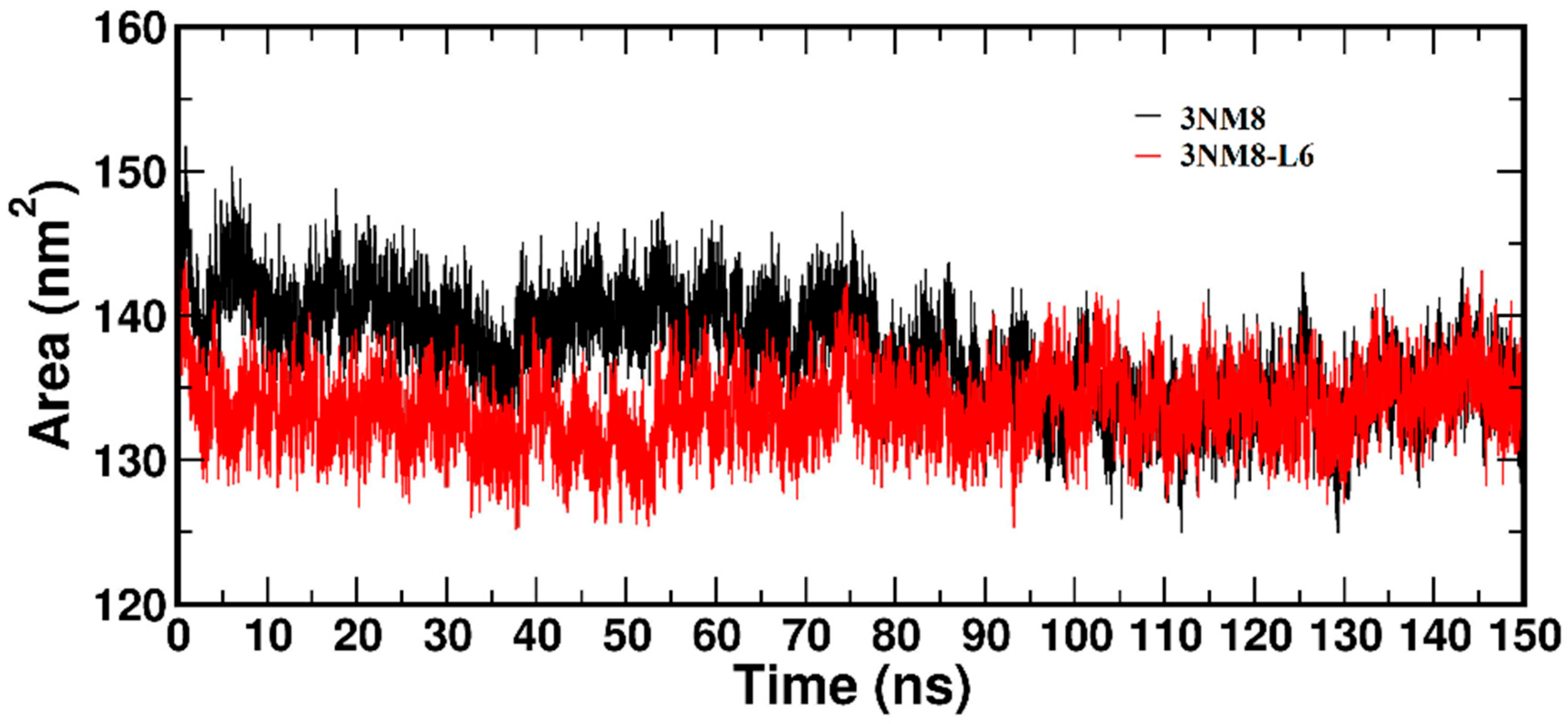
3.3.2. Molecular Dynamics and Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bailly, C.; Hecquet, P.-E.; Kouach, M.; Thuru, X.; Goossens, J.-F. Chemical Reactivity and Uses of 1-Phenyl-3-Methyl-5-Pyrazolone (PMP), Also Known as Edaravone. Bioorganic Med. Chem. 2020, 28, 115463. [Google Scholar] [CrossRef] [PubMed]
- Mohan, D.; Kulkarni, N. Recent Developments in the Design of Functional Derivatives of Edaravone and Exploration of Their Antioxidant Activities. Mol. Divers. 2024. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Tahara, M.; Todo, S. The Novel Antioxidant Edaravone: From Bench to Bedside. Cardiovasc. Ther. 2008, 26, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Abe, K. Update on Antioxidant Therapy with Edaravone: Expanding Applications in Neurodegenerative Diseases. Int. J. Mol. Sci. 2024, 25, 2945. [Google Scholar] [CrossRef] [PubMed]
- Canadian Agency for Drugs and Technologies in Health. Pharmacoeconomic Review Report: Edaravone (Radicava): (Mitsubishi Tanabe Pharma Corporation): Indication: For the treatment of Amyotrophic Lateral Sclerosis (ALS) [Internet]; Canadian Agency for Drugs and Technologies in Health: Ottawa, ON, Canada, 2019. [Google Scholar] [PubMed]
- Nourelden, A.Z.; Kamal, I.; Hagrass, A.I.; Tawfik, A.G.; Elhady, M.M.; Fathallah, A.H.; Eshag, M.M.E.; Zaazouee, M.S. Safety and Efficacy of Edaravone in Patients with Amyotrophic Lateral Sclerosis: A Systematic Review and Meta-Analysis. Neurol. Sci. 2023, 44, 3429–3442. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Shukla, S. Role of Edaravone as a Treatment Option for Patients with Amyotrophic Lateral Sclerosis. Pharmaceuticals 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Potential Use of Edaravone to Reduce Specific Side Effects of Chemo-, Radio- and Immuno-Therapy of Cancers. Int. Immunopharmacol. 2019, 77, 105967. [Google Scholar] [CrossRef]
- Cha, S.J.; Kim, K. Effects of the Edaravone, a Drug Approved for the Treatment of Amyotrophic Lateral Sclerosis, on Mitochondrial Function and Neuroprotection. Antioxidants 2022, 11, 195. [Google Scholar] [CrossRef]
- Chen, C.; Li, M.; Lin, L.; Chen, S.; Chen, Y.; Hong, L. Clinical Effects and Safety of Edaravone in Treatment of Acute Ischaemic Stroke: A Meta-analysis of Randomized Controlled Trials. J. Clin. Pharm. Ther. 2021, 46, 907–917. [Google Scholar] [CrossRef]
- Jayasinghe, M.; Jena, R.; Singhal, M.; Jain, S.; Karnakoti, S.; Silva, M.S.; Kayani, A.M.A. Ethnical Disparities in Response to Edaravone in Patients With Amyotrophic Lateral Sclerosis. Curēus 2022, 14, e25960. [Google Scholar] [CrossRef]
- Rahim, M.; Madi, F.; Nouar, L.; Haiahem, S.; Fateh, D.; Khatmi, D. β-Cyclodextrin Interaction with Edaravone: Molecular Modeling Study. In Advances in Quantum Chemistry; Academic Press: Cambridge, MA, USA, 2014; pp. 269–278. [Google Scholar]
- Pérez-González, A.; Galano, A. On the Outstanding Antioxidant Capacity of Edaravone Derivatives through Single Electron Transfer Reactions. J. Phys. Chem. B 2012, 116, 1180–1188. [Google Scholar] [CrossRef] [PubMed]
- Barajas-Carrillo, V.W.; Estolano-Cobián, A.; Díaz-Rubio, L.; Ayllón-Gutiérrez, R.R.; Salazar-Aranda, R.; Díaz-Molina, R.; García-González, V.; Almanza-Reyes, H.; Rivero, I.A.; Marrero, J.G.; et al. Antioxidant and Acetylcholinesterase Inhibition Activity of Aliphatic and Aromatic Edaravone Derivatives. Med. Chem. Res. 2020, 30, 610–623. [Google Scholar] [CrossRef]
- Minnelli, C.; Laudadio, E.; Galeazzi, R.; Rusciano, D.; Armeni, T.; Stipa, P.; Cantarini, M.; Mobbili, G. Synthesis, Characterization and Antioxidant Properties of a New Lipophilic Derivative of Edaravone. Antioxidants 2019, 8, 258. [Google Scholar] [CrossRef] [PubMed]
- Polkam, N.; Ramaswamy, V.R.; Rayam, P.; Allaka, T.R.; Anantaraju, H.S.; Dharmarajan, S.; Perumal, Y.; Gandamalla, D.; Yellu, N.R.; Balasubramanian, S.; et al. Synthesis, Molecular Properties Prediction and Anticancer, Antioxidant Evaluation of New Edaravone Derivatives. Bioorganic Med. Chem. Lett. 2016, 26, 2562–2568. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, G.; Zia-Ur-Rehman, M.; Sumrra, S.H.; Ashfaq, M.; Zafar, W.; Ashfaq, M. A Critical Review on Recent Trends on Pharmacological Applications of Pyrazolone Endowed Derivatives. J. Mol. Struct. 2022, 1262, 133044. [Google Scholar] [CrossRef]
- Minnelli, C.; Laudadio, E.; Fiorini, R.; Galeazzi, R.; Armeni, T.; Stipa, P.; Rusciano, D.; Mobbili, G. Influence of a Lipophilic Edaravone on Physical State and Activity of Antioxidant Liposomes: An Experimental and in Silico Study. Colloids Surf. B Biointerfaces 2022, 210, 112217. [Google Scholar] [CrossRef]
- Laudadio, E.; Minnelli, C.; Mobbili, G.; Sabbatini, G.; Stipa, P.; Rusciano, D.; Galeazzi, R. Salt Effects on Mixed Composition Membranes Containing an Antioxidant Lipophilic Edaravone Derivative: A Computational-Experimental Study. Org. Biomol. Chem. 2022, 20, 5784–5795. [Google Scholar] [CrossRef]
- Sheng, X.; Hua, K.; Yang, C.; Wang, X.; Ji, H.; Xu, J.; Huang, Z.; Zhang, Y. Novel Hybrids of 3-n-Butylphthalide and Edaravone: Design, Synthesis and Evaluations as Potential Anti-Ischemic Stroke Agents. Bioorganic Med. Chem. Lett. 2015, 25, 3535–3540. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Miao, L.; Guo, Y.; Yuan, R.; Tian, H. Design, Synthesis, and Neuroprotective Effects of Novel Hybrid Compounds Containing Edaravone Analogue and 3-n-Butylphthalide Ring-Opened Derivatives. Biochem. Biophys. Res. Commun. 2021, 556, 99–105. [Google Scholar] [CrossRef]
- Qiang, X.; Li, Y.; Yang, X.; Luo, L.; Xu, R.; Zheng, Y.; Cao, Z.; Tan, Z.; Deng, Y. DL -3- n -Butylphthalide-Edaravone Hybrids as Novel Dual Inhibitors of Amyloid- β Aggregation and Monoamine Oxidases with High Antioxidant Potency for Alzheimer’s Therapy. Bioorganic Med. Chem. Lett. 2017, 27, 718–722. [Google Scholar] [CrossRef]
- Hua, K.; Sheng, X.; Li, T.-T.; Wang, L.-N.; Zhang, Y.-H.; Huang, Z.-J.; Ji, H. The Edaravone and 3-n-Butylphthalide Ring-Opening Derivative 10b Effectively Attenuates Cerebral Ischemia Injury in Rats. Acta Pharmacol. Sin. 2015, 36, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Wu, J.; Ji, D.; Jiao, W.; Wang, X.; Huang, Z.; Zhang, Y. Synthesis and Biological Evaluation of Hybrids from Optically Active Ring-Opened 3-N-Butylphthalide Derivatives and 4-Fluro-Edaravone as Potential Anti-Acute Ischemic Stroke Agents. Bioorganic Med. Chem. 2022, 69, 116891. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Lyu, Y.; Jiang, M.; Tang, B.; Nie, T.; Lu, S. Human Urinary Kallidinogenase or Edaravone Combined with Butylphthalide in the Treatment of Acute Ischemic Stroke. Brain Behav. 2019, 9, e01438. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Li, P.; Liu, Y.; Guo, L.; Wu, Q.; Cheng, Y. Protective Multi-target Effects of DL-3-n-butylphthalide Combined with 3-methyl-1-phenyl-2-pyrazolin-5-one in Mice with Ischemic Stroke. Mol. Med. Rep. 2021, 24, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lapshina, M.A.; Shevtsova, E.F.; Grigoriev, V.V.; Aksinenko, A.Y.; Ustyugov, A.A.; Steinberg, D.A.; Maleev, G.V.; Dubrovskaya, E.S.; Goreva, T.V.; Epishina, T.A.; et al. New Adamantane-Containing Edaravone Conjugates as Potential Neuroprotective Agents for ALS Treatments. Molecules 2023, 28, 7567. [Google Scholar] [CrossRef]
- Zondagh, L.S.; Malan, S.F.; Joubert, J. Design, Synthesis and Biological Evaluation of Edaravone Derivatives Bearing the N-Benzyl Pyridinium Moiety as Multifunctional Anti-Alzheimer’s Agents. J. Enzym. Inhib. Med. Chem. 2020, 35, 1596–1605. [Google Scholar] [CrossRef]
- Thawabteh, A.M.; Jibreen, A.; Karaman, D.; Thawabteh, A.; Karaman, R. Skin Pigmentation Types, Causes and Treatment—A Review. Molecules 2023, 28, 4839. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Namasivayam, V.; Manickam, M.; Jung, S.-H. Inhibitors of Melanogenesis: An Updated Review. J. Med. Chem. 2018, 61, 7395–7418. [Google Scholar] [CrossRef]
- Maddaleno, A.S.; Camargo, J.; Mitjans, M.; Vinardell, M.P. Melanogenesis and Melasma Treatment. Cosmetics 2021, 8, 82. [Google Scholar] [CrossRef]
- Gillbro, J.M.; Olsson, M.J. The Melanogenesis and Mechanisms of Skin-lightening Agents—Existing and New Approaches. Int. J. Cosmet. Sci. 2011, 33, 210–221. [Google Scholar] [CrossRef]
- Kao, Y.-Y.; Chuang, T.-F.; Chao, S.-H.; Yang, J.-H.; Lin, Y.-C.; Huang, H.-Y. Evaluation of the Antioxidant and Melanogenesis Inhibitory Properties of Pracparatum Mungo (Lu-Do Huang). J. Tradit. Complement. Med. 2013, 3, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Mermer, A.; Demirci, S. Recent Advances in Triazoles as Tyrosinase Inhibitors. Eur. J. Med. Chem. 2023, 259, 115655. [Google Scholar] [CrossRef] [PubMed]
- Denat, L.; Kadekaro, A.L.; Marrot, L.; Leachman, S.A.; Abdel-Malek, Z.A. Melanocytes as Instigators and Victims of Oxidative Stress. J. Investig. Dermatol. 2014, 134, 1512–1518. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, P.R.; Starner, R.J.; Swope, V.B.; Wakamatsu, K.; Ito, S.; Abdel-Malek, Z.A. Differential Induction of Reactive Oxygen Species and Expression of Antioxidant Enzymes in Human Melanocytes Correlate with Melanin Content: Implications on the Response to Solar UV and Melanoma Susceptibility. Antioxidants 2022, 11, 1204. [Google Scholar] [CrossRef] [PubMed]
- Snyman, M.; Walsdorf, R.E.; Wix, S.N.; Gill, J.G. The Metabolism of Melanin Synthesis—From Melanocytes to Melanoma. Pigment. Cell Melanoma Res. 2024, 37, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Samsonowicz, M.; Kalinowska, M.; Gryko, K. Enhanced Antioxidant Activity of Ursolic Acid by Complexation with Copper (II): Experimental and Theoretical Study. Materials 2021, 14, 264. [Google Scholar] [CrossRef]
- Hamadouche, S.; Merouani, H.; May, A.A.; Ouddai, N.; Alam, M.; Micoli, L.; Erto, A.; Benguerba, Y. Theoretical Exploration of Enhanced Antioxidant Activity in Copper Complexes of Tetrahydroxystilbenes: Insights into Mechanisms and Molecular Interactions. ACS Omega 2024, 9, 9076–9089. [Google Scholar] [CrossRef]
- Rodríguez-Arce, E.; Saldías, M. Antioxidant Properties of Flavonoid Metal Complexes and Their Potential Inclusion in the Development of Novel Strategies for the Treatment against Neurodegenerative Diseases. Biomed. Pharmacother. 2021, 143, 112236. [Google Scholar] [CrossRef]
- El-Lateef, H.M.A.; El-Dabea, T.; Khalaf, M.M.; Abu-Dief, A.M. Recent Overview of Potent Antioxidant Activity of Coordination Compounds. Antioxidants 2023, 12, 213. [Google Scholar] [CrossRef]
- Williams, D.B.G.; Lawton, M. Drying of Organic Solvents: Quantitative Evaluation of the Efficiency of Several Desiccants. J. Org. Chem. 2010, 75, 8351–8354. [Google Scholar] [CrossRef]
- Vogel, A.I. A Textbook of Quantitative Inorganic Analysis; John Wiley & Sons Inc.: New York, NY, USA, 1966. [Google Scholar]
- Ruch, R.J.; Cheng, S.-J.; Klaunig, J.E. Prevention of Cytotoxicity and Inhibition of Intercellular Communication by Antioxidant Catechins Isolated from Chinese Green Tea. Carcinogenesis 1989, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Manolov, S.; Ivanov, I.; Bojilov, D. Synthesis of New 1,2,3,4-Tetrahydroquinoline Hybrid of Ibuprofen and Its Biological Evaluation. Molbank 2022, 2022, M1350. [Google Scholar] [CrossRef]
- Docheva, M.; Dagnon, S.; Statkova-Abeghe, S. Flavonoid Content and Radical Scavenging Potential of Extracts Prepared from Tobacco Cultivars and Waste. Nat. Prod. Res. 2014, 28, 1328–1334. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant Activity Applying an Improved ABTS Radical Cation Decolorization Assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision A.03; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView, Version 6.1; Semichem Inc.: Shawnee Mission, KS, USA, 2016. [Google Scholar]
- Ullah, Z.; Sattar, F.; Kim, H.J.; Jang, S.; Mary, Y.S.; Zhan, X.; Kwon, H.W. Computational Study of Pd–Cd Bimetallic Crystals: Spectroscopic Properties, Hirshfeld Surface Analysis, Non-Covalent Interaction, and Sensor Activity. J. Mol. Liq. 2022, 365, 120111. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Sendovski, M.; Kanteev, M.; Ben-Yosef, V.S.; Adir, N.; Fishman, A. First Structures of an Active Bacterial Tyrosinase Reveal Copper Plasticity. J. Mol. Biol. 2011, 405, 227–237. [Google Scholar] [CrossRef]
- Schüttelkopf, A.W.; Van Aalten, D.M.F. PRODRG: A Tool for High-Throughput Crystallography of Protein–Ligand Complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef]
- Mary, Y.S.; Mary, Y.S.; Rad, A.S.; Yadav, R.; Celik, I.; Sarala, S. Theoretical Investigation on the Reactive and Interaction Properties of Sorafenib—DFT, AIM, Spectroscopic and Hirshfeld Analysis, Docking and Dynamics Simulation. J. Mol. Liq. 2021, 330, 115652. [Google Scholar] [CrossRef]
- Prasanth, D.S.N.B.K.; Murahari, M.; Chandramohan, V.; Panda, S.P.; Atmakuri, L.R.; Guntupalli, C. In Silicoidentification of Potential Inhibitors fromCinnamonagainst Main Protease and Spike Glycoprotein of SARS CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 4618–4632. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. G_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Nakum, K.; Jadeja, R.N. Synthesis, Characterization, and Electrochemical Study of a Mononuclear Cu(II) Complex with a 4-Acyl Pyrazolone Ligand. Z. Für Naturforschung B 2018, 73, 713–718. [Google Scholar] [CrossRef]
- Patel, J.D.; Shah, P.J. Synthesis, Characterization of Some Metal Complexes of 4-Acetylsemicarbazone-1-Phenyl-3-Methyl-2-Pyrazolin-5-One. Asian J. Chem. 2010, 22, 3069–3075. [Google Scholar]
- Mao, X.; Ni, J.; Xu, B.; Ding, C. K2S2O8-Promoted Direct Thiocyanation of Pyrazolin-5-Ones with Ammonium Thiocyanate at Room Temperature. Org. Chem. Front. 2020, 7, 350–354. [Google Scholar] [CrossRef]
- Prajuli, R.; Banerjee, J.; Khanal, H. Synthesis of Some Pyrazolone Derivatives and Evaluation of Its Antibacterial and Cytotoxic Activity. Orient. J. Chem. 2015, 31, 2099–2106. [Google Scholar] [CrossRef]
- Reshma, G.; Padmanabhan, V.; Varma, A.R.; Gouri; Nair, U.R.; Parvathy, P.B.; Kulkarni, N.V.; Senthurpandi, D. Synthesis and Structure of Mono and Bis {1,3-Bis(2- Pyridylimino)Isoindoline} Supported 3d Transition Metal Complexes. J. Mol. Struct. 2021, 1226, 129344. [Google Scholar] [CrossRef]
- Hegde, V.; Sreekala, C.O.; Kulkarni, N.V.; Senthurpandi, D.; Mathew, J. Synthesis and Characterization of a Series of Cobalt Complexes: Investigation of Their Efficacy as Sensitizers in Dye-Sensitized Solar Cell Applications. J. Mol. Struct. 2022, 1266, 133512. [Google Scholar] [CrossRef]
- Swathy, S.; Chandran, H.; Reshma, G.; Nakul, S.; Kumar, M.; Krishnan, M.A.; Kulkarni, N.V.; Senthurpandi, D.; Contractor, S.S.; Arakera, S.B. First Row Transition Metal Complexes of Bis(3,5-Dimethyl Pyrazolyl)Methane: Synthesis, Molecular Structure and Antibacterial Properties. J. Mol. Struct. 2022, 1251, 132018. [Google Scholar] [CrossRef]
- Hegde, V.; Sreekala, C.O.; Kulkarni, N.V.; Mathew, J. Bis(Pyrazolyl)Methane Supported Cobalt (II) Complexes as Sensitizers in Dye-Sensitized Solar Cells. J. Photochem. Photobiology. A Chem. 2024, 449, 115389. [Google Scholar] [CrossRef]
- Kulkarni, N.V.; Revankar, V.K.; Kirasur, B.N.; Hugar, M.H. Transition Metal Complexes of Thiosemicarbazones with Quinoxaline Hub: An Emphasis on Antidiabetic Property. Med. Chem. Res. 2011, 21, 663–671. [Google Scholar] [CrossRef]
- Kulkarni, N.V.; Kamath, A.; Budagumpi, S.; Revankar, V.K. Pyrazole Bridged Binuclear Transition Metal Complexes: Synthesis, Characterization, Antimicrobial Activity and DNA Binding/Cleavage Studies. J. Mol. Struct. 2011, 1006, 580–588. [Google Scholar] [CrossRef]
- Chalana, A.; Rai, R.K.; Karri, R.; Jha, K.K.; Kumar, B.; Roy, G. Interplay of the Intermolecular and Intramolecular Interactions in Stabilizing the Thione-Based Copper(I) Complexes and Their Significance in Protecting the Biomolecules against Metal-Mediated Oxidative Damage. Polyhedron 2022, 215, 115647. [Google Scholar] [CrossRef]
- Galano, A.; Macías-Ruvalcaba, N.A.; Campos, O.N.M.; Pedraza-Chaverri, J. Mechanism of the OH Radical Scavenging Activity of Nordihydroguaiaretic Acid: A Combined Theoretical and Experimental Study. J. Phys. Chem. B 2010, 114, 6625–6635. [Google Scholar] [CrossRef] [PubMed]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized Methods for the Determination of Antioxidant Capacity and Phenolics in Foods and Dietary Supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, A.; Selga, A.; Torres, J.L.; Julià, L. Reducing Activity of Polyphenols with Stable Radicals of the TTM Series. Electron Transfer versus H-Abstraction Reactions in Flavan-3-Ols. Org. Lett. 2004, 6, 4583–4586. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M. Free Radicals in Biology and Medicine; Oxford University Press: Cary, NC, USA, 1985. [Google Scholar]
- Ravichandran, H.; Sivalingam, S.; Elumalai; Pramodh, A. Characterization of in Vitro Antioxidant Potential ofAzadirachta Indica and Abutilon Indicum by Different Assay Methods. J. Pharm. Res. 2012, 5, 3227–3231. [Google Scholar]
- Cano, A.; Maestre, A.B.; Hernández-Ruiz, J.; Arnao, M.B. ABTS/TAC Methodology: Main Milestones and Recent Applications. Processes 2023, 11, 185. [Google Scholar] [CrossRef]
- Roy, D.R.; Parthasarathi, R.; Padmanabhan, J.; Sarkar, U.; Subramanian, V.; Chattaraj, P.K. Careful Scrutiny of the Philicity Concept. J. Phys. Chem. A 2006, 110, 1084–1093. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Zolghadri, S.; Bahrami, A.; Khan, M.T.H.; Munoz-Munoz, J.; Garcia-Molina, F.; Garcia-Canovas, F.; Saboury, A.A. A Comprehensive Review on Tyrosinase Inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 279–309. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, A.; Chisvert, A.; Salvador, A. Environmentally Friendly LC for the Simultaneous Determination of Ascorbic Acid and Its Derivatives in Skin-whitening Cosmetics. J. Sep. Sci. 2008, 31, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Al-Otaibi, J.S.; Mary, Y.S.; Mary, S.; Trivedi, R.; Chakraborty, B.; Yadav, R.; Celik, I.; Soman, S. DFT and MD Investigations of the Biomolecules of Phenothiazine Derivatives: Interactions with Gold and Water Molecules and Investigations in Search of Effective Drug for SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 41, 4522–4533. [Google Scholar] [CrossRef] [PubMed]
- Al-Otaibi, J.S.; Mary, Y.S.; Mary, Y.S.; Yadav, R. Structural and Reactivity Studies of Pravadoline –An Ionic Liquid, with Reference to Its Wavefunction-Relative Properties Using DFT and MD Simulation. J. Mol. Struct. 2021, 1245, 131074. [Google Scholar] [CrossRef]
- Mary, Y.S.; Mary, Y.S.; Armaković, S.; Armaković, S.J.; Yadav, R.; Celik, I.; Mane, P.; Chakraborty, B. Stability and Reactivity Study of Bio-Molecules Brucine and Colchicine towards Electrophile and Nucleophile Attacks: Insight from DFT and MD Simulations. J. Mol. Liq. 2021, 335, 116192. [Google Scholar] [CrossRef]
- Smitha, M.; Mary, Y.S.; Mary, Y.S.; Serdaroglu, G.; Chowdhury, P.; Rana, M.; Umamahesvari, H.; Sarojini, B.K.; Mohan, B.J.; Pavithran, R. Modeling the DFT Structural and Reactivity Studies of a Pyrimidine -6-Carboxylate Derivative with Reference to Its Wavefunction-Dependent, MD Simulations and Evaluation for Potential Antimicrobial Activity. J. Mol. Struct. 2021, 1237, 130397. [Google Scholar] [CrossRef]

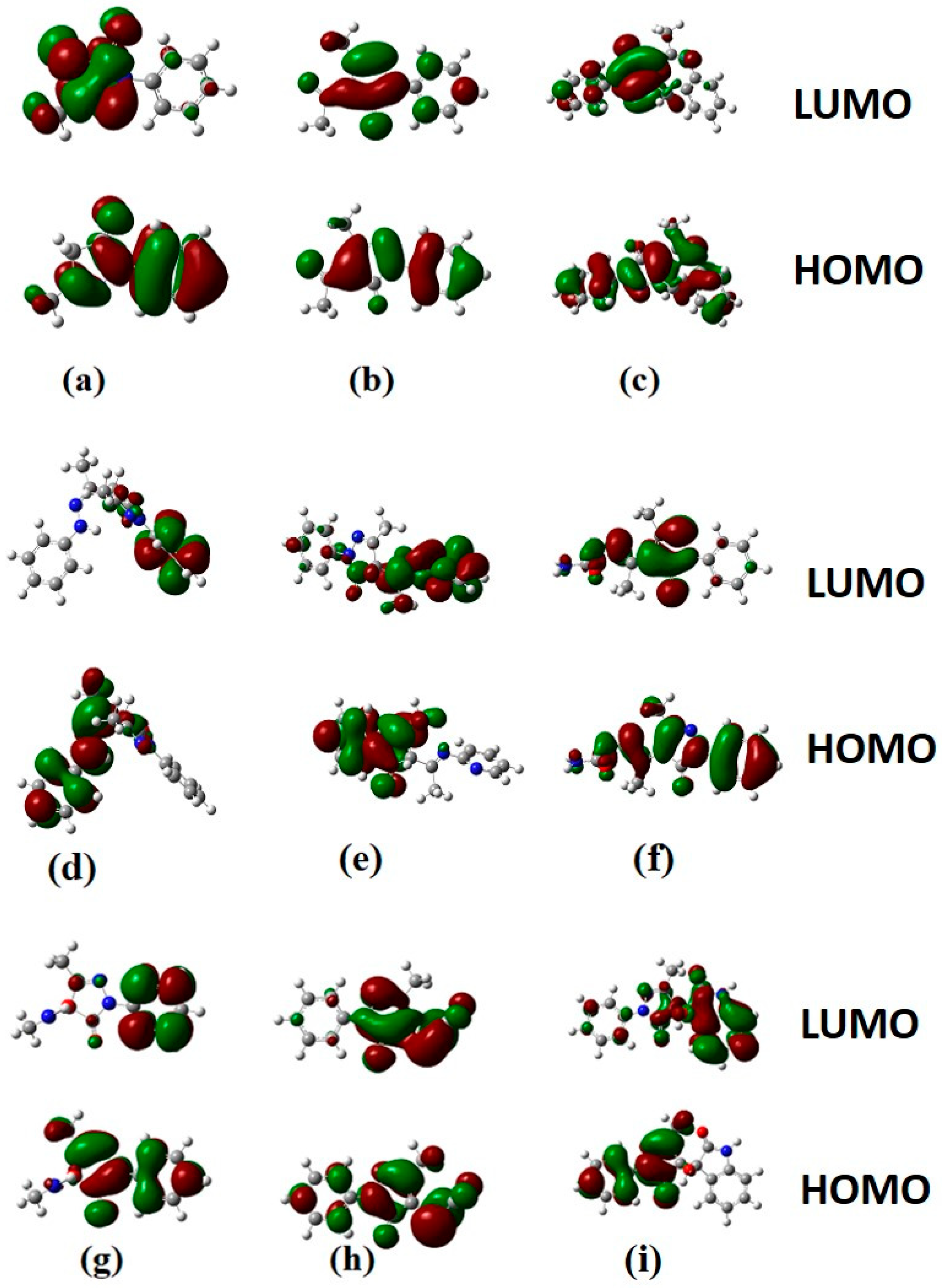
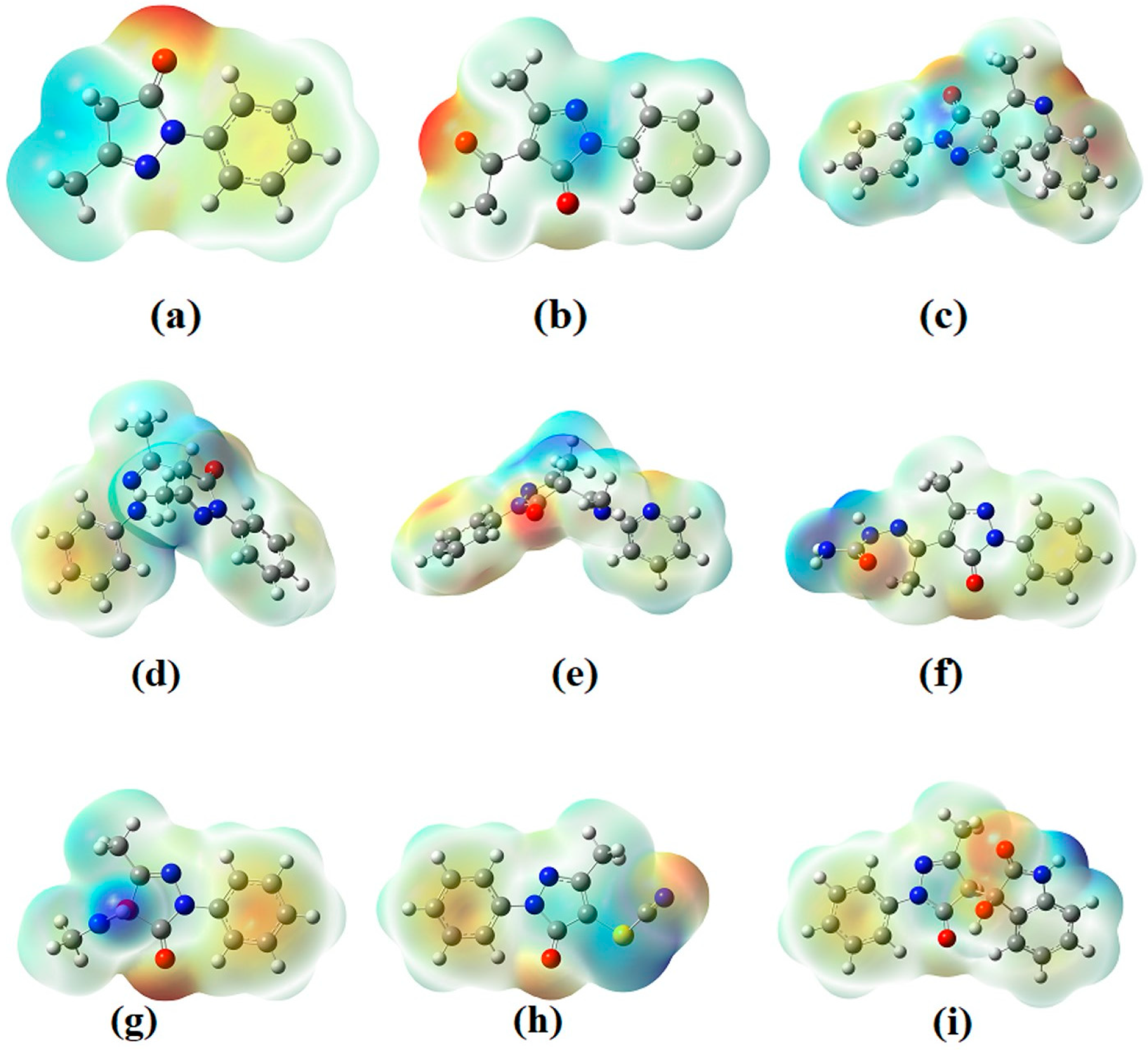



| Compound | EHOMO (eV) | ELUMO (eV) | ELUMO–EHOMO Gap (eV) | Hardness | Chemical Potential | Electrophilicity Index |
|---|---|---|---|---|---|---|
| L1 | −5.8392 | −1.0781 | 4.7611 | 2.3806 | −3.4587 | 2.5124 |
| L2 | −6.1799 | −0.9448 | 5.2351 | 2.6176 | −3.5624 | 2.4240 |
| L3 | −5.8819 | −1.0182 | 4.8637 | 2.4319 | −3.4501 | 2.4472 |
| L4 | −5.3959 | −1.3848 | 4.0111 | 2.0056 | −3.3904 | 2.8656 |
| L5 | −5.7557 | −1.1924 | 4.5633 | 2.2817 | −3.4741 | 2.6447 |
| L6 | −5.7461 | −0.7129 | 5.0332 | 2.5166 | −3.2295 | 2.0722 |
| L7 | −5.6623 | −0.7377 | 4.9246 | 2.4623 | −3.2000 | 2.0794 |
| L8 | −6.3094 | −1.0776 | 5.2318 | 2.6159 | −3.6935 | 2.6075 |
| L9 | −5.9097 | −1.1684 | 4.7413 | 2.3707 | −3.5391 | 2.6416 |
| Compounds | Binding Energy (kcal/mol) |
|---|---|
| L1 | −7.8 |
| L2 | −9.2 |
| L3 | −7.6 |
| L4 | −7.3 |
| L5 | −6.9 |
| L6 | −9.4 |
| L7 | −8.9 |
| L8 | −8.0 |
| L9 | −11.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Divya Mohan, R.; Anaswara, S.A.; Kulkarni, N.V.; Bojilov, D.G.; Manolov, S.P.; Ivanov, I.I.; Al-Otaibi, J.S.; Sheena Mary, Y. Synthesis, Characterization and Assessment of Antioxidant and Melanogenic Inhibitory Properties of Edaravone Derivatives. Antioxidants 2024, 13, 1148. https://doi.org/10.3390/antiox13091148
Divya Mohan R, Anaswara SA, Kulkarni NV, Bojilov DG, Manolov SP, Ivanov II, Al-Otaibi JS, Sheena Mary Y. Synthesis, Characterization and Assessment of Antioxidant and Melanogenic Inhibitory Properties of Edaravone Derivatives. Antioxidants. 2024; 13(9):1148. https://doi.org/10.3390/antiox13091148
Chicago/Turabian StyleDivya Mohan, R., S. A. Anaswara, Naveen V. Kulkarni, Dimitar G. Bojilov, Stanimir P. Manolov, Iliyan I. Ivanov, Jamelah S. Al-Otaibi, and Y. Sheena Mary. 2024. "Synthesis, Characterization and Assessment of Antioxidant and Melanogenic Inhibitory Properties of Edaravone Derivatives" Antioxidants 13, no. 9: 1148. https://doi.org/10.3390/antiox13091148
APA StyleDivya Mohan, R., Anaswara, S. A., Kulkarni, N. V., Bojilov, D. G., Manolov, S. P., Ivanov, I. I., Al-Otaibi, J. S., & Sheena Mary, Y. (2024). Synthesis, Characterization and Assessment of Antioxidant and Melanogenic Inhibitory Properties of Edaravone Derivatives. Antioxidants, 13(9), 1148. https://doi.org/10.3390/antiox13091148








