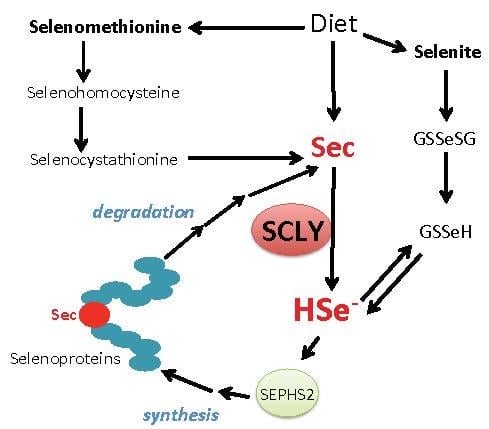
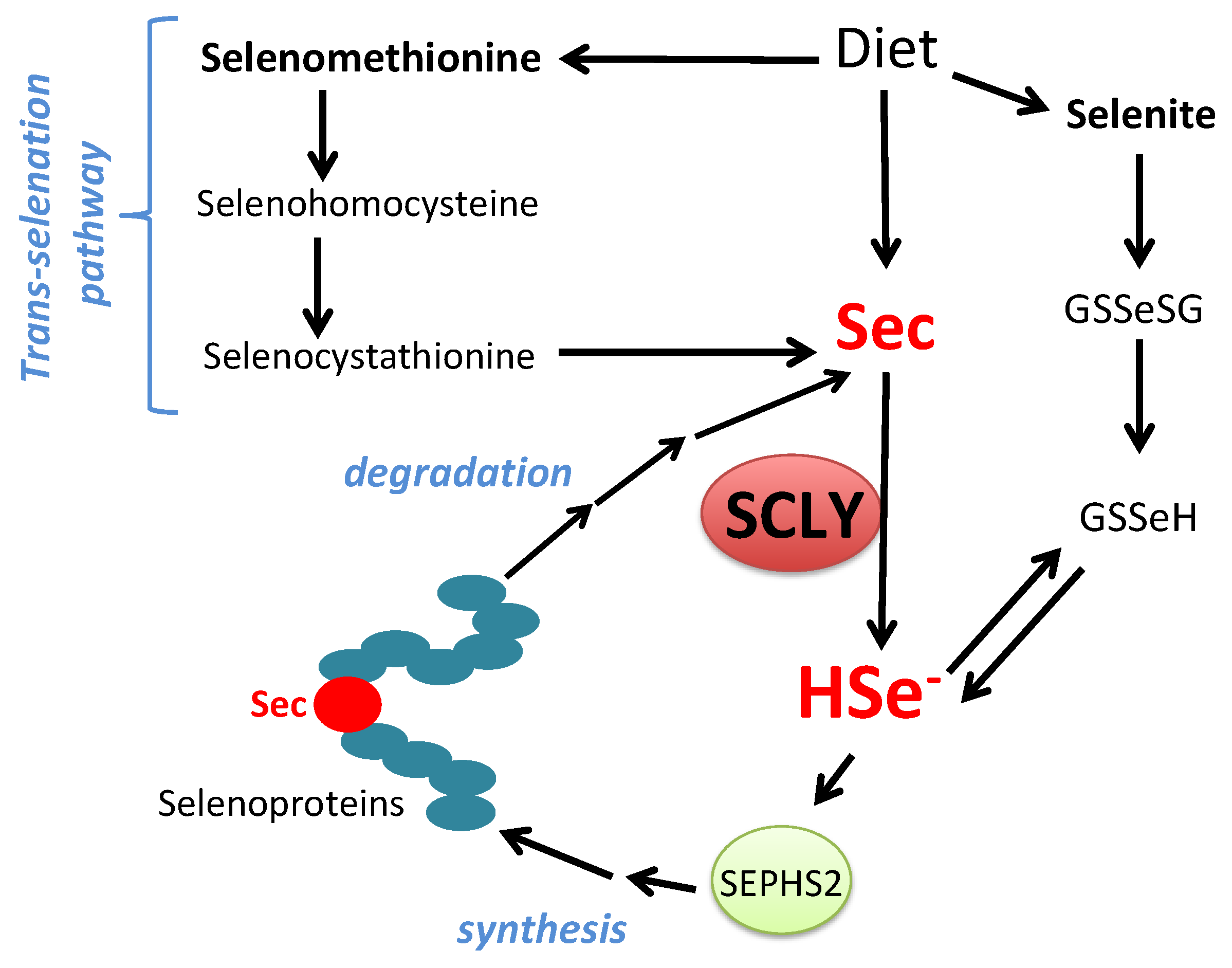
Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme
Abstract
:1. Introduction
1.1. Selenocysteine β-lyase Identification
1.2. Brief Overview of Selenocysteine and Selenoproteins
1.3. Biosynthesis of Selenocysteine
2. Selenocysteine β-Lyase
2.1. Biochemical Characteristics and Mechanism of Action
2.2. Phylogenetic Distribution
2.3. Subcellular Localization
2.4. Tissue Distribution of SCLY
2.5. Regulation of Gene Expression and Protein Levels
2.6. Physiological Role
2.7. SCLY Gene Polymorphisms
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Esaki, N.; Nakamura, T.; Tanaka, H.; Soda, K. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine. Mammalian distribution and purification and properties of pig liver enzyme. J. Biol. Chem. 1982, 257, 4386–4391. [Google Scholar] [PubMed]
- Esaki, N.; Nakamura, T.; Tanaka, H.; Suzuki, T.; Morino, Y.; Soda, K. Enzymatic synthesis of selenocysteine in rat liver. Biochemistry 1981, 20, 4492–4496. [Google Scholar] [CrossRef] [PubMed]
- Copeland, P.R.; Fletcher, J.E.; Carlson, B.A.; Hatfield, D.L.; Driscoll, D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000, 19, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Low, S.C.; Grundner-Culemann, E.; Harney, J.W.; Berry, M.J. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000, 19, 6882–6890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, M.J.; Tujebajeva, R.M.; Copeland, P.R.; Xu, X.M.; Carlson, B.A.; Martin, G.W., III; Low, S.C.; Mansell, J.B.; Grundner-Culemann, E.; Harney, J.W.; et al. Selenocysteine incorporation directed from the 3′UTR: Characterization of eukaryotic EFsec and mechanistic implications. Biofactors 2001, 14, 17–24. [Google Scholar] [CrossRef]
- Chavatte, L.; Brown, B.A.; Driscoll, D.M. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 2005, 12, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef]
- Mix, H.; Lobanov, A.V.; Gladyshev, V.N. SECIS elements in the coding regions of selenoprotein transcripts are functional in higher eukaryotes. Nucleic Acids Res. 2007, 35, 414–423. [Google Scholar] [CrossRef]
- Simonovic, M.; Puppala, A.K. On elongation factor eEFSec, its role and mechanism during selenium incorporation into nascent selenoproteins. Biochim. Biophys. Acta Gen Subj. 2018, 1862, 2463–2472. [Google Scholar] [CrossRef]
- Fradejas-Villar, N.; Seeher, S.; Anderson, C.B.; Doengi, M.; Carlson, B.A.; Hatfield, D.L.; Schweizer, U.; Howard, M.T. The RNA-binding protein Secisbp2 differentially modulates UGA codon reassignment and RNA decay. Nucleic Acids Res. 2017, 45, 4094–4107. [Google Scholar] [CrossRef]
- Latreche, L.; Jean-Jean, O.; Driscoll, D.M.; Chavatte, L. Novel structural determinants in human SECIS elements modulate the translational recoding of UGA as selenocysteine. Nucleic Acids Res. 2009, 37, 5868–5880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budiman, M.E.; Bubenik, J.L.; Miniard, A.C.; Middleton, L.M.; Gerber, C.A.; Cash, A.; Driscoll, D.M. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell 2009, 35, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Miniard, A.C.; Middleton, L.M.; Budiman, M.E.; Gerber, C.A.; Driscoll, D.M. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res. 2010, 38, 4807–4820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamond, A.M.; Choi, I.S.; Crain, P.F.; Hashizume, T.; Pomerantz, S.C.; Cruz, R.; Steer, C.J.; Hill, K.E.; Burk, R.F.; McCloskey, J.A.; et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem. 1993, 268, 14215–14223. [Google Scholar] [PubMed]
- Carlson, B.A.; Schweizer, U.; Perella, C.; Shrimali, R.K.; Feigenbaum, L.; Shen, L.; Speransky, S.; Floss, T.; Jeong, S.J.; Watts, J.; et al. The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice. Biochem. J. 2009, 418, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, B.A.; Lee, B.J.; Tsuji, P.A.; Copeland, P.R.; Schweizer, U.; Gladyshev, V.N.; Hatfield, D.L. Selenocysteine tRNA([Ser]Sec), the Central Component of Selenoprotein Biosynthesis: Isolation, Identification, Modification, and Sequencing. Methods Mol. Biol. 2018, 1661, 43–60. [Google Scholar] [CrossRef]
- Howard, M.T.; Copeland, P.R. New Directions for Understanding the Codon Redefinition Required for Selenocysteine Incorporation. Biol. Trace Elem. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Vindry, C.; Ohlmann, T.; Chavatte, L. Translation regulation of mammalian selenoproteins. Biochim. Biophys. Acta Gen Subj. 2018, 1862, 2480–2492. [Google Scholar] [CrossRef]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigo, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef]
- Burk, R.F.; Hill, K.E. Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta 2009, 1790, 1441–1447. [Google Scholar] [CrossRef]
- Cupp-Sutton, K.A.; Ashby, M.T. Biological Chemistry of Hydrogen Selenide. Antioxidants 2016, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Mousa, R.; Notis Dardashti, R.; Metanis, N. Selenium and Selenocysteine in Protein Chemistry. Angew. Chem. Int. Ed. 2017, 56, 15818–15827. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.M.; Mugesh, G. Modelling the Inhibition of Selenoproteins by Small Molecules Using Cysteine and Selenocysteine Derivatives. Chemistry 2019, 25, 8875–8883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Palioura, S.; Salazar, J.C.; Su, D.; O’Donoghue, P.; Hohn, M.J.; Cardoso, A.M.; Whitman, W.B.; Soll, D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. USA 2006, 103, 18923–18927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holman, K.M.; Puppala, A.K.; Lee, J.W.; Lee, H.; Simonovic, M. Insights into substrate promiscuity of human seryl-tRNA synthetase. RNA 2017, 23, 1685–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.M.; Carlson, B.A.; Mix, H.; Zhang, Y.; Saira, K.; Glass, R.S.; Berry, M.J.; Gladyshev, V.N.; Hatfield, D.L. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007, 5, e4. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Carlson, B.A.; Irons, R.; Mix, H.; Zhong, N.; Gladyshev, V.N.; Hatfield, D.L. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 2007, 404, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Na, J.; Jung, J.; Bang, J.; Lu, Q.; Carlson, B.A.; Guo, X.; Gladyshev, V.N.; Kim, J.; Hatfield, D.L.; Lee, B.J. Selenophosphate synthetase 1 and its role in redox homeostasis, defense and proliferation. Free Radic. Biol. Med. 2018, 127, 190–197. [Google Scholar] [CrossRef]
- Tobe, R.; Carlson, B.A.; Huh, J.H.; Castro, N.P.; Xu, X.M.; Tsuji, P.A.; Lee, S.G.; Bang, J.; Na, J.W.; Kong, Y.Y.; et al. Selenophosphate synthetase 1 is an essential protein with roles in regulation of redox homoeostasis in mammals. Biochem. J. 2016, 473, 2141–2154. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.M.; Turanov, A.A.; Carlson, B.A.; Yoo, M.H.; Everley, R.A.; Nandakumar, R.; Sorokina, I.; Gygi, S.P.; Gladyshev, V.N.; Hatfield, D.L. Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc. Natl. Acad. Sci. USA 2010, 107, 21430–21434. [Google Scholar] [CrossRef] [Green Version]
- Turanov, A.A.; Everley, R.A.; Hybsier, S.; Renko, K.; Schomburg, L.; Gygi, S.P.; Hatfield, D.L.; Gladyshev, V.N. Regulation of Selenocysteine Content of Human Selenoprotein P by Dietary Selenium and Insertion of Cysteine in Place of Selenocysteine. PLoS ONE 2015, 10, e0140353. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E. Regulation of Selenium Metabolism and Transport. Annu. Rev. Nutr. 2015, 35, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, S.; Takehashi, M.; Tanaka, H.; Mihara, H.; Kurihara, T.; Tanaka, S.; Hill, K.; Burk, R.; Esaki, N. Mammalian selenocysteine lyase is involved in selenoprotein biosynthesis. J. Nutr. Sci. Vitam. 2011, 57, 298–305. [Google Scholar] [CrossRef]
- Mihara, H.; Kurihara, T.; Watanabe, T.; Yoshimura, T.; Esaki, N. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase. Candidate for selenium delivery protein in selenoprotein synthesis. J. Biol. Chem. 2000, 275, 6195–6200. [Google Scholar] [CrossRef] [PubMed]
- Ouzounis, C.; Sander, C. Homology of the NifS family of proteins to a new class of pyridoxal phosphate-dependent enzymes. FEBS Lett. 1993, 322, 159–164. [Google Scholar] [CrossRef] [Green Version]
- Soda, K.; Esaki, N.; Nakamura, T.; Karai, N.; Chocat, P.; Tanaka, H. Selenocysteine beta-lyase: A novel pyridoxal enzyme. Prog. Clin. Biol. Res. 1984, 144, 319–328. [Google Scholar]
- Daher, R.; Van, F.L. Characterization of selenocysteine lyase in human tissues and its relationship to tissue selenium concentrations. J. Trace Elem. Electrolytes Health Dis. 1992, 6, 189–194. [Google Scholar] [PubMed]
- Stadtman, T.C. Selenocysteine Lyase. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Suzuki, K.T.; Kurasaki, K.; Suzuki, N. Selenocysteine beta-lyase and methylselenol demethylase in the metabolism of Se-methylated selenocompounds into selenide. Biochim. Biophys. Acta 2007, 1770, 1053–1061. [Google Scholar] [CrossRef]
- Omi, R.; Kurokawa, S.; Mihara, H.; Hayashi, H.; Goto, M.; Miyahara, I.; Kurihara, T.; Hirotsu, K.; Esaki, N. Reaction mechanism and molecular basis for selenium/sulfur discrimination of selenocysteine lyase. J. Biol. Chem. 2010, 285, 12133–12139. [Google Scholar] [CrossRef]
- Collins, R.; Johansson, A.L.; Karlberg, T.; Markova, N.; van den Berg, S.; Olesen, K.; Hammarstrom, M.; Flores, A.; Schuler, H.; Schiavone, L.H.; et al. Biochemical discrimination between selenium and sulfur 1: A single residue provides selenium specificity to human selenocysteine lyase. PLoS ONE 2012, 7, e30581. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.L.; Collins, R.; Arner, E.S.; Brzezinski, P.; Hogbom, M. Biochemical discrimination between selenium and sulfur 2: Mechanistic investigation of the selenium specificity of human selenocysteine lyase. PLoS ONE 2012, 7, e30528. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCBI. SCLY Selenocysteine Lyase [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/51540/ (accessed on 22 August 2019).
- Mihara, H.; Esaki, N. Selenocysteine lyase: Mechanism, structure, and biological role. In Selenium—Its Molecular Biology and Role in Human Health, 1st ed.; Hatfield, D.L., Gladyshev, V.N., Berry, M.J., Eds.; Springer: New York, NY, USA, 2012; pp. 95–105. [Google Scholar]
- Poliak, P.; Van Hoewyk, D.; Obornik, M.; Zikova, A.; Stuart, K.D.; Tachezy, J.; Pilon, M.; Lukes, J. Functions and cellular localization of cysteine desulfurase and selenocysteine lyase in Trypanosoma brucei. FEBS J. 2010, 277, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Wei, T.; Xu, Y.; Li, L.; Kumar Sahu, S.; Wang, H.; Li, H.; Fu, X.; Zhang, G.; Melkonian, M.; et al. Phylogenomics Provides New Insights into Gains and Losses of Selenoproteins among Archaeplastida. Int. J. Mol. Sci. 2019, 20, 3020. [Google Scholar] [CrossRef]
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94. [Google Scholar] [CrossRef] [PubMed]
- Stock, T.; Rother, M. Selenoproteins in Archaea and Gram-positive bacteria. Biochim. Biophys. Acta 2009, 1790, 1520–1532. [Google Scholar] [CrossRef]
- Schiavon, M.; Ertani, A.; Parrasia, S.; Vecchia, F.D. Selenium accumulation and metabolism in algae. Aquat. Toxicol. 2017, 189, 1–8. [Google Scholar] [CrossRef]
- Eswayah, A.S.; Smith, T.J.; Gardiner, P.H. Microbial Transformations of Selenium Species of Relevance to Bioremediation. Appl. Environ. Microbiol. 2016, 82, 4848–4859. [Google Scholar] [CrossRef] [Green Version]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef]
- Estevam, E.C.; Griffin, S.; Nasim, M.J.; Denezhkin, P.; Schneider, R.; Lilischkis, R.; Dominguez-Alvarez, E.; Witek, K.; Latacz, G.; Keck, C.; et al. Natural selenium particles from Staphylococcus carnosus: Hazards or particles with particular promise? J. Hazard. Mater. 2017, 324, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.S.; Debieux, C.M.; Dridge, E.J.; Splatt, P.; Wright, M. Biomineralization of selenium by the selenate-respiring bacterium Thauera selenatis. Biochem. Soc. Trans. 2012, 40, 1239–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Okeke, B.C.; Frankenberger, W.T., Jr. Bacterial reduction of selenate to elemental selenium utilizing molasses as a carbon source. Bioresour. Technol. 2008, 99, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Chocat, P.; Esaki, N.; Nakamura, T.; Tanaka, H.; Soda, K. Microbial distribution of selenocysteine lyase. J. Bacteriol. 1983, 156, 455–457. [Google Scholar] [PubMed]
- Mihara, H.; Kurihara, T.; Yoshimura, T.; Soda, K.; Esaki, N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J. Biol. Chem. 1997, 272, 22417–22424. [Google Scholar] [CrossRef] [PubMed]
- Lacourciere, G.M.; Stadtman, T.C. The NIFS protein can function as a selenide delivery protein in the biosynthesis of selenophosphate. J. Biol. Chem. 1998, 273, 30921–30926. [Google Scholar] [CrossRef] [PubMed]
- Lacourciere, G.M.; Mihara, H.; Kurihara, T.; Esaki, N.; Stadtman, T.C. Escherichia coli NifS-like proteins provide selenium in the pathway for the biosynthesis of selenophosphate. J. Biol. Chem. 2000, 275, 23769–23773. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, C.; Mangiapane, E.; Pessione, A.; Mazzoli, R.; Giunta, C.; Pessione, E. Proteomic characterization of a selenium-metabolizing probiotic Lactobacillus reuteri Lb2 BM for nutraceutical applications. Proteomics 2011, 11, 2212–2221. [Google Scholar] [CrossRef]
- Mihara, H.; Kurihara, T.; Yoshimura, T.; Esaki, N. Kinetic and mutational studies of three NifS homologs from Escherichia coli: Mechanistic difference between L-cysteine desulfurase and L-selenocysteine lyase reactions. J. Biochem. 2000, 127, 559–567. [Google Scholar] [CrossRef]
- Tobe, R.; Mihara, H. Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis. Biochim. Biophys. Acta Gen Subj. 2018. [Google Scholar] [CrossRef]
- Seale, L.A.; Ha, H.Y.; Hashimoto, A.C.; Berry, M.J. Relationship between selenoprotein P and selenocysteine lyase: Insights into selenium metabolism. Free Radic. Biol. Med. 2018, 127, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Seale, L.A.; Hashimoto, A.C.; Kurokawa, S.; Gilman, C.L.; Seyedali, A.; Bellinger, F.P.; Raman, A.V.; Berry, M.J. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol. Cell. Biol. 2012, 32, 4141–4154. [Google Scholar] [CrossRef] [PubMed]
- Deagen, J.T.; Butler, J.A.; Beilstein, M.A.; Whanger, P.D. Effects of dietary selenite, selenocystine and selenomethionine on selenocysteine lyase and glutathione peroxidase activities and on selenium levels in rat tissues. J. Nutr. 1987, 117, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Fagerberg, L.; Hallstrom, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Duff, M.O.; Olson, S.; Wei, X.; Garrett, S.C.; Osman, A.; Bolisetty, M.; Plocik, A.; Celniker, S.E.; Graveley, B.R. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature 2015, 521, 376–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, L.A.; Gilman, C.L.; Moorman, B.P.; Berry, M.J.; Grau, E.G.; Seale, A.P. Effects of acclimation salinity on the expression of selenoproteins in the tilapia, Oreochromis mossambicus. J. Trace Elem. Med. Biol. 2014, 28, 284–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafari, C.; Panzer, U.; Steinmetz, O.M.; Zahner, G.; Stahl, R.A.; Harendza, S. Enhanced expression of selenocysteine lyase in acute glomerulonephritis and its regulation by AP-1. Cell. Mol. Biol. Lett. 2006, 11, 424–437. [Google Scholar] [CrossRef]
- Yun, J.W.; Lum, K.; Lei, X.G. A novel upregulation of glutathione peroxidase 1 by knockout of liver-regenerating protein Reg3beta aggravates acetaminophen-induced hepatic protein nitration. Free Radic. Biol. Med. 2013. [Google Scholar] [CrossRef]
- Becker, N.P.; Martitz, J.; Renko, K.; Stoedter, M.; Hybsier, S.; Cramer, T.; Schomburg, L. Hypoxia reduces and redirects selenoprotein biosynthesis. Metallomics 2014, 6, 1079–1086. [Google Scholar] [CrossRef] [Green Version]
- Yepes, J.O.; Gunturiz, M.L.; Henao, L.F.; Navas, M.C.; Balcazar, N.; Gomez, L.A. Differential display of messenger RNA and identification of selenocysteine lyase gene in hepatocellular carcinoma cells transiently expressing hepatitis C virus core protein. Biomedica 2006, 26, 194–205. [Google Scholar] [CrossRef]
- Yu, M.W.; Horng, I.S.; Hsu, K.H.; Chiang, Y.C.; Liaw, Y.F.; Chen, C.J. Plasma selenium levels and risk of hepatocellular carcinoma among men with chronic hepatitis virus infection. Am. J. Epidemiol. 1999, 150, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Wray, J.R.; Davies, A.; Sefton, C.; Allen, T.J.; Adamson, A.; Chapman, P.; Lam, B.Y.H.; Yeo, G.S.H.; Coll, A.P.; Harno, E.; et al. Global transcriptomic analysis of the arcuate nucleus following chronic glucocorticoid treatment. Mol. Metab. 2019, 26, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, T.L.; Correa-Medina, M.; Campos, M.P.; Wittmann, G.; Werneck-de-Castro, J.P.; Arrojo e Drigo, R.; Mora-Garzon, M.; Ueta, C.B.; Caicedo, A.; Fekete, C.; et al. Coordination of hypothalamic and pituitary T3 production regulates TSH expression. J. Clin. Investig. 2013, 123, 1492–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luongo, C.; Dentice, M.; Salvatore, D. Deiodinases and their intricate role in thyroid hormone homeostasis. Nat. Rev. Endocrinol. 2019, 15, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Franca, M.R.; da Silva, M.I.S.; Pugliesi, G.; Van Hoeck, V.; Binelli, M. Evidence of endometrial amino acid metabolism and transport modulation by peri-ovulatory endocrine profiles driving uterine receptivity. J. Anim. Sci. Biotechnol. 2017, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Dalto, D.B.; Roy, M.; Audet, I.; Palin, M.F.; Guay, F.; Lapointe, J.; Matte, J.J. Interaction between vitamin B6 and source of selenium on the response of the selenium-dependent glutathione peroxidase system to oxidative stress induced by oestrus in pubertal pig. J. Trace Elem. Med. Biol. 2015, 32, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Seale, A.P.; Riley, L.G.; Leedom, T.A.; Kajimura, S.; Dores, R.M.; Hirano, T.; Grau, E.G. Effects of environmental osmolality on release of prolactin, growth hormone and ACTH from the tilapia pituitary. Gen. Comp. Endocrinol 2002, 128, 91–101. [Google Scholar] [CrossRef]
- Tamura, T.; Yamamoto, S.; Takahata, M.; Sakaguchi, H.; Tanaka, H.; Stadtman, T.C.; Inagaki, K. Selenophosphate synthetase genes from lung adenocarcinoma cells: Sps1 for recycling L-selenocysteine and Sps2 for selenite assimilation. Proc. Natl. Acad. Sci. USA 2004, 101, 16162–16167. [Google Scholar] [CrossRef]
- Tobe, R.; Mihara, H.; Kurihara, T.; Esaki, N. Identification of proteins interacting with selenocysteine lyase. Biosci. Biotechnol. Biochem. 2009, 73, 1230–1232. [Google Scholar] [CrossRef]
- Byrns, C.N.; Pitts, M.W.; Gilman, C.A.; Hashimoto, A.C.; Berry, M.J. Mice lacking selenoprotein P and selenocysteine lyase exhibit severe neurological dysfunction, neurodegeneration, and audiogenic seizures. J. Biol. Chem. 2014, 289, 9662–9674. [Google Scholar] [CrossRef]
- Pitts, M.W.; Kremer, P.M.; Hashimoto, A.C.; Torres, D.J.; Byrns, C.N.; Williams, C.S.; Berry, M.J. Competition between the Brain and Testes under Selenium-Compromised Conditions: Insight into Sex Differences in Selenium Metabolism and Risk of Neurodevelopmental Disease. J. Neurosci. 2015, 35, 15326–15338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raman, A.V.; Pitts, M.W.; Seyedali, A.; Hashimoto, A.C.; Seale, L.A.; Bellinger, F.P.; Berry, M.J. Absence of selenoprotein P but not selenocysteine lyase results in severe neurological dysfunction. Genes Brain Behav. 2012, 11, 601–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, L.A.; Gilman, C.L.; Hashimoto, A.C.; Ogawa-Wong, A.N.; Berry, M.J. Diet-Induced Obesity in the Selenocysteine Lyase Knockout Mouse. Antioxid. Redox Signal. 2015, 23, 761–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa-Wong, A.N.; Hashimoto, A.C.; Ha, H.; Pitts, M.W.; Seale, L.A.; Berry, M.J. Sexual Dimorphism in the Selenocysteine Lyase Knockout Mouse. Nutrients 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Schomburg, L. Sex-specific differences in biological effects and metabolism of selenium. In Selenium—Its Molecular Biology and Role in Human Health, 4th ed.; Hatfield, D.L., Schweizer, U., Tsuji, P.A., Gladyshev, V.N., Eds.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Seale, L.A.; Ogawa-Wong, A.N.; Berry, M.J. Sexual Dimorphism in Selenium Metabolism and Selenoproteins. Free Radic. Biol. Med. 2018, 127, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Labbe, S.M.; Caron, A.; Lanfray, D.; Monge-Rofarello, B.; Bartness, T.J.; Richard, D. Hypothalamic control of brown adipose tissue thermogenesis. Front. Syst. Neurosci. 2015, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Oswal, A.; Yeo, G. Leptin and the control of body weight: A review of its diverse central targets, signaling mechanisms, and role in the pathogenesis of obesity. Obesity 2010, 18, 221–229. [Google Scholar] [CrossRef]
- Coppola, A.; Liu, Z.W.; Andrews, Z.B.; Paradis, E.; Roy, M.C.; Friedman, J.M.; Ricquier, D.; Richard, D.; Horvath, T.L.; Gao, X.B.; et al. A central thermogenic-like mechanism in feeding regulation: An interplay between arcuate nucleus T3 and UCP2. Cell Metab. 2007, 5, 21–33. [Google Scholar] [CrossRef]
- Torres, D.J.; Pitts, M.W.; Hashimoto, A.C.; Berry, M.J. Agrp-Specific Ablation of Scly Protects against Diet-Induced Obesity and Leptin Resistance. Nutrients 2019, 11, 1693. [Google Scholar] [CrossRef]
- Kwak, M.S.; Mihara, H.; Esaki, N. A novel regulatory function of selenocysteine lyase, a unique catalyst to modulate major urinary protein. J. Mol. Catal. B Enzym. 2003, 23, 367–372. [Google Scholar] [CrossRef]
- Gao, C.; Hsu, F.C.; Dimitrov, L.M.; Okut, H.; Chen, Y.I.; Taylor, K.D.; Rotter, J.I.; Langefeld, C.D.; Bowden, D.W.; Palmer, N.D. A genome-wide linkage and association analysis of imputed insertions and deletions with cardiometabolic phenotypes in Mexican Americans: The Insulin Resistance Atherosclerosis Family Study. Genet. Epidemiol. 2017, 41, 353–362. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seale, L.A. Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme. Antioxidants 2019, 8, 357. https://doi.org/10.3390/antiox8090357
Seale LA. Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme. Antioxidants. 2019; 8(9):357. https://doi.org/10.3390/antiox8090357
Chicago/Turabian StyleSeale, Lucia A. 2019. "Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme" Antioxidants 8, no. 9: 357. https://doi.org/10.3390/antiox8090357
APA StyleSeale, L. A. (2019). Selenocysteine β-Lyase: Biochemistry, Regulation and Physiological Role of the Selenocysteine Decomposition Enzyme. Antioxidants, 8(9), 357. https://doi.org/10.3390/antiox8090357