The Role of Polyphenols in Abiotic Stress Tolerance and Their Antioxidant Properties to Scavenge Reactive Oxygen Species and Free Radicals
Abstract
:1. Introduction
2. The MYB Family Emerges as Particularly Significant in Phenolic Metabolism Regulation and Abiotic Stress Tolerance
3. The Role of Polyphenolic Compounds in Abiotic Stress Tolerance
3.1. Temperature
3.2. Light
UV Radiation
3.3. Nanoparticles and Agrochemical Applications
3.4. Drought Stress
3.5. Salt Stress
3.6. Heavy Metals Toxicity
3.7. Polyphenols Pathway Responds to Multiple Environmental Stressors
4. Polyphenols Antioxidant Properties to Scavenge Free Radicals and Reactive Oxygen Species
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.; Tilman, D.; Jin, Z.; Smith, P.; Barrett, C.B.; Zhu, Y.-G.; Burney, J.; D’Odorico, P.; Fantke, P.; Fargione, J. Climate Change Exacerbates the Environmental Impacts of Agriculture. Science 2024, 385, eadn3747. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, S.; Chen, J.; Yu, H.; Yang, T.; Wang, C.; Huang, S.; Chen, H.; Ao, X. Impacts of Global Climate Change on Agricultural Production: A Comprehensive Review. Agronomy 2024, 14, 1360. [Google Scholar] [CrossRef]
- Paul, A.; Acharya, K.; Chakraborty, N. Involvement of Phenylpropanoid Pathway and Shikimic Acid Pathway in Environmental Stress Response. In Biology and Biotechnology of Environmental Stress Tolerance in Plants; Apple Academic Press: Williston, VT, USA, 2023; pp. 27–66. [Google Scholar]
- Saini, N.; Anmol, A.; Kumar, S.; Bakshi, M.; Dhiman, Z. Exploring Phenolic Compounds as Natural Stress Alleviators in Plants-a Comprehensive Review. Physiol. Mol. Plant Pathol. 2024, 133, 102383. [Google Scholar] [CrossRef]
- Zagoskina, N.V.; Zubova, M.Y.; Nechaeva, T.L.; Kazantseva, V.V.; Goncharuk, E.A.; Katanskaya, V.M.; Baranova, E.N.; Aksenova, M.A. Polyphenols in Plants: Structure, Biosynthesis, Abiotic Stress Regulation, and Practical Applications. Int. J. Mol. Sci. 2023, 24, 13874. [Google Scholar] [CrossRef]
- El-Saadony, M.T.; Yang, T.; Saad, A.M.; Alkafaas, S.S.; Elkafas, S.S.; Eldeeb, G.S.; Mohammed, D.M.; Salem, H.M.; Korma, S.A.; Loutfy, S.A. Chemistry, Bioavailability, Bioactivity, Nutritional Aspects and Human Health Benefits of Polyphenols: A Comprehensive Review. Int. J. Biol. Macromol. 2024, 277, 134223. [Google Scholar] [CrossRef]
- Silva, A.; Silva, V.; Igrejas, G.; Aires, A.; Falco, V.; Valentão, P.; Poeta, P. Phenolic Compounds Classification and Their Distribution in Winemaking By-Products. Eur. Food Res. Technol. 2023, 249, 207–239. [Google Scholar] [CrossRef]
- Singla, R.K.; Dubey, A.K.; Garg, A.; Sharma, R.K.; Fiorino, M.; Ameen, S.M.; Haddad, M.A.; Al-Hiary, M. Natural Polyphenols: Chemical Classification, Definition of Classes, Subcategories, and Structures. J. AOAC Int. 2019, 102, 1397–1400. [Google Scholar] [CrossRef]
- Salam, U.; Ullah, S.; Tang, Z.-H.; Elateeq, A.A.; Khan, Y.; Khan, J.; Khan, A.; Ali, S. Plant Metabolomics: An Overview of the Role of Primary and Secondary Metabolites against Different Environmental Stress Factors. Life 2023, 13, 706. [Google Scholar] [CrossRef]
- Nehra, A.; Kalwan, G.; Gill, R.; Nehra, K.; Agarwala, N.; Jain, P.K.; Naeem, M.; Tuteja, N.; Pudake, R.N.; Gill, S.S. Status of Impact of Abiotic Stresses on Global Agriculture. In Nanotechnology for Abiotic Stress Tolerance and Management in Crop Plants; Elsevier: Amsterdam, The Netherlands, 2024; pp. 1–21. [Google Scholar]
- Scalbert, A.; Johnson, I.T.; Saltmarsh, M. Polyphenols: Antioxidants and beyond. Am. J. Clin. Nutr. 2005, 81, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Dangles, O. A Comprehensive Review on Flavanones, the Major Citrus Polyphenols. J. Food Compos. Anal. 2014, 33, 85–104. [Google Scholar] [CrossRef]
- Lattanzio, V. Bioactive Polyphenols: Their Role in Quality and Storability of Fruit and Vegetables. J. Appl. Bot. 2003, 77, 128–146. [Google Scholar]
- Yang, S.; Wang, C.; Li, X.; Wu, C.; Liu, C.; Xue, Z.; Kou, X. Investigation on the Biological Activity of Anthocyanins and Polyphenols in Blueberry. J. Food Sci. 2021, 86, 614–627. [Google Scholar] [CrossRef] [PubMed]
- Ben-Abdallah, S.; Zorrig, W.; Amyot, L.; Renaud, J.; Hannoufa, A.; Lachâal, M.; Karray-Bouraoui, N. Potential Production of Polyphenols, Carotenoids and Glycoalkaloids in Solanum villosum Mill. under Salt Stress. Biologia 2019, 74, 309–324. [Google Scholar] [CrossRef]
- Lacikova, L.; Jancova, M.; Muselik, J.; Masterova, I.; Grancai, D.; Fickova, M. Antiproliferative, Cytotoxic, Antioxidant Activity and Polyphenols Contents in Leaves of Four Staphylea L. Species. Molecules 2009, 14, 3259–3267. [Google Scholar] [CrossRef]
- Griesser, M.; Weingart, G.; Schoedl-Hummel, K.; Neumann, N.; Becker, M.; Varmuza, K.; Liebner, F.; Schuhmacher, R.; Forneck, A. Severe Drought Stress Is Affecting Selected Primary Metabolites, Polyphenols, and Volatile Metabolites in Grapevine Leaves (Vitis vinifera cv. Pinot Noir). Plant Physiol. Biochem. 2015, 88, 17–26. [Google Scholar] [CrossRef]
- Zhang, B.; Xia, T.; Duan, W.; Zhang, Z.; Li, Y.; Fang, B.; Xia, M.; Wang, M. Effects of Organic Acids, Amino Acids and Phenolic Compounds on Antioxidant Characteristic of Zhenjiang Aromatic Vinegar. Molecules 2019, 24, 3799. [Google Scholar] [CrossRef]
- Naikoo, M.I.; Dar, M.I.; Raghib, F.; Jaleel, H.; Ahmad, B.; Raina, A.; Khan, F.A.; Naushin, F. Role and Regulation of Plants Phenolics in Abiotic Stress Tolerance: An Overview. In Plant Signaling Molecules; Woodhead Publishing: Amsterdam, The Netherlands, 2019; pp. 157–168. [Google Scholar]
- Ahmed, U.; Rao, M.J.; Qi, C.; Xie, Q.; Noushahi, H.A.; Yaseen, M.; Shi, X.; Zheng, B. Expression Profiling of Flavonoid Biosynthesis Genes and Secondary Metabolites Accumulation in Populus under Drought Stress. Molecules 2021, 26, 5546. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; Zhao, Q.; Gallego-Giraldo, L.; Dai, X.; Zhao, P.X.; Dixon, R.A. Genome-wide Analysis of Phenylpropanoid Defence Pathways. Mol. Plant Pathol. 2010, 11, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.; Chen, L.; Xiao, Y.; Wu, J.; Zeng, L.; Li, H.; Wu, Q.; Hu, Z. Recent Advanced Metabolic and Genetic Engineering of Phenylpropanoid Biosynthetic Pathways. Int. J. Mol. Sci. 2021, 22, 9544. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef]
- Alami, M.M.; Guo, S.; Mei, Z.; Yang, G.; Wang, X. Environmental Factors on Secondary Metabolism in Medicinal Plants: Exploring Accelerating Factors. Med. Plant Biol. 2024, 3, e016. [Google Scholar] [CrossRef]
- Tang, H.; Wang, Q.; Xie, H.; Li, W. The Function of Secondary Metabolites in Resisting Stresses in Horticultural Plants. Fruit Res. 2024, 4, e021. [Google Scholar] [CrossRef]
- Oyebamiji, Y.O.; Adigun, B.A.; Shamsudin, N.A.A.; Ikmal, A.M.; Salisu, M.A.; Malike, F.A.; Lateef, A.A. Recent Advancements in Mitigating Abiotic Stresses in Crops. Horticulturae 2024, 10, 156. [Google Scholar] [CrossRef]
- Kashyap, A.K.; Dubey, S.K.; Shah, S.; Kumar, A. A Short Review on Genes Regulating Biosynthesis of Major Secondary Metabolites. In Phytochemical Genomics Plant Metabolomics and Medicinal Plant Genomics; Springer: Singapore, 2023; pp. 501–519. [Google Scholar]
- Rao, M.J.; Duan, M.; Yang, M.; Fan, H.; Shen, S.; Hu, L.; Wang, L. Novel Insights into Anthocyanin Metabolism and Molecular Characterization of Associated Genes in Sugarcane Rinds Using the Metabolome and Transcriptome. Int. J. Mol. Sci. 2022, 23, 338. [Google Scholar] [CrossRef]
- Rao, M.J.; Duan, M.; Wang, J.; Han, S.; Ma, L.; Mo, X.; Li, M.; Hu, L.; Wang, L. Transcriptomic and Widely Targeted Metabolomic Approach Identified Diverse Group of Bioactive Compounds, Antiradical Activities, and Their Associated Genes in Six Sugarcane Varieties. Antioxidants 2022, 11, 1319. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Ng, M.-S.; Cheng, S.-S.; Lo, A.W.-Y.; Xiao, Z.; Shin, T.-S.; Chung, G.; Lam, H.-M. Understanding the Composition, Biosynthesis, Accumulation and Transport of Flavonoids in Crops for the Promotion of Crops as Healthy Sources of Flavonoids for Human Consumption. Nutrients 2020, 12, 1717. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.; Li, H.; Chiang, V.L.; Fu, Y. Cooperative Regulation of Flavonoid and Lignin Biosynthesis in Plants. Crit. Rev. Plant Sci. 2021, 40, 109–126. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. Overexpression of MusaMYB31, a R2R3 Type MYB Transcription Factor Gene Indicate Its Role as a Negative Regulator of Lignin Biosynthesis in Banana. PLoS ONE 2017, 12, e0172695. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ran, L.; Hou, Y.; Tian, Q.; Li, C.; Liu, R.; Fan, D.; Luo, K. The Transcription Factor MYB115 Contributes to the Regulation of Proanthocyanidin Biosynthesis and Enhances Fungal Resistance in Poplar. New Phytol. 2017, 215, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Kong, W.; Wong, G.; Fu, L.; Peng, R.; Li, Z.; Yao, Q. AtMYB12 Regulates Flavonoids Accumulation and Abiotic Stress Tolerance in Transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Mitra, M.; Roy, S. Study of Changes in Folding/Unfolding Properties and Stability of Arabidopsis thaliana MYB12 Transcription Factor Following UV-B Exposure in Vitro. Heliyon 2024, 10, 13. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Meng, Z.; Jia, Z.; Fu, F.; Jin, B.; Cao, F.; Wang, L. LncNAT11-GbMYB11-GbF3′H/GbFLS Module Mediates Flavonol Biosynthesis to Regulate Salt Stress Tolerance in Ginkgo biloba. J. Exp. Bot. 2024, erae438. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Zhou, L.J.; Gao, H.N.; Wang, X.F.; Li, Z.W.; Li, Y.Y. The Transcription Factor MdMYB2 Influences Cold Tolerance and Anthocyanin Accumulation by Activating SUMO E3 Ligase MdSIZ1 in Apple. Plant Physiol. 2022, 189, 2044–2060. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Zhang, L.; Shu, J.; Court, M.H.; Sun, Z.; Jiang, L.; Zheng, C.; Shu, H.; Ji, L.; et al. Genome-Wide Analysis of the Apple Family 1 Glycosyltransferases Identified a Flavonoid-Modifying UGT, MdUGT83L3, Which Is Targeted by MdMYB88 and Contributes to Stress Adaptation. Plant Sci. 2022, 321, 111314. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Bi, S.Q.; You, C.X.; Hao, Y.J. An Apple MYB Transcription Factor Regulates Cold Tolerance and Anthocyanin Accumulation and Undergoes MIEL1-Mediated Degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Quan, R.; Chen, G.; Yu, G.; Li, X.; Han, Z.; Xu, W.; Li, G.; Shi, J.; Li, B. An R2R3-MYB Transcription Factor VyMYB24, Isolated from Wild Grape Vitis yanshanesis JX Chen., Regulates the Plant Development and Confers the Tolerance to Drought. Front. Plant Sci. 2022, 13, 966641. [Google Scholar]
- Wang, J.; Wang, F.; Jin, C.; Tong, Y.; Wang, T. A R2R3-MYB Transcription Factor VvMYBF1 from Grapevine (Vitis vinifera L.) Regulates Flavonoids Accumulation and Abiotic Stress Tolerance in Transgenic Arabidopsis. J. Hortic. Sci. Biotechnol. 2020, 95, 147–161. [Google Scholar] [CrossRef]
- Han, J.; Dai, J.; Chen, Z.; Li, W.; Li, X.; Zhang, L.; Yao, A.; Zhang, B.; Han, D. Overexpression of a ‘Beta’MYB Factor Gene, VhMYB15, Increases Salinity and Drought Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 1534. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Agarwal, P.; Choudhury, S.R.; Roy, S. MYB4, a Member of R2R3-Subfamily of MYB Transcription Factor Functions as a Repressor of Key Genes Involved in Flavonoid Biosynthesis and Repair of UV-B Induced DNA Double Strand Breaks in Arabidopsis. Plant Physiol. Biochem. 2024, 211, 108698. [Google Scholar] [CrossRef]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 Interaction Affects MYB-BHLH-WD40 Complex Formation and Anthocyanin Accumulation in Arabidopsis. J. Plant Physiol. 2021, 268, 153558. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gu, X.; Jiang, Y.; Wang, L.; Xiao, N.; Chen, Y.; Jin, B.; Wang, L.; Li, W. UV-B Promotes Flavonoid Biosynthesis in Ginkgo Biloba by Inducing the GbHY5-GbMYB1-GbFLS Module. Hortic. Res. 2023, 10, uhad118. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Wang, Z.; Su, L.; Gong, L.; Xin, H. Vitis Myb14 Confer Cold and Drought Tolerance by Activating Lipid Transfer Protein Genes Expression and Reactive Oxygen Species Scavenge. Gene 2024, 890, 147792. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhao, W.; Zhou, R.; Wang, B.; Liu, Y.; Ge, W.; Liang, J.; Zhao, Q.; Wen, P. VviMYB24 Positively Regulates Flavonol Biosynthesis in Response to Moderate Drought Stress in ‘Cabernet Sauvignon’ Grape Seedlings. Sci. Hortic. 2024, 338, 113769. [Google Scholar] [CrossRef]
- Raziq, A.; Zhang, K.; Sun, W.; Ahmad, N.; Zhao, H.; Raza, M.A.; Ahmed, S.; Din, A.M.U.; Zhao, S.; Pan, J.; et al. Transcriptome Profiling of MYB-Overexpressed Transgenic Lines Provides Crucial Molecular Insights into Anthocyanin and Remodel the Biosynthesis Regulatory Network in Nicotiana tabacum. Ind. Crops Prod. 2024, 213, 118374. [Google Scholar] [CrossRef]
- Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. [Google Scholar] [CrossRef]
- Yang, J.; Guo, C.; Chen, F.; Lv, B.; Song, J.; Ning, G.; He, Y.; Lin, J.; He, H.; Yang, Y.; et al. Heat-Induced Modulation of Flavonoid Biosynthesis via a LhMYBC2-Mediated Regulatory Network in Oriental Hybrid Lily. Plant Physiol. Biochem. 2024, 214, 108966. [Google Scholar] [CrossRef]
- Zhao, X.; Li, P.; Zuo, H.; Peng, A.; Lin, J.; Li, P.; Wang, K.; Tang, Q.; Tadege, M.; Liu, Z. CsMYBL2 Homologs Modulate the Light and Temperature Stress-regulated Anthocyanin and Catechins Biosynthesis in Tea Plants (Camellia sinensis). Plant J. 2023, 115, 1051–1070. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.N.; Naing, A.H.; Yun, B.-W.; Lim, S.H.; Kim, C.K. Overexpression of RsMYB1 Enhances Anthocyanin Accumulation and Heavy Metal Stress Tolerance in Transgenic Petunia. Front. Plant Sci. 2018, 9, 01388. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, R.; Yao, Q. SlMYB14 Promotes Flavonoids Accumulation and Confers Higher Tolerance to 2,4,6-Trichlorophenol in Tomato. Plant Sci. 2021, 303, 110796. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yu, Z.; Yao, B.; Teixeira da Silva, J.A.; Wen, D. SsMYB113, a Schima superba MYB Transcription Factor, Regulates the Accumulation of Flavonoids and Functions in Drought Stress Tolerance by Modulating ROS Generation. Plant Soil. 2022, 478, 427–444. [Google Scholar] [CrossRef]
- Chen, Q.; Peng, L.; Wang, A.; Yu, L.; Liu, Y.; Zhang, X.; Wang, R.; Li, X.; Yang, Y.; Li, X.; et al. An R2R3-MYB FtMYB11 from Tartary Buckwheat Has Contrasting Effects on Abiotic Tolerance in Arabidopsis. J. Plant Physiol. 2023, 280, 153842. [Google Scholar] [CrossRef]
- Mahmoud, L.M.; Killiny, N.; Dutt, M. Physiological and Molecular Responses of ‘Hamlin’ Sweet Orange Trees Expressing the VvmybA1 Gene under Cold Stress Conditions. Planta 2024, 260, 67. [Google Scholar] [CrossRef]
- D’Amelia, V.; Aversano, R.; Ruggiero, A.; Batelli, G.; Appelhagen, I.; Dinacci, C.; Hill, L.; Martin, C.; Carputo, D. Subfunctionalization of Duplicate MYB Genes in Solanum commersonii Generated the Cold-induced ScAN2 and the Anthocyanin Regulator ScAN1. Plant Cell Environ. 2018, 41, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, S.; Li, Y.; Zhang, L.; Song, W.; Chen, C. Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza. Int. J. Mol. Sci. 2024, 25, 4111. [Google Scholar] [CrossRef]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.; Kim, H.S.; Kim, S. MYB3 Plays an Important Role in Lignin and Anthocyanin Biosynthesis under Salt Stress Condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Dixon, R.A. Plant Phenylalanine/Tyrosine Ammonia-Lyases. Trends Plant Sci. 2020, 25, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Marchiosi, R.; dos Santos, W.D.; Constantin, R.P.; de Lima, R.B.; Soares, A.R.; Finger-Teixeira, A.; Mota, T.R.; de Oliveira, D.M.; Foletto-Felipe, M.D.P.; Abrahão, J.; et al. Biosynthesis and Metabolic Actions of Simple Phenolic Acids in Plants. Phytochem. Rev. 2020, 19, 865–906. [Google Scholar] [CrossRef]
- Kołton, A.; Długosz-Grochowska, O.; Wojciechowska, R.; Czaja, M. Biosynthesis Regulation of Folates and Phenols in Plants. Sci. Hortic. 2022, 291, 110561. [Google Scholar] [CrossRef]
- Saigo, T.; Wang, T.; Watanabe, M.; Tohge, T. Diversity of Anthocyanin and Proanthocyanin Biosynthesis in Land Plants. Curr. Opin. Plant Biol. 2020, 55, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W. The Proanthocyanidin-specific Transcription Factor Md MYBPA 1 Initiates Anthocyanin Synthesis under Low-temperature Conditions in Red-fleshed Apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Patel, S.; Pan, X.; Naz, S.; Silva, A.S.; Saeed, F.; Suleria, H.A.R. Proanthocyanidins: A Comprehensive Review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive Oxygen Species, Abiotic Stress and Stress Combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic Stress and Reactive Oxygen Species: Generation, Signaling, and Defense Mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple Abiotic Stresses Occurring with Salinity Stress in Citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- Nawaz, M.; Sun, J.; Shabbir, S.; Khattak, W.A.; Ren, G.; Nie, X.; Bo, Y.; Javed, Q.; Du, D.; Sonne, C. A Review of Plants Strategies to Resist Biotic and Abiotic Environmental Stressors. Sci. Total Environ. 2023, 900, 165832. [Google Scholar] [CrossRef]
- Mayor, R. Oxidative Stress and Antioxidant Defense System. J. Inst. Med. Trop. 2010, 5, 23–39. [Google Scholar]
- Scagel, C.F.; Lee, J.; Mitchell, J.N. Salinity from NaCl Changes the Nutrient and Polyphenolic Composition of Basil Leaves. Ind. Crops Prod. 2019, 127, 119–128. [Google Scholar] [CrossRef]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Talebi, M.; Matkowski, A. The Effect of Drought Stress on Polyphenolic Compounds and Expression of Flavonoid Biosynthesis Related Genes in Achillea pachycephala Rech. f. Phytochemistry 2019, 162, 90–98. [Google Scholar] [CrossRef]
- Rao, M.J.; Ding, F.; Wang, N.; Deng, X.; Xu, Q. Metabolic Mechanisms of Host Species Against Citrus Huanglongbing (Greening Disease). Crit. Rev. Plant Sci. 2018, 37, 1544843. [Google Scholar] [CrossRef]
- Rao, M.J.; Xu, Y.; Huang, Y.; Tang, X.; Deng, X.; Xu, Q. Ectopic Expression of Citrus UDP-GLUCOSYL TRANSFERASE Gene Enhances Anthocyanin and Proanthocyanidins Contents and Confers High Light Tolerance in Arabidopsis. BMC Plant Biol. 2019, 19, 603. [Google Scholar] [CrossRef]
- Rao, M.J.; Ahmed, U.; Ahmed, M.H.; Duan, M.; Wang, J.; Wang, Y.; Wang, L. Comparison and Quantification of Metabolites and Their Antioxidant Activities in Young and Mature Leaves of Sugarcane. ACS Food Sci. Technol. 2021, 1, 362–373. [Google Scholar] [CrossRef]
- Kerbler, S.M.; Wigge, P.A. Temperature Sensing in Plants. Annu. Rev. Plant Biol. 2023, 74, 341–366. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zhang, Z.; Lv, X.; Cheng, N.; Peng, B.; Cao, W. Effect of 24-Epibrassinolide on Chilling Injury of Peach Fruit in Relation to Phenolic and Proline Metabolisms. Postharvest Biol. Technol. 2016, 111, 390–397. [Google Scholar] [CrossRef]
- Wang, J.; Yuan, B.; Huang, B. Differential Heat-induced Changes in Phenolic Acids Associated with Genotypic Variations in Heat Tolerance for Hard Fescue. Crop Sci. 2019, 59, 667–674. [Google Scholar] [CrossRef]
- Semmar, N. Temperature-Linked Constraints and Plant Protection Responses. In Secondary Metabolites in Plant Stress Adaptation: Analytic Space of Secondary Metabolites; Springer: Berlin/Heidelberg, Germany, 2024; pp. 155–219. [Google Scholar]
- He, J.; Yao, L.; Pecoraro, L.; Liu, C.; Wang, J.; Huang, L.; Gao, W. Cold Stress Regulates Accumulation of Flavonoids and Terpenoids in Plants by Phytohormone, Transcription Process, Functional Enzyme, and Epigenetics. Crit. Rev. Biotechnol. 2023, 43, 680–697. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Hur, Y.; Nou, I.-S. Anthocyanin Biosynthesis for Cold and Freezing Stress Tolerance and Desirable Color in Brassica Rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Ahmed, A.; Tariq, A.; Habib, S. Interactive Biology of Auxins and Phenolics in Plant Environment. In Plant Phenolics in Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1, pp. 117–133. [Google Scholar]
- Kavi, K.P.B.; Srinivas, B.; Prashant, S.; Sahitya, G.; Tulya, R.S.V.; Rajasheker, G.; Prashanth, S. Modulation of Lignin and Its Implications in Salt, Drought and Temperature Stress Tolerance. Curr. Chem. Biol. 2023, 17, 2–12. [Google Scholar] [CrossRef]
- Singh, A.K.; Dhanapal, S.; Yadav, B.S. The Dynamic Responses of Plant Physiology and Metabolism during Environmental Stress Progression. Mol. Biol. Rep. 2020, 47, 1459–1470. [Google Scholar] [CrossRef]
- Gupta, R.; Deswal, R. Antifreeze Proteins Enable Plants to Survive in Freezing Conditions. J. Biosci. 2014, 39, 931–944. [Google Scholar] [CrossRef]
- Zhou, P.; Li, Q.; Liu, G.; Xu, N.; Yang, Y.; Zeng, W.; Chen, A.; Wang, S. Integrated Analysis of Transcriptomic and Metabolomic Data Reveals Critical Metabolic Pathways Involved in Polyphenol Biosynthesis in Nicotiana tabacum under Chilling Stress. Funct. Plant Biol. 2018, 46, 30–43. [Google Scholar] [CrossRef]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low Temperature Promotes Anthocyanin Biosynthesis and Related Gene Expression in the Seedlings of Purple Head Chinese Cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef]
- Muhlemann, J.K.; Younts, T.L.B.; Muday, G.K. Flavonols Control Pollen Tube Growth and Integrity by Regulating ROS Homeostasis during High-Temperature Stress. Proc. Natl. Acad. Sci. USA 2018, 115, 11188–11197. [Google Scholar] [CrossRef]
- Chebrolu, K.K.; Fritschi, F.B.; Ye, S.; Krishnan, H.B.; Smith, J.R.; Gillman, J.D. Impact of Heat Stress during Seed Development on Soybean Seed Metabolome. Metabolomics 2016, 12, 28. [Google Scholar] [CrossRef]
- Gouot, J.C.; Smith, J.P.; Holzapfel, B.P.; Walker, A.R.; Barril, C. Grape Berry Flavonoids: A Review of Their Biochemical Responses to High and Extreme High Temperatures. J. Exp. Bot. 2019, 70, 397–423. [Google Scholar] [CrossRef]
- Zhou, L.-J.; Geng, Z.; Wang, Y.; Wang, Y.; Liu, S.; Chen, C.; Song, A.; Jiang, J.; Chen, S.; Chen, F. A Novel Transcription Factor CmMYB012 Inhibits Flavone and Anthocyanin Biosynthesis in Response to High Temperatures in Chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- Yamagishi, M. Mechanisms by Which High Temperatures Suppress Anthocyanin Coloration in Flowers and Fruits, and Discovery of Floricultural Crops That Exhibit High-Temperature-Tolerant Flower Pigmentation. Hortic. J. 2024, 93, 203–215. [Google Scholar] [CrossRef]
- Cingoz, G.S.; Gurel, E. Effects of Salicylic Acid on Thermotolerance and Cardenolide Accumulation under High Temperature Stress in Digitalis trojana Ivanina. Plant Physiol. Biochem. 2016, 105, 145–149. [Google Scholar] [CrossRef]
- Pasternak, T.P.; Steinmacher, D. Plant Growth Regulation in Cell and Tissue Culture in Vitro. Plants 2024, 13, 327. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qi, Z.; Ren, X.; Li, Y.; Zhang, N.; Liu, Q. Effects of Red and Blue Light on Red Clover (Trifolium pratense L.) Growth and Secondary Metabolism. Plant Growth Regul. 2024, 104, 1087–1106. [Google Scholar] [CrossRef]
- Hong, J.; Meng, K.; Thomas, H.R.; Yang, Y.; Williams, B.; Kang, H.; Zhou, Y. Reframing Agriculture by Light: The Role of Light-Mediated Jasmonates/Salicylic Acid Regulation in Plant Defense, Development and Beyond. Veg. Res. 2024, 4, e027. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light Induced Regulation Pathway of Anthocyanin Biosynthesis in Plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef]
- Guo, X.L.; Hu, J.B.; Wang, D.L. Effect of Light Intensity on Blueberry Fruit Coloration, Anthocyanin Synthesis Pathway Enzyme Activity, and Gene Expression. Russ. J. Plant Physiol. 2023, 70, 136. [Google Scholar] [CrossRef]
- Zhu, W.; Wu, H.; Yang, C.; Shi, B.; Zheng, B.; Ma, X.; Zhou, K.; Qian, M. Postharvest Light-Induced Flavonoids Accumulation in Mango (Mangifera indica L.) Peel Is Associated with the up-Regulation of Flavonoids-Related and Light Signal Pathway Genes. Front. Plant Sci. 2023, 14, 1136281. [Google Scholar] [CrossRef]
- Balarynová, J.; Danihlík, J.; Fellner, M. Changes in Plasma Membrane Aquaporin Gene Expression under Osmotic Stress and Blue Light in Tomato. Acta Physiol. Plant 2018, 40, 27. [Google Scholar] [CrossRef]
- Hussain, S.; Shuxian, L.; Mumtaz, M.; Shafiq, I.; Iqbal, N.; Brestic, M.; Shoaib, M.; Sisi, Q.; Li, W.; Mei, X. Foliar Application of Silicon Improves Stem Strength under Low Light Stress by Regulating Lignin Biosynthesis Genes in Soybean (Glycine max (L.) Merr.). J. Hazard. Mater. 2021, 401, 123256. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, L.; Shan, Y.; Liu, Y.; Tian, Y.; Xia, T. Influence of Shade on Flavonoid Biosynthesis in Tea (Camellia sinensis (L.) O. Kuntze). Sci. Hortic. 2012, 141, 7–16. [Google Scholar] [CrossRef]
- Zubova, M.Y.; Nechaeva, T.L.; Kartashov, A.V.; Zagoskina, N.V. Regulation of the Phenolic Compounds Accumulation in the Tea-Plant Callus Culture with a Separate and Combined Effect of Light and Cadmium Ions. Biol. Bull. 2020, 47, 593–604. [Google Scholar] [CrossRef]
- Jaakola, L.; Määttä, K.; Pirttilä, A.M.; Törrönen, R.; Kärenlampi, S.; Hohtola, A. Expression of Genes Involved in Anthocyanin Biosynthesis in Relation to Anthocyanin, Proanthocyanidin, and Flavonol Levels during Bilberry Fruit Development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef]
- Wang, Y.; Fong, S.K.; Singh, A.P.; Vorsa, N.; Johnson-Cicalese, J. Variation of Anthocyanins, Proanthocyanidins, Flavonols, and Organic Acids in Cultivated and Wild Diploid Blueberry Species. HortScience 2019, 54, 576–585. [Google Scholar] [CrossRef]
- Park, W.T.; Yeo, S.K.; Sathasivam, R.; Park, J.S.; Kim, J.K.; Park, S.U. Influence of Light-Emitting Diodes on Phenylpropanoid Biosynthetic Gene Expression and Phenylpropanoid Accumulation in Agastache Rugosa. Appl. Biol. Chem. 2020, 63, 25. [Google Scholar] [CrossRef]
- Jang, E.B.; Ho, T.-T.; Park, S.-Y. Effect of Light Quality and Tissue Origin on Phenolic Compound Accumulation and Antioxidant Activity in Camellia japonica Calli. In Vitro Cell. Dev. Biol.-Plant 2020, 56, 567–577. [Google Scholar] [CrossRef]
- Yao, P.; Huang, Y.; Dong, Q.; Wan, M.; Wang, A.; Chen, Y.; Li, C.; Wu, Q.; Chen, H.; Zhao, H. FtMYB6, a Light-Induced SG7 R2R3-MYB Transcription Factor, Promotes Flavonol Biosynthesis in Tartary Buckwheat (Fagopyrum tataricum). J. Agric. Food Chem. 2020, 68, 13685–13696. [Google Scholar] [CrossRef]
- Voitsekhovskaja, O.V. Phytochromes and Other (Photo) Receptors of Information in Plants. Russ. J. Plant Physiol. 2019, 66, 351–364. [Google Scholar] [CrossRef]
- Artés-Hernández, F.; Castillejo, N.; Martínez-Zamora, L. UV and Visible Spectrum Led Lighting as Abiotic Elicitors of Bioactive Compounds in Sprouts, Microgreens, and Baby Leaves—A Comprehensive Review Including Their Mode of Action. Foods 2022, 11, 265. [Google Scholar] [CrossRef]
- Skowron, E.; Trojak, M.; Pacak, I. Effects of UV-B and UV-C Spectrum Supplementation on the Antioxidant Properties and Photosynthetic Activity of Lettuce Cultivars. Int. J. Mol. Sci. 2024, 25, 9298. [Google Scholar] [CrossRef] [PubMed]
- Gąstoł, M.; Błaszczyk, U. Effect of Magnetic Field and UV-C Radiation on Postharvest Fruit Properties. Agriculture 2024, 14, 1167. [Google Scholar] [CrossRef]
- Liaqat, W.; Altaf, M.T.; Barutçular, C.; Nawaz, H.; Ullah, I.; Basit, A.; Mohamed, H.I. Ultraviolet-B Radiation in Relation to Agriculture in the Context of Climate Change: A Review. Cereal Res. Commun. 2024, 52, 1–24. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.; Zu, Y.; He, Y.; Li, Z.; Li, Y. Effects of UV-B Radiation Exposure on Transgenerational Plasticity in Grain Morphology and Proanthocyanidin Content in Yuanyang Red Rice. Int. J. Mol. Sci. 2024, 25, 4766. [Google Scholar] [CrossRef]
- Leonardelli, M.; Tissot, N.; Podolec, R.; Ares-Orpel, F.; Glauser, G.; Ulm, R.; Demarsy, E. Photoreceptor-Induced Sinapate Synthesis Contributes to Photoprotection in Arabidopsis. Plant Physiol. 2024, 196, 1518–1533. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Kaur, P.; Soni, R.; Madan, A.; Agarwal, P.; Singh, G. Decoding the Photoprotection Strategies and Manipulating Cyanobacterial Photoprotective Metabolites, Mycosporine-like Amino Acids, for next-Generation Sunscreens. Plant Physiol. Biochem. 2024, 212, 108744. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.M.; Hamilton, C.M.; Street, H.E. A Strain of Rosa Damascena Cultured Cells Resistant to Ultraviolet Light. Plant Physiol. 1979, 64, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; Pinto, D.C.G.A.; Freitas, H.; Santos, C.; Silva, A.M.S. The Antioxidant System in Olea Europaea to Enhanced UV-B Radiation Also Depends on Flavonoids and Secoiridoids. Phytochemistry 2020, 170, 112199. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Shi, B.; Zhu, W.; Zheng, B.; Zhou, K.; Qian, M.; Wu, H. Genome-Wide Identification, Characterization and Expression Analysis of Mango (Mangifera indica L.) Chalcone Synthase (CHS) Genes in Response to Light. Horticulturae 2022, 8, 968. [Google Scholar] [CrossRef]
- Dini, I.; Grumetto, L. Recent Advances in Natural Polyphenol Research. Molecules 2022, 27, 8777. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple Functional Roles of Anthocyanins in Plant-Environment Interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as Antioxidants in Plants: Location and Functional Significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef]
- Ghasemi, S.; Kumleh, H.H.; Kordrostami, M. Changes in the Expression of Some Genes Involved in the Biosynthesis of Secondary Metabolites in Cuminum cyminum L. under UV Stress. Protoplasma 2019, 256, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Charles, M.T.; Luo, Z.; Mimee, B.; Veronneau, P.-Y.; Rolland, D.; Roussel, D. Preharvest Ultraviolet C Irradiation Increased the Level of Polyphenol Accumulation and Flavonoid Pathway Gene Expression in Strawberry Fruit. J. Agric. Food Chem. 2017, 65, 9970–9979. [Google Scholar] [CrossRef]
- Demkura, P.V.; Abdala, G.; Baldwin, I.T.; Ballareݩ, C.L. Jasmonate-Dependent and-Independent Pathways Mediate Specific Effects of Solar Ultraviolet B Radiation on Leaf Phenolics and Antiherbivore Defense. Plant Physiol. 2010, 152, 1084–1095. [Google Scholar] [CrossRef]
- Berli, F.J.; Fanzone, M.; Piccoli, P.; Bottini, R. Solar UV-B and ABA Are Involved in Phenol Metabolism of Vitis vinifera L. Increasing Biosynthesis of Berry Skin Polyphenols. J. Agric. Food Chem. 2011, 59, 4874–4884. [Google Scholar] [CrossRef]
- Nenadis, N.; Llorens, L.; Koufogianni, A.; Diaz, L.; Font, J.; Gonzalez, J.A.; Verdaguer, D. Interactive Effects of UV Radiation and Reduced Precipitation on the Seasonal Leaf Phenolic Content/Composition and the Antioxidant Activity of Naturally Growing Arbutus Unedo Plants. J. Photochem. Photobiol. B 2015, 153, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Moreira-Rodríguez, M.; Nair, V.; Benavides, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. UVA, UVB Light, and Methyl Jasmonate, Alone or Combined, Redirect the Biosynthesis of Glucosinolates, Phenolics, Carotenoids, and Chlorophylls in Broccoli Sprouts. Int. J. Mol. Sci. 2017, 18, 2330. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Cao, B.; Zhou, S.; Liu, Y. Responses of the Flavonoid Pathway to UV-B Radiation Stress and the Correlation with the Lipid Antioxidant Characteristics in the Desert Plant Caryopteris mongolica. Acta Ecol. Sin. 2012, 32, 150–155. [Google Scholar] [CrossRef]
- Mariz-Ponte, N.; Mendes, R.J.; Sario, S.; De Oliveira, J.M.P.F.; Melo, P.; Santos, C. Tomato Plants Use Non-Enzymatic Antioxidant Pathways to Cope with Moderate UV-A/B Irradiation: A Contribution to the Use of UV-A/B in Horticulture. J. Plant Physiol. 2018, 221, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Nascimento, L.B.; Leal-Costa, M.V.; Menezes, E.A.; Lopes, V.R.; Muzitano, M.F.; Costa, S.S.; Tavares, E.S. Ultraviolet-B Radiation Effects on Phenolic Profile and Flavonoid Content of Kalanchoe Pinnata. J. Photochem. Photobiol. B 2015, 148, 73–81. [Google Scholar] [CrossRef]
- Sytar, O.; Zivcak, M.; Bruckova, K.; Brestic, M.; Hemmerich, I.; Rauh, C.; Simko, I. Shift in Accumulation of Flavonoids and Phenolic Acids in Lettuce Attributable to Changes in Ultraviolet Radiation and Temperature. Sci. Hortic. 2018, 239, 193–204. [Google Scholar] [CrossRef]
- Lee, M.; Son, J.E.; Oh, M. Growth and Phenolic Compounds of Lactuca sativa L. Grown in a Closed-type Plant Production System with UV-A,-B, Or-C Lamp. J. Sci. Food Agric. 2014, 94, 197–204. [Google Scholar] [CrossRef]
- Huyskens-Keil, S.; Eichholz, I.; Kroh, L.W.; Rohn, S. UV-B Induced Changes of Phenol Composition and Antioxidant Activity in Black Currant Fruit (Ribes nigrum L.). J. Appl. Bot. Food Qual. 2012, 81, 140–144. [Google Scholar]
- Chen, Z.; Ma, Y.; Yang, R.; Gu, Z.; Wang, P. Effects of Exogenous Ca2+ on Phenolic Accumulation and Physiological Changes in Germinated Wheat (Triticum aestivum L.) under UV-B Radiation. Food Chem. 2019, 288, 368–376. [Google Scholar] [CrossRef]
- Chen, Z.; Ma, Y.; Weng, Y.; Yang, R.; Gu, Z.; Wang, P. Effects of UV-B Radiation on Phenolic Accumulation, Antioxidant Activity and Physiological Changes in Wheat (Triticum aestivum L.) Seedlings. Food Biosci. 2019, 30, 100409. [Google Scholar] [CrossRef]
- Goyal, A.; Siddiqui, S.; Upadhyay, N.; Soni, J. Effects of Ultraviolet Irradiation, Pulsed Electric Field, Hot Water and Ethanol Vapours Treatment on Functional Properties of Mung Bean Sprouts. J. Food Sci. Technol. 2014, 51, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Alonso, R.; Berli, F.J.; Fontana, A.; Piccoli, P.; Bottini, R. Malbec Grape (Vitis vinifera L.) Responses to the Environment: Berry Phenolics as Influenced by Solar UV-B, Water Deficit and Sprayed Abscisic Acid. Plant Physiol. Biochem. 2016, 109, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Singh, O.S.; Pant, N.C.; Laishram, L.; Tewari, M.; Dhoundiyal, R.; Joshi, K.; Pandey, C.S. Effect of CuO Nanoparticles on Polyphenols Content and Antioxidant Activity in Ashwagandha (Withania somnifera L. Dunal). J. Pharmacogn. Phytochem. 2018, 7, 3433–3439. [Google Scholar]
- Nourozi, E.; Hosseini, B.; Maleki, R.; Mandoulakani, B.A. Pharmaceutical Important Phenolic Compounds Overproduction and Gene Expression Analysis in Dracocephalum kotschyi Hairy Roots Elicited by SiO2 Nanoparticles. Ind. Crops Prod. 2019, 133, 435–446. [Google Scholar] [CrossRef]
- Kőrösi, L.; Bouderias, S.; Csepregi, K.; Bognár, B.; Teszlák, P.; Scarpellini, A.; Castelli, A.; Hideg, É.; Jakab, G. Nanostructured TiO2-Induced Photocatalytic Stress Enhances the Antioxidant Capacity and Phenolic Content in the Leaves of Vitis Vinifera on a Genotype-Dependent Manner. J. Photochem. Photobiol. B 2019, 190, 137–145. [Google Scholar] [CrossRef]
- Girilal, M.; Fayaz, A.M.; Elumalai, L.K.; Sathiyaseelan, A.; Gandhiappan, J.; Kalaichelvan, P.T. Comparative Stress Physiology Analysis of Biologically and Chemically Synthesized Silver Nanoparticles on Solanum lycopersicum L. Colloid. Interface Sci. Commun. 2018, 24, 1–6. [Google Scholar] [CrossRef]
- Raigond, P.; Raigond, B.; Kaundal, B.; Singh, B.; Joshi, A.; Dutt, S. Effect of Zinc Nanoparticles on Antioxidative System of Potato Plants. J. Environ. Biol. 2017, 38, 435. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Thukral, A.K.; Bhardwaj, R. Epibrassinolide-Imidacloprid Interaction Enhances Non-Enzymatic Antioxidants in Brassica juncea L. Indian J. Plant Physiol. 2016, 21, 70–75. [Google Scholar] [CrossRef]
- Sharma, A.; Thakur, S.; Kumar, V.; Kanwar, M.K.; Kesavan, A.K.; Thukral, A.K.; Bhardwaj, R.; Alam, P.; Ahmad, P. Pre-Sowing Seed Treatment with 24-Epibrassinolide Ameliorates Pesticide Stress in Brassica juncea L. through the Modulation of Stress Markers. Front. Plant Sci. 2016, 7, 1569. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Yuan, H.; Kumar, V.; Ramakrishnan, M.; Kohli, S.K.; Kaur, R.; Thukral, A.K.; Bhardwaj, R.; Zheng, B. Castasterone Attenuates Insecticide Induced Phytotoxicity in Mustard. Ecotoxicol. Environ. Saf. 2019, 179, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, V.; Farimani, M.M.; Fathi, F.; Ghassempour, A. A Targeted Metabolomics Approach toward Understanding Metabolic Variations in Rice under Pesticide Stress. Anal. Biochem. 2015, 478, 65–72. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of Oxidative and Drought Tolerance in Arabidopsis by Overaccumulation of Antioxidant Flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Duan, M.; Eman, M.; Yuan, H.; Sharma, A.; Zheng, B. Comparative Analysis of Citrus Species’ Flavonoid Metabolism, Gene Expression Profiling, and Their Antioxidant Capacity under Drought Stress. Antioxidants 2024, 13, 1149. [Google Scholar] [CrossRef]
- Rao, M.J.; Feng, B.; Ahmad, M.H.; Tahir Ul Qamar, M.; Aslam, M.Z.; Khalid, M.F.; Hussain, S.B.; Zhong, R.; Ali, Q.; Xu, Q.; et al. LC-MS/MS-Based Metabolomics Approach Identified Novel Antioxidant Flavonoids Associated with Drought Tolerance in Citrus Species. Front. Plant Sci. 2023, 14, 1150854. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X. CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Rezayian, M.; Niknam, V.; Ebrahimzadeh, H. Differential Responses of Phenolic Compounds of Brassica napus under Drought Stress. Iran. J. Plant Physiol. 2018, 8, 2417–2425. [Google Scholar]
- Li, M.; Li, Y.; Zhang, W.; Li, S.; Gao, Y.; Ai, X.; Zhang, D.; Liu, B.; Li, Q. Metabolomics Analysis Reveals That Elevated Atmospheric CO2 Alleviates Drought Stress in Cucumber Seedling Leaves. Anal. Biochem. 2018, 559, 71–85. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, E.; Moreno, D.A.; Ferreres, F.; del Mar Rubio-Wilhelmi, M.; Ruiz, J.M. Differential Responses of Five Cherry Tomato Varieties to Water Stress: Changes on Phenolic Metabolites and Related Enzymes. Phytochemistry 2011, 72, 723–729. [Google Scholar] [CrossRef]
- Gharibi, S.; Tabatabaei, B.E.S.; Saeidi, G.; Goli, S.A.H. Effect of Drought Stress on Total Phenolic, Lipid Peroxidation, and Antioxidant Activity of Achillea Species. Appl. Biochem. Biotechnol. 2016, 178, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Khalil, N.; Fekry, M.; Bishr, M.; El-Zalabani, S.; Salama, O. Foliar Spraying of Salicylic Acid Induced Accumulation of Phenolics, Increased Radical Scavenging Activity and Modified the Composition of the Essential Oil of Water Stressed Thymus vulgaris L. Plant Physiol. Biochem. 2018, 123, 65–74. [Google Scholar] [CrossRef]
- Hodaei, M.; Rahimmalek, M.; Arzani, A.; Talebi, M. The Effect of Water Stress on Phytochemical Accumulation, Bioactive Compounds and Expression of Key Genes Involved in Flavonoid Biosynthesis in Chrysanthemum morifolium L. Ind. Crops Prod. 2018, 120, 295–304. [Google Scholar] [CrossRef]
- Perin, E.C.; da Silva Messias, R.; Borowski, J.M.; Crizel, R.L.; Schott, I.B.; Carvalho, I.R.; Rombaldi, C.V.; Galli, V. ABA-Dependent Salt and Drought Stress Improve Strawberry Fruit Quality. Food Chem. 2019, 271, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Galieni, A.; Di Mattia, C.; De Gregorio, M.; Speca, S.; Mastrocola, D.; Pisante, M.; Stagnari, F. Effects of Nutrient Deficiency and Abiotic Environmental Stresses on Yield, Phenolic Compounds and Antiradical Activity in Lettuce (Lactuca sativa L.). Sci. Hortic. 2015, 187, 93–101. [Google Scholar] [CrossRef]
- Varela, M.C.; Arslan, I.; Reginato, M.A.; Cenzano, A.M.; Luna, M.V. Phenolic Compounds as Indicators of Drought Resistance in Shrubs from Patagonian Shrublands (Argentina). Plant Physiol. Biochem. 2016, 104, 81–91. [Google Scholar] [CrossRef] [PubMed]
- García-Calderón, M.; Pons-Ferrer, T.; Mrazova, A.; Pal’ove-Balang, P.; Vilkova, M.; Pérez-Delgado, C.M.; Vega, J.M.; Eliášová, A.; Repčák, M.; Márquez, A.J. Modulation of Phenolic Metabolism under Stress Conditions in a Lotus japonicus Mutant Lacking Plastidic Glutamine Synthetase. Front. Plant Sci. 2015, 6, 760. [Google Scholar] [CrossRef]
- Silva, F.L.B.; Vieira, L.G.E.; Ribas, A.F.; Moro, A.L.; Neris, D.M.; Pacheco, A.C. Proline Accumulation Induces the Production of Total Phenolics in Transgenic Tobacco Plants under Water Deficit without Increasing the G6PDH Activity. Theor. Exp. Plant Physiol. 2018, 30, 251–260. [Google Scholar] [CrossRef]
- Pirbalouti, A.G.; Malekpoor, F.; Salimi, A.; Golparvar, A. Exogenous Application of Chitosan on Biochemical and Physiological Characteristics, Phenolic Content and Antioxidant Activity of Two Species of Basil (Ocimum ciliatum and Ocimum basilicum) under Reduced Irrigation. Sci. Hortic. 2017, 217, 114–122. [Google Scholar] [CrossRef]
- Ma, D.; Sun, D.; Wang, C.; Li, Y.; Guo, T. Expression of Flavonoid Biosynthesis Genes and Accumulation of Flavonoid in Wheat Leaves in Response to Drought Stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; Di Gaspero, G. Transcriptional Regulation of Anthocyanin Biosynthesis in Ripening Fruits of Grapevine under Seasonal Water Deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Taïbi, K.; Taïbi, F.; Abderrahim, L.A.; Ennajah, A.; Belkhodja, M.; Mulet, J.M. Effect of Salt Stress on Growth, Chlorophyll Content, Lipid Peroxidation and Antioxidant Defence Systems in Phaseolus vulgaris L. S. Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- De Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; de Abreu, C.E.B.; Gomes-Filho, E. Effect of Salt Stress on Antioxidative Enzymes and Lipid Peroxidation in Leaves and Roots of Salt-Tolerant and Salt-Sensitive Maize Genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]
- Martinez, V.; Mestre, T.C.; Rubio, F.; Girones-Vilaplana, A.; Moreno, D.A.; Mittler, R.; Rivero, R.M. Accumulation of Flavonols over Hydroxycinnamic Acids Favors Oxidative Damage Protection under Abiotic Stress. Front. Plant Sci. 2016, 7, 838. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Wang, Y.; Wu, Z.; Fu, C.; Lin, H. NtMYB4 and NtCHS1 Are Critical Factors in the Regulation of Flavonoid Biosynthesis and Are Involved in Salinity Responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef]
- Bistgani, Z.E.; Hashemi, M.; DaCosta, M.; Craker, L.; Maggi, F.; Morshedloo, M.R. Effect of Salinity Stress on the Physiological Characteristics, Phenolic Compounds and Antioxidant Activity of Thymus vulgaris L. and Thymus daenensis Celak. Ind. Crops Prod. 2019, 135, 311–320. [Google Scholar] [CrossRef]
- Rossi, L.; Borghi, M.; Francini, A.; Lin, X.; Xie, D.-Y.; Sebastiani, L. Salt Stress Induces Differential Regulation of the Phenylpropanoid Pathway in Olea europaea Cultivars Frantoio (Salt-Tolerant) and Leccino (Salt-Sensitive). J. Plant Physiol. 2016, 204, 8–15. [Google Scholar] [CrossRef]
- Al-Ghamdi, A.A.; Elansary, H.O. Synergetic Effects of 5-Aminolevulinic Acid and Ascophyllum Nodosum Seaweed Extracts on Asparagus Phenolics and Stress Related Genes under Saline Irrigation. Plant Physiol. Biochem. 2018, 129, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Golkar, P.; Taghizadeh, M. In Vitro Evaluation of Phenolic and Osmolite Compounds, Ionic Content, and Antioxidant Activity in Safflower (Carthamus tinctorius L.) under Salinity Stress. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 134, 357–368. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, H.; Chen, D.; Li, Z.; Peng, R.; Yao, Q. A Grape BHLH Transcription Factor Gene, VvbHLH1, Increases the Accumulation of Flavonoids and Enhances Salt and Drought Tolerance in Transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Yan, J.; Wang, B.; Jiang, Y.; Cheng, L.; Wu, T. GmFNSII-Controlled Soybean Flavone Metabolism Responds to Abiotic Stresses and Regulates Plant Salt Tolerance. Plant Cell Physiol. 2014, 55, 74–86. [Google Scholar] [CrossRef]
- Sarker, U.; Oba, S. Augmentation of Leaf Color Parameters, Pigments, Vitamins, Phenolic Acids, Flavonoids and Antioxidant Activity in Selected Amaranthus Tricolor under Salinity Stress. Sci. Rep. 2018, 8, 12349. [Google Scholar] [CrossRef] [PubMed]
- Ben Dkhil, B.; Denden, M. Effect of Salt Stress on Growth, Anthocyanins, Membrane Permeability and Chlorophyll Fluorescence of Okra (Abelmoschus esculentus L.) Seedlings. Am. J. Plant Physiol. 2012, 7, 174–183. [Google Scholar] [CrossRef]
- Eryılmaz, F. The Relationships between Salt Stress and Anthocyanin Content in Higher Plants. Biotechnol. Biotechnol. Equip. 2006, 20, 47–52. [Google Scholar] [CrossRef]
- Aloisi, I.; Parrotta, L.; Ruiz, K.B.; Landi, C.; Bini, L.; Cai, G.; Biondi, S.; Del Duca, S. New Insight into Quinoa Seed Quality under Salinity: Changes in Proteomic and Amino Acid Profiles, Phenolic Content, and Antioxidant Activity of Protein Extracts. Front. Plant Sci. 2016, 7, 656. [Google Scholar] [CrossRef]
- Lucini, L.; Borgognone, D.; Rouphael, Y.; Cardarelli, M.; Bernardi, J.; Colla, G. Mild Potassium Chloride Stress Alters the Mineral Composition, Hormone Network, and Phenolic Profile in Artichoke Leaves. Front. Plant Sci. 2016, 7, 948. [Google Scholar] [CrossRef] [PubMed]
- Valifard, M.; Mohsenzadeh, S.; Kholdebarin, B.; Rowshan, V. Effects of Salt Stress on Volatile Compounds, Total Phenolic Content and Antioxidant Activities of Salvia mirzayanii. S. Afr. J. Bot. 2014, 93, 92–97. [Google Scholar] [CrossRef]
- Valifard, M.; Mohsenzadeh, S.; Niazi, A.; Moghadam, A. Phenylalanine Ammonia Lyase Isolation and Functional Analysis of Phenylpropanoid Pathway under Salinity Stress in ’Salvia’ species. Aust. J. Crop Sci. 2015, 9, 656–665. [Google Scholar]
- Ma, Y.; Wang, P.; Gu, Z.; Tao, Y.; Shen, C.; Zhou, Y.; Han, Y.; Yang, R. Ca2+ Involved in GABA Signal Transduction for Phenolics Accumulation in Germinated Hulless Barley under NaCl Stress. Food Chem. X 2019, 2, 100023. [Google Scholar] [CrossRef] [PubMed]
- Çoban, Ö.; Baydar, N.G. Brassinosteroid Effects on Some Physical and Biochemical Properties and Secondary Metabolite Accumulation in Peppermint (Mentha piperita L.) under Salt Stress. Ind. Crops Prod. 2016, 86, 251–258. [Google Scholar] [CrossRef]
- Kaur, L.; Zhawar, V.K. Phenolic Parameters under Exogenous ABA, Water Stress, Salt Stress in Two Wheat Cultivars Varying in Drought Tolerance. Indian J. Plant Physiol. 2015, 20, 151–156. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Nisha, N.; Ejaz, B.; Khan, M.I.R.; Kumar, M.; Ramteke, P.W.; Gupta, R. A Comprehensive Review on the Heavy Metal Toxicity and Sequestration in Plants. Biomolecules 2021, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Ghori, N.-H.; Ghori, T.; Hayat, M.Q.; Imadi, S.R.; Gul, A.; Altay, V.; Ozturk, M. Heavy Metal Stress and Responses in Plants. Int. J. Environ. Sci. Technol. 2019, 16, 1807–1828. [Google Scholar] [CrossRef]
- Zhang, H.; Reynolds, M. Cadmium Exposure in Living Organisms: A Short Review. Sci. Total Environ. 2019, 678, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Muszyńska, E.; Labudda, M. Dual Role of Metallic Trace Elements in Stress Biology—From Negative to Beneficial Impact on Plants. Int. J. Mol. Sci. 2019, 20, 3117. [Google Scholar] [CrossRef] [PubMed]
- Renu, K.; Mukherjee, A.G.; Gopalakrishnan, A.V.; Wanjari, U.R.; Kannampuzha, S.; Murali, R.; Veeraraghavan, V.P.; Vinayagam, S.; Paz-Montelongo, S.; George, A. Protective Effects of Macromolecular Polyphenols, Metal (Zinc, Selenium, and Copper)-Polyphenol Complexes, and Pectin in Different Organs with an Emphasis on Arsenic Poisoning: A Review. Int. J. Biol. Macromol. 2023, 23, 126715. [Google Scholar]
- Kováčik, J.; Klejdus, B.; Hedbavny, J.; Štork, F.; Bačkor, M. Comparison of Cadmium and Copper Effect on Phenolic Metabolism, Mineral Nutrients and Stress-Related Parameters in Matricaria chamomilla Plants. Plant Soil 2009, 320, 231–242. [Google Scholar] [CrossRef]
- Mishra, B.; Singh Sangwan, N. Amelioration of Cadmium Stress in Withania somnifera by ROS Management: Active Participation of Primary and Secondary Metabolism. Plant Growth Regul. 2019, 87, 403–412. [Google Scholar] [CrossRef]
- Mishra, B.; Sangwan, R.S.; Mishra, S.; Jadaun, J.S.; Sabir, F.; Sangwan, N.S. Effect of Cadmium Stress on Inductive Enzymatic and Nonenzymatic Responses of ROS and Sugar Metabolism in Multiple Shoot Cultures of Ashwagandha (Withania somnifera Dunal). Protoplasma 2014, 251, 1031–1045. [Google Scholar] [CrossRef]
- Kaur, R.; Bhardwaj, R.; Sirhindi, G. Castasterone Regulated Polyphenolic Metabolism and Photosynthetic System in Brassica juncea Plants under Copper Stress. J. Pharmacogn. Phytochem. 2015, 4, 282–289. [Google Scholar]
- Chang, X.; Li, J.; Wei, S.; Ying, J.; Nevill, P.; Qi, Z.; Lu, Q.; You, Z. Integrated Comparative Transcriptome and Weighted Gene Co-Expression Network Analysis Provide Valuable Insights into the Response Mechanisms of Alisma Orientale to Cadmium Stress. Sci. Total Environ. 2024, 957, 177401. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, A.S.; Feng, X.; Gao, G.; Yu, C.; Chen, J.; Chen, K.; Wang, X.; Mou, P.; Shao, D.; Chen, P. Genome Wide Characterization of R2R3 MYB Transcription Factor from Apocynum venetum Revealed Potential Stress Tolerance and Flavonoid Biosynthesis Genes. Genomics 2022, 114, 110275. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Abubakar, A.S.; Chen, K.; Yu, C.; Zhu, A.; Chen, J.; Gao, G.; Wang, X.; Mou, P.; Chen, P. Genome-Wide Analysis of R2R3-MYB Transcription Factors in Boehmeria nivea (L.) Gaudich Revealed Potential Cadmium Tolerance and Anthocyanin Biosynthesis Genes. Front. Genet. 2023, 14, 1080909. [Google Scholar] [CrossRef]
- Villiers, F.; Ducruix, C.; Hugouvieux, V.; Jarno, N.; Ezan, E.; Garin, J.; Junot, C.; Bourguignon, J. Investigating the Plant Response to Cadmium Exposure by Proteomic and Metabolomic Approaches. Proteomics 2011, 11, 1650–1663. [Google Scholar] [CrossRef] [PubMed]
- Kohli, S.K.; Handa, N.; Sharma, A.; Gautam, V.; Arora, S.; Bhardwaj, R.; Wijaya, L.; Alyemeni, M.N.; Ahmad, P. Interaction of 24-Epibrassinolide and Salicylic Acid Regulates Pigment Contents, Antioxidative Defense Responses, and Gene Expression in Brassica juncea L. Seedlings under Pb Stress. Environ. Sci. Pollut. Res. 2018, 25, 15159–15173. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Yadav, P.; Sharma, A.; Thukral, A.K.; Kumar, V.; Kohli, S.K.; Bhardwaj, R. Castasterone and Citric Acid Treatment Restores Photosynthetic Attributes in Brassica juncea L. under Cd (II) Toxicity. Ecotoxicol. Environ. Saf. 2017, 145, 466–475. [Google Scholar] [CrossRef]
- Kejík, Z.; Kaplánek, R.; Masařík, M.; Babula, P.; Matkowski, A.; Filipenský, P.; Veselá, K.; Gburek, J.; Sýkora, D.; Martásek, P. Iron Complexes of Flavonoids-Antioxidant Capacity and Beyond. Int. J. Mol. Sci. 2021, 22, 646. [Google Scholar] [CrossRef] [PubMed]
- Mira, L.; Tereza Fernandez, M.; Santos, M.; Rocha, R.; Helena Florêncio, M.; Jennings, K.R. Interactions of Flavonoids with Iron and Copper Ions: A Mechanism for Their Antioxidant Activity. Free Radic. Res. 2002, 36, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, M.M.; Erxleben, A.; Ochocki, J. Properties and Applications of Flavonoid Metal Complexes. RSC Adv. 2015, 5, 45853–45877. [Google Scholar] [CrossRef]
- Anjitha, K.S.; Sameena, P.P.; Puthur, J.T. Functional Aspects of Plant Secondary Metabolites in Metal Stress Tolerance and Their Importance in Pharmacology. Plant Stress 2021, 2, 100038. [Google Scholar] [CrossRef]
- Kaur, P.; Bali, S.; Sharma, A.; Vig, A.P.; Bhardwaj, R. Effect of Earthworms on Growth, Photosynthetic Efficiency and Metal Uptake in Brassica juncea L. Plants Grown in Cadmium-Polluted Soils. Environ. Sci. Pollut. Res. 2017, 24, 13452–13465. [Google Scholar] [CrossRef]
- Kaur, P.; Bali, S.; Sharma, A.; Vig, A.P.; Bhardwaj, R. Role of Earthworms in Phytoremediation of Cadmium (Cd) by Modulating the Antioxidative Potential of Brassica juncea L. Appl. Soil Ecol. 2018, 124, 306–316. [Google Scholar] [CrossRef]
- Kohli, S.K.; Handa, N.; Sharma, A.; Kumar, V.; Kaur, P.; Bhardwaj, R. Synergistic Effect of 24-Epibrassinolide and Salicylic Acid on Photosynthetic Efficiency and Gene Expression in Brassica juncea L. under Pb Stress. Turk. J. Biol. 2017, 41, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.-M.; Rekha, K.; Rajakumar, G.; Thiruvengadam, M. Production of Bioactive Compounds and Gene Expression Alterations in Hairy Root Cultures of Chinese Cabbage Elicited by Copper Oxide Nanoparticles. Plant Cell Tissue Organ Cult. (PCTOC) 2018, 134, 95–106. [Google Scholar] [CrossRef]
- Jańczak-Pieniążek, M.; Cichoński, J.; Michalik, P.; Chrzanowski, G. Effect of Heavy Metal Stress on Phenolic Compounds Accumulation in Winter Wheat Plants. Molecules 2022, 28, 241. [Google Scholar] [CrossRef] [PubMed]
- Handa, N.; Kohli, S.K.; Sharma, A.; Thukral, A.K.; Bhardwaj, R.; Alyemeni, M.N.; Wijaya, L.; Ahmad, P. Selenium Ameliorates Chromium Toxicity through Modifications in Pigment System, Antioxidative Capacity, Osmotic System, and Metal Chelators in Brassica juncea Seedlings. S. Afr. J. Bot. 2018, 119, 1–10. [Google Scholar] [CrossRef]
- Kısa, D.; Elmastaş, M.; Öztürk, L.; Kayır, Ö. Responses of the Phenolic Compounds of Zea mays under Heavy Metal Stress. Appl. Biol. Chem. 2016, 59, 813–820. [Google Scholar] [CrossRef]
- Smirnov, O.E.; Kosyan, A.M.; Kosyk, O.I.; Taran, N.Y. Response of Phenolic Metabolism Induced by Aluminium Toxicity in Fagopyrum Esculentum Moench. Plants. Ukr. Biochem. J. 2015, 87, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, Q.; Lu, H.; Li, J.; Yang, D.; Liu, J.; Yan, C. Phenolic Metabolism and Related Heavy Metal Tolerance Mechanism in Kandelia obovata under Cd and Zn Stress. Ecotoxicol. Environ. Saf. 2019, 169, 134–143. [Google Scholar] [CrossRef]
- Leng, X.; Jia, H.; Sun, X.; Shangguan, L.; Mu, Q.; Wang, B.; Fang, J. Comparative Transcriptome Analysis of Grapevine in Response to Copper Stress. Sci. Rep. 2015, 5, 17749. [Google Scholar] [CrossRef]
- Ling, J.; Tan, J.; Chen, H.; Yang, Z.; Luo, Q.; Jia, J. Physiology, Transcriptome and Root Exudates Analysis of Response to Aluminum Stress in Pinus massoniana. Forests 2023, 14, 1410. [Google Scholar] [CrossRef]
- Lu, H.; Zhu, F.; Xu, H.; Liu, J.; Wu, Y. Insights of Mechanism into Enhanced Removal of Cr (VI) by Ginkgo Biloba Leaves Synthesized Bimetallic Nano-Zero-Valent Iron/Copper. Colloids Surf. A Physicochem. Eng. Asp. 2023, 675, 132094. [Google Scholar] [CrossRef]
- Zafari, S.; Sharifi, M.; Chashmi, N.A.; Mur, L.A.J. Modulation of Pb-Induced Stress in Prosopis Shoots through an Interconnected Network of Signaling Molecules, Phenolic Compounds and Amino Acids. Plant Physiol. Biochem. 2016, 99, 11–20. [Google Scholar] [CrossRef]
- Correia, B.; Hancock, R.D.; Amaral, J.; Gomez-Cadenas, A.; Valledor, L.; Pinto, G. Combined Drought and Heat Activates Protective Responses in Eucalyptus Globulus That Are Not Activated When Subjected to Drought or Heat Stress Alone. Front. Plant Sci. 2018, 9, 819. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.C.; de Oliveira, J.M.P.F.; Marum, L.; Pereira, V.; Almeida, T.; Nunes, S.; Araujo, M.; Moutinho-Pereira, J.; Correia, C.M.; Santos, C. Pinus elliottii and P. elliottii x P. caribaea Hybrid Differently Cope with Combined Drought and Heat Episodes. Ind. Crops Prod. 2022, 176, 114428. [Google Scholar] [CrossRef]
- Valente, S.; Machado, B.; Pinto, D.C.G.A.; Santos, C.; Silva, A.M.S.; Dias, M.C. Modulation of Phenolic and Lipophilic Compounds of Olive Fruits in Response to Combined Drought and Heat. Food Chem. 2020, 329, 127191. [Google Scholar] [CrossRef]
- Peres, F.; Martins, L.L.; Mourato, M.; Vitorino, C.; Antunes, P.; Ferreira-Dias, S. Phenolic Compounds of ‘Galega Vulgar’ and ‘Cobrançosa’ Olive Oils along Early Ripening Stages. Food Chem. 2016, 211, 51–58. [Google Scholar] [CrossRef]
- Martinelli, F.; Remorini, D.; Saia, S.; Massai, R.; Tonutti, P. Metabolic Profiling of Ripe Olive Fruit in Response to Moderate Water Stress. Sci. Hortic. 2013, 159, 52–58. [Google Scholar] [CrossRef]
- Cizmarova, B.; Hubkova, B.; Birkova, A. Quercetin as an Effective Antioxidant against Superoxide Radical. Funct. Food Sci. 2023, 3, 15–25. [Google Scholar] [CrossRef]
- Parcheta, M.; Świsłocka, R.; Orzechowska, S.; Akimowicz, M.; Choińska, R.; Lewandowski, W. Recent Developments in Effective Antioxidants: The Structure and Antioxidant Properties. Materials 2021, 14, 1984. [Google Scholar] [CrossRef]
- Jan, R.; Khan, M.; Asaf, S.; Lubna; Asif, S.; Kim, K.-M. Bioactivity and Therapeutic Potential of Kaempferol and Quercetin: New Insights for Plant and Human Health. Plants 2022, 11, 2623. [Google Scholar] [CrossRef] [PubMed]
- Erlund, I. Review of the Flavonoids Quercetin, Hesperetin, and Naringenin. Dietary Sources, Bioactivities, Bioavailability, and Epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Saviranta, N.M.M.; Veeroos, L.; Granlund, L.J.; Hassinen, V.H.; Kaarniranta, K.; Karjalainen, R.O. Plant Flavonol Quercetin and Isoflavone Biochanin A Differentially Induce Protection against Oxidative Stress and Inflammation in ARPE-19 Cells. Food Res. Int. 2011, 44, 109–113. [Google Scholar] [CrossRef]
- Wang, F.; Ren, X.; Zhang, F.; Qi, M.; Zhao, H.; Chen, X.; Ye, Y.; Yang, J.; Li, S.; Zhang, Y. A R2R3-Type MYB Transcription Factor Gene from Soybean, GmMYB12, Is Involved in Flavonoids Accumulation and Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Biotechnol. Rep. 2019, 13, 219–233. [Google Scholar] [CrossRef]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant Flavonoids: Classification, Distribution, Biosynthesis, and Antioxidant Activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Hudecova, L.; Lauro, P.; Simunková, M.; Barbierikova, Z.; Malcek, M.; Alwasel, S.H.; Alhazza, I.M.; Rhodes, C.J.; Valko, M. The Effect of Luteolin on DNA Damage Mediated by a Copper Catalyzed Fenton Reaction. J. Inorg. Biochem. 2022, 226, 111635. [Google Scholar] [CrossRef] [PubMed]
- Vafadar, F.; Amooaghaie, R.; Ehsanzadeh, P.; Ghanadian, M.; Talebi, M.; Ghanati, F. Melatonin and Calcium Modulate the Production of Rosmarinic Acid, Luteolin, and Apigenin in Dracocephalum kotschyi under Salinity Stress. Phytochemistry 2020, 177, 112422. [Google Scholar] [CrossRef]
- Borges Bubols, G.; da Rocha Vianna, D.; Medina-Remon, A.; von Poser, G.; Maria Lamuela-Raventos, R.; Lucia Eifler-Lima, V.; Cristina Garcia, S. The Antioxidant Activity of Coumarins and Flavonoids. Mini Rev. Med. Chem. 2013, 13, 318–334. [Google Scholar]
- Dędek, K.; Rosicka-Kaczmarek, J.; Nebesny, E.; Kowalska, G. Characteristics and Biological Properties of Ferulic Acid. Biotechnol. Food Sci. 2019, 83, 71–85. [Google Scholar]
- Kaurinovic, B.; Vastag, D. Flavonoids and Phenolic Acids as Potential Natural Antioxidants; IntechOpen: London, UK, 2019; ISBN 1789239206. [Google Scholar]
- Xu, J.-G.; Hu, Q.-P.; Liu, Y. Antioxidant and DNA-Protective Activities of Chlorogenic Acid Isomers. J. Agric. Food Chem. 2012, 60, 11625–11630. [Google Scholar] [CrossRef] [PubMed]
- Mughal, A.; Jabeen, N.; Ashraf, K.; Sultan, K.; Farhan, M.; Hussain, M.I.; Deng, G.; Alsudays, I.M.; Saleh, M.A.; Tariq, S. Exploring the Role of Caffeic Acid in Mitigating Abiotic Stresses in Plants: A Review. Plant Stress 2024, 12, 100487. [Google Scholar] [CrossRef]

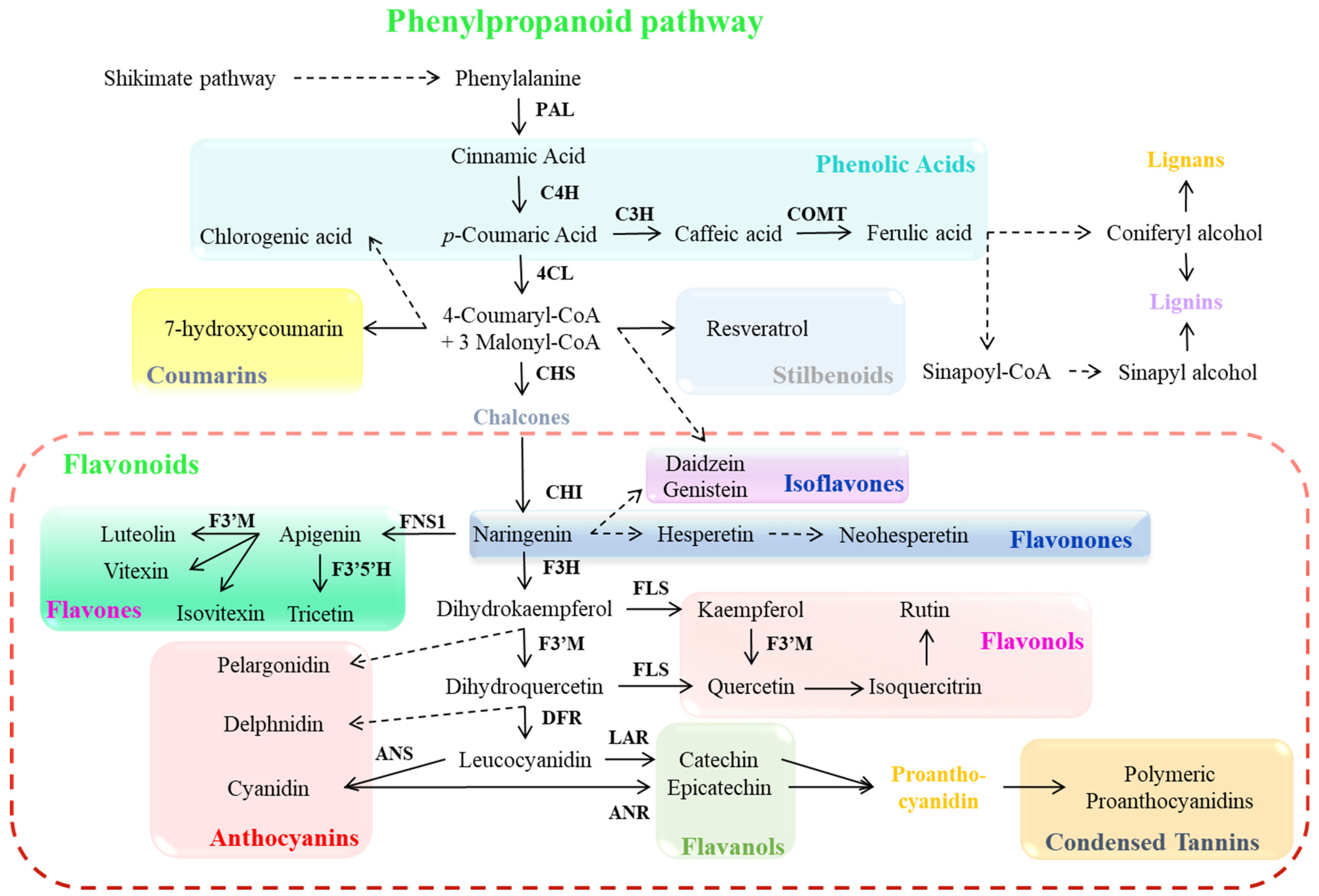
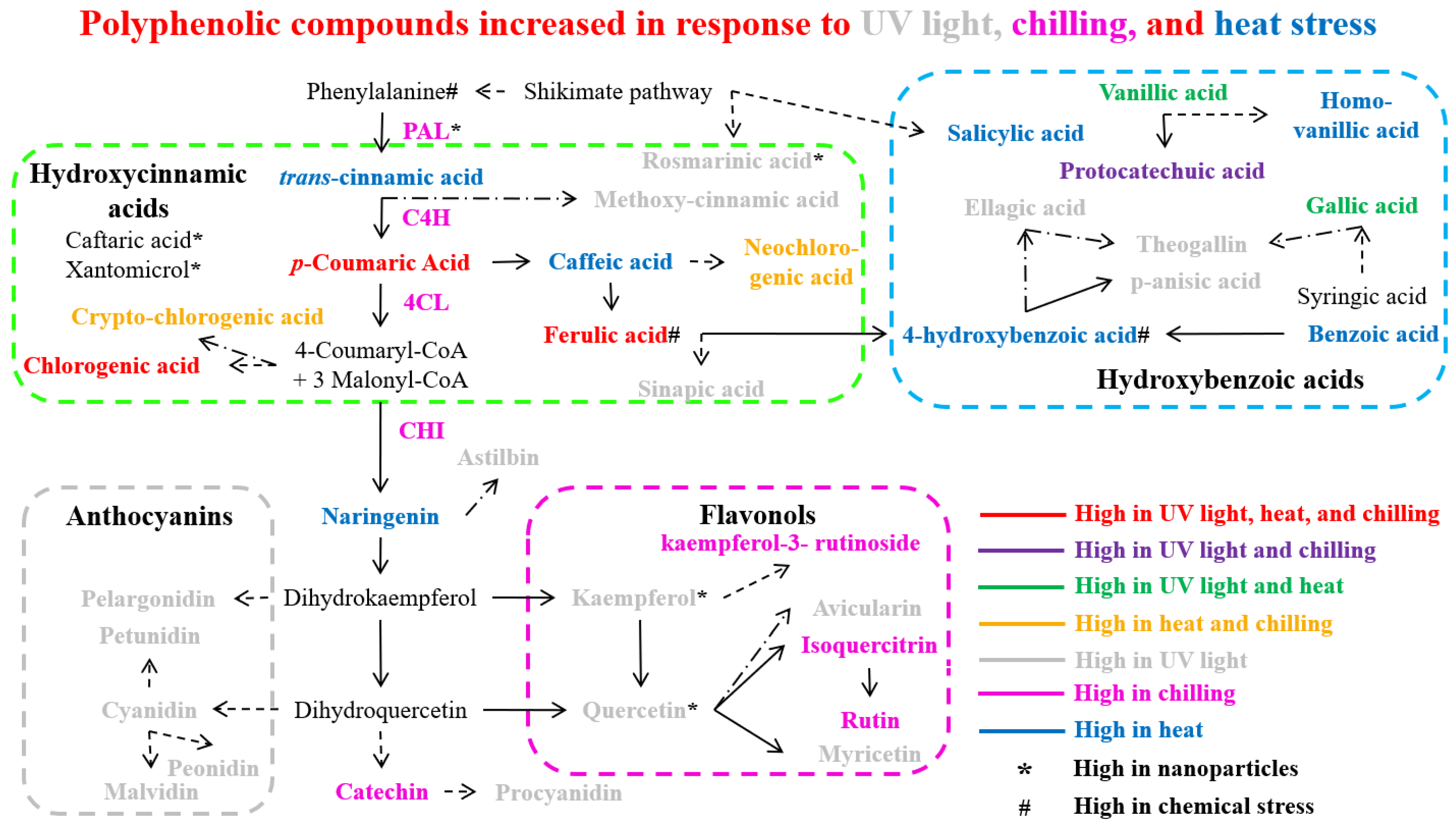
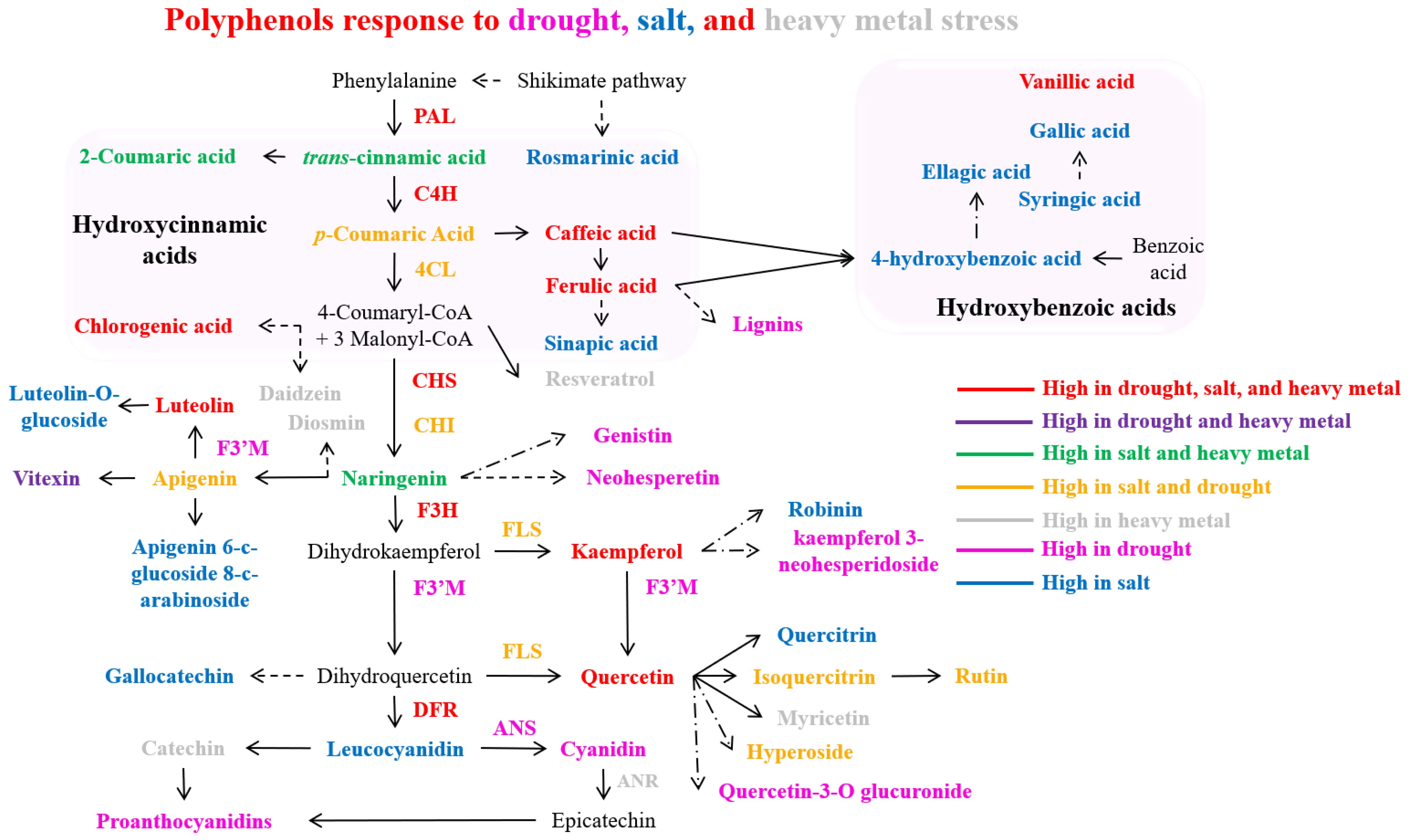
| Serial No. | Transcription Factor | Plant Species | Regulated Compounds | Stress Tolerance | Regulatory Mechanism | Stress-Protective Mechanism | References |
|---|---|---|---|---|---|---|---|
| 1 | AtMYB12 | Arabidopsis thaliana | Flavonoids and flavonols | Salt and drought | Activation of CHS, CHI, and F3H genes | Neutralization of ROS | [35] |
| 2 | AtMYB12 | Arabidopsis thaliana | Flavonoid and flavones | UV-B | Upregulates FLS and CHS genes | Protect photosynthetic apparatus | [36] |
| 3 | GbMYB11 | Ginkgo biloba | Flavonol | Salt | F3′H and FLS | Neutralize ROS | [37] |
| 4 | MdMYB2 | Apple Borkh | Anthocyanins | Cold tolerance | Anthocyanin biosynthesis genes | Interact MdSIZ1 promoter | [38] |
| 5 | MdMYB88 | Malus domestica | Anthocyanidins, kaemferol, and quercetin | Salt and cold stress | Regulates MdUGT83L3 | Decreased ROS | [39] |
| 6 | MdMYB308L | Malus domestica | Accumulates anthocyanins | Cold | Binds to promoters of the DFR gene | Interaction with MdbHLH33 | [40] |
| 7 | VyMYB24 | Vitis yanshanesis | Regulates proline, gibberellin, and metabolites | Drought tolerance | Enhances SOD, POD, and CAT | Decreased ROS | [41] |
| 8 | VvMYBF1 | Vitis vinifera L. | Flavonoid | Drought and salt | Regulates PAL, CHI, FLS, and DFR. | Reduction of H2O2 and MDA | [42] |
| 9 | VhMYB15 | hybrid V. riparia L. and V. labrusca | Proline and antioxidant enzymes | Salinity and drought | Regulates AtSOS1-3 and AtNHX1 | Less H2O2 and MDA, high SOD, POD, CAT | [43] |
| 10 | MYB4 | Arabidopsis thaliana | Negative regulator of sinapate ester and flavonoids | UV-B stress | Represses cinnamate 4-hydroxylase (C4H) | Repair UV-B-induced DNA breaks | [44] |
| 11 | AtMYB111 | Arabidopsis thaliana | Increases quercetin production | Salt stress response | Regulates CHS, F3H, and FLS1 genes | Salt resistance | [45] |
| 12 | GbMYB1 | Ginkgo biloba | Activates flavonoid biosynthesis | UV-B radiation | Regulates FLS module | Photoprotection by anthocyanins | [46] |
| 13 | VaMyb14 | Vitis amurensis | Activating lipid transfer protein | Cold and drought tolerance | Controls stilbene synthase expression | Increased POD activity and reduced MDA | [47] |
| 14 | VviMYB24 | Cabernet Sauvignon’ grape | Positively regulates flavonol biosynthesis | Drought stress | Activate VviFLS5 expression | Forms MYB24-iMYC2b-FLS5 module | [48] |
| 15 | MYB-Arahy.J3K16K | Arachis hypogaea | Increases anthocyanins | Abiotic stress tolerance | Regulates PAL, CHS, CHI, F3H, DFR, and ANR | MAPK cascade, ethylene, auxin, and abscisic acid | [49] |
| 16 | LlMYB3 | Lilium lancifolium L. | Anthocyanin accumulation | Chilling, drought, and salt | Directly binds with LlCHS2 | Upregulates APX2 and LEA14 | [50] |
| 17 | LhMYBC2 | Lilium cv. ‘Sunny Martinique’ | Modulates flavonoid biosynthesis | Heat stress | Regulates isoflavonoid biosynthesis genes | Increased isoflavonoid | [51] |
| 18 | CsMYBL2 | Camellia sinensis | Upregulates anthocyanin and catechins | Light and temperature | Activates phenylpropanoid biosynthesis genes | Enhances cold and UV-B tolerance | [52] |
| 19 | RsMYB1 | Raphanus sativus | Enhances anthocyanin production | Heavy metal stress | Activates anthocyanin biosynthesis genes | Increased activity of GST, PCS, SOD, POD, and CAT | [53] |
| 20 | SlMYB14 | Solanum lycopersicum | Promotes flavonoid biosynthesis | Pesticide stress | Binds with SlPAL gene | Maintain ROS homeostasis | [54] |
| 21 | SsMYB113 | Schima superba | Accumulates flavonoids | Drought | Bind directly to SsCHS gene | Modulates ROS generation | [55] |
| 22 | FtMYB11 | Fagopyrum tataricum | Flavonoids | Drought, Salt, and ABA | Regulates AtC4H, AtF3H, AtANS, AtFLS, and At4CL | Regulating MDA and proline | [56] |
| 23 | VvmybA1 from Vitis vinifera | Overexpressed in Hamlin’s citrus trees | Anthocyanin accumulation | Cold stress | Regulates anthocyanin biosynthesis genes | Enhanced ROS scavenging capacity | [57] |
| 24 | GmMYB12 | Glycine max | Flavonoids accumulation | Salt and drought | Upregulates flavonoid genes | Increase proline content, SOD, and POD, less MDA and H2O2 | |
| 25 | ScAN2 (MYB) | Solanum commersonii | Activates hydroxycinnamic acid derivatives | Cold stress | Regulates phenylpropanoid genes | Quench ROS and induce cold tolerance | [58] |
| 26 | AtMYB2 | Arabidopsis thaliana | Triggers phenolic acids synthesis | Salt stress | Regulates phenolic acid genes | Decrease ROS accumulation | [59] |
| 27 | AtMYB3 | Arabidopsis thaliana | Stimulates lignin and anthocyanins | Salt stress | Promote expression of PAL1, C4H, COMT, DFR, etc. | Reduces ROS | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, M.J.; Zheng, B. The Role of Polyphenols in Abiotic Stress Tolerance and Their Antioxidant Properties to Scavenge Reactive Oxygen Species and Free Radicals. Antioxidants 2025, 14, 74. https://doi.org/10.3390/antiox14010074
Rao MJ, Zheng B. The Role of Polyphenols in Abiotic Stress Tolerance and Their Antioxidant Properties to Scavenge Reactive Oxygen Species and Free Radicals. Antioxidants. 2025; 14(1):74. https://doi.org/10.3390/antiox14010074
Chicago/Turabian StyleRao, Muhammad Junaid, and Bingsong Zheng. 2025. "The Role of Polyphenols in Abiotic Stress Tolerance and Their Antioxidant Properties to Scavenge Reactive Oxygen Species and Free Radicals" Antioxidants 14, no. 1: 74. https://doi.org/10.3390/antiox14010074
APA StyleRao, M. J., & Zheng, B. (2025). The Role of Polyphenols in Abiotic Stress Tolerance and Their Antioxidant Properties to Scavenge Reactive Oxygen Species and Free Radicals. Antioxidants, 14(1), 74. https://doi.org/10.3390/antiox14010074







