Anti-Inflammatory Properties and Gut Microbiota Modulation of Porphyra tenera Extracts in Dextran Sodium Sulfate-Induced Colitis in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Porphyra tenera (PT) Extracts
2.2. Determination of Total Polyphenols Content
2.3. Determination of 1,1-Diphenyl-2-Picrylhydrazyl (DPPH) Free Radical Scavenging Activity
2.4. Animal Experiment
2.5. Preparation of Splenocytes and Fluorescence-Activated Cell Sorting (FACS) Analysis
2.6. Disease Activity Index
2.7. RNA Extraction and Real-Time PCR
2.8. Histological Analysis
2.9. Gut Microbiota Analysis
2.10. Statistical Analysis
3. Results and Discussion
3.1. In Vitro Antioxidant Activity of PT Extracts
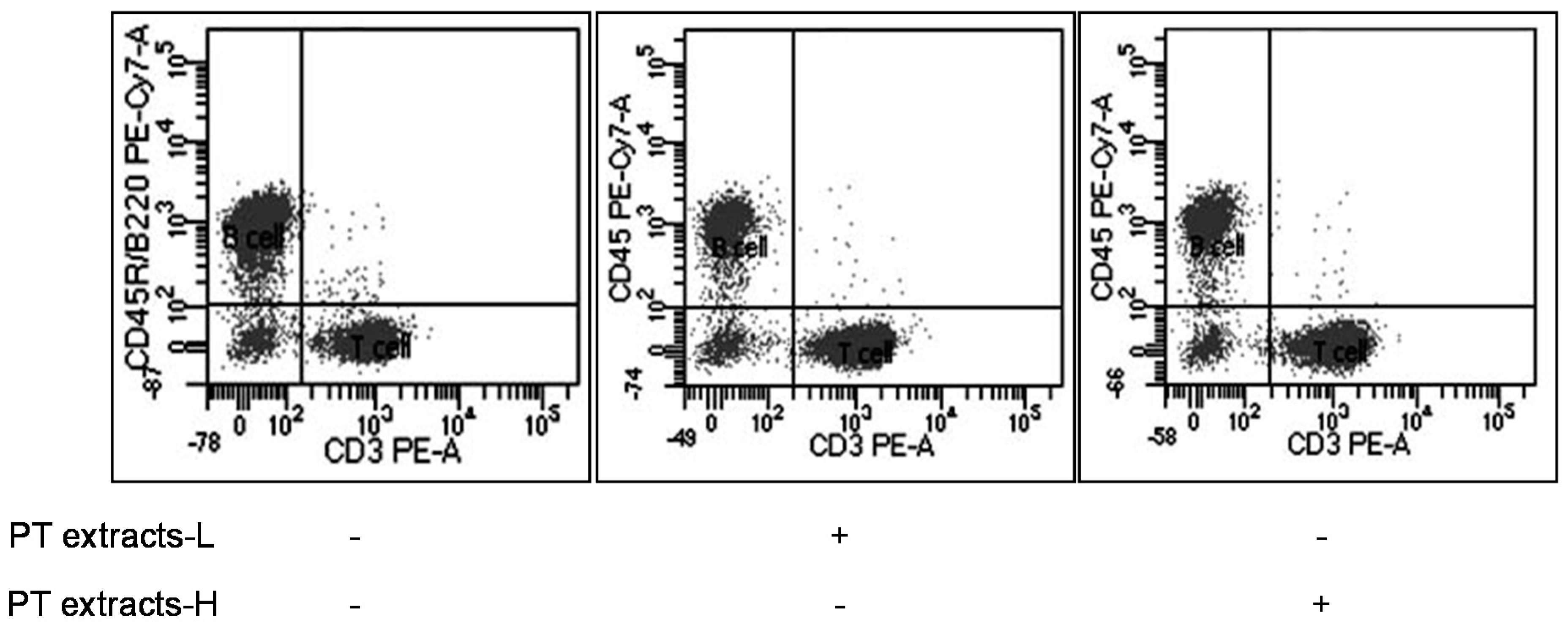
3.2. Effects of Differentiation of T and B Cells by PT Extracts
3.3. Inhibitory Effects of PT Extracts on DSS-Reduced Colitis Symptoms
3.4. Inhibitory Effects of PT Extracts on DSS-Induced Inflammatory Markers
3.5. Alteration of Gut Microbiota by PT Extracts on Colitis-Induced Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sittipo, P.; Shim, J.W.; Lee, Y.K. Microbial metabolites determine host health and the status of some diseases. Int. J. Mol. Sci. 2019, 20, 5296. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Lee, B.; Zaragoza, J.; Marco, M.L. Dietary perturbations alter the ecological significance of ingested Lactobacillus plantarum in the digestive tract. Sci. Rep. 2017, 7, 7267. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.J.; Naik, N.N.; Wild, L.E.; Patterson, W.B.; Alderete, T.L. Exposure to air pollutants and the gut microbiota: A potential link between exposure, obesity, and type 2 diabetes. Gut Microbes 2020, 11, 1188–1202. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.Y.; Yoon, S.J.; Han, D.H.; Gupta, H.; Youn, G.S.; Shin, M.J.; Ham, Y.L.; Kwak, M.J.; Kim, B.Y.; Yu, J.S.; et al. Lactobacillus and Pediococcus ameliorate progression of non-alcoholic fatty liver disease through modulation of the gut microbiome. Gut Microbes 2020, 11, 882–899. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z. Gut microbiome and cardiovascular disease. Curr. Opin. Cardiol. 2020, 35, 207–218. [Google Scholar] [CrossRef]
- Nair, A.T.; Ramachandran, V.; Joghee, N.M.; Antony, S.; Ramalingam, G. Gut microbiota dysfunction as reliable non-invasive early diagnostic biomarkers in the pathophysiology of Parkinson’s disease: A critical review. J. Neurogastroenterol. Motil. 2018, 24, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Verdugo-Meza, A.; Ye, J.; Dadlani, H.; Ghosh, S.; Gibson, D.L. Connecting the dots between inflammatory bowel disease and metabolic syndrome: A focus on gut-derived metabolites. Nutrients 2020, 12, 1434. [Google Scholar] [CrossRef]
- Shamoon, M.; Martin, N.M.; O’Brien, C.L. Recent advances in gut microbiota mediated therapeutic targets in inflammatory bowel diseases: Emerging modalities for future pharmacological implications. Pharmacol. Res. 2019, 148, 104344. [Google Scholar] [CrossRef]
- Leonard, S.G.; Sweeney, T.; Bahar, B.; Lynch, B.P.; O’Doherty, J.V. Effect of dietary seaweed extracts and fish oil supplementation in sows on performance, intestinal microflora, intestinal morphology, volatile fatty acid concentrations and immune status of weaned pigs. Br. J. Nutr. 2011, 105, 549–560. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Wang, X.M.; Han, Z.P.; Yin, L.; Zhao, M.X.; Yu, S.C. Physicochemical properties and inhibition effect on iron deficiency anemia of a novel polysaccharide-iron complex (LPPC). Bioorg. Med. Chem. Lett. 2012, 22, 489–492. [Google Scholar] [CrossRef]
- Hwang, E.S.; Thi, N.D. Effects of extraction and processing methods on antioxidant compound contents and radical scavenging activities of laver (Porphyra tenera). Prev. Nutr. Food Sci. 2014, 19, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Oyamada, C.; Matsushima, R.; Murata, M.; Muraoka, T. Inhibitory effect of porphyran, prepared from dried “Nori”, on contact hypersensitivity in mice. Biosci. Biotechnol. Biochem. 2005, 69, 1824–1830. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Lee, S.W.; Kwon, Y.S.; Park, J.G.; Baek, K.H. Chemical characterization and antioxidant potential of volatile oil from an edible seaweed Porphyra tenera (Kjellman, 1897). Chem. Cent. J. 2017, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Zhaohui, Z.; Wenshan, S.; Bafang, L.; Hu, H. Protective effect of MAAs extracted from Porphyra tenera against UV irradiation-induced photoaging in mouse skin. J. Photochem. Photobiol. B 2019, 192, 26–33. [Google Scholar] [CrossRef]
- Ichihara, T.; Wanibuchi, H.; Taniyama, T.; Okai, Y.; Yano, Y.; Otani, S.; Imaoka, S.; Funae, Y.; Fukushima, S. Inhibition of liver glutathione S-transferase placental form-positive foci development in the rat hepatocarcinogenesis by Porphyra tenera (Asakusa-nori). Cancer Lett. 1999, 141, 211–218. [Google Scholar] [CrossRef]
- Song, J.H.; Kang, H.B.; Park, S.H.; Jeong, J.H.; Park, J.; You, Y.; Lee, Y.H.; Lee, J.; Kim, E.; Choi, K.C.; et al. Extracts of Porphyra tenera (Nori seaweed) activate the immune response in mouse RAW264.7 macrophages via NF-kappaB signaling. J. Med. Food 2017, 20, 1152–1159. [Google Scholar] [CrossRef]
- Jung, S.J.; Jang, H.Y.; Jung, E.S.; Noh, S.O.; Shin, S.W.; Ha, K.C.; Baek, H.I.; Ahn, B.J.; Oh, T.H.; Chae, S.W. Effects of Porphyra tenera supplementation on the immune system: A randomized, double-blind, and placebo-controlled clinical trial. Nutrients 2020, 12, 1642. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Dudonne, S.; Vitrac, X.; Coutiere, P.; Woillez, M.; Merillon, J.M. Comparative study of antioxidant properties and total phenolic content of 30 plant extracts of industrial interest using DPPH, ABTS, FRAP, SOD, and ORAC assays. J. Agric. Food Chem. 2009, 57, 1768–1774. [Google Scholar] [CrossRef]
- Kang, H.-B.; Song, J.; You, Y.; Park, J.; Kwak, S.; Lee, Y.; Lee, J.; Kim, S.-I.; Choi, K.; Jun, W. Porphyra tenera extracts have immune stimulation activity via increasing cytokines in mouse primary splenocytes and RAW264.7 macrophages. J. Food Nutr. Res. 2016, 4, 558–565. [Google Scholar]
- Best, W.R.; Becktel, J.M.; Singleton, J.W.; Kern, F., Jr. Development of a Crohn’s disease activity index. National cooperative Crohn’s disease study. Gastroenterology 1976, 70, 439–444. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam, S.A.; McGarrell, D.M.; Garrity, G.M.; Tiedje, J.M. The ribosomal database project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005, 33, D294–D296. [Google Scholar] [CrossRef] [PubMed]
- Westcott, S.L.; Schloss, P.D. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. MSphere 2017, 2, e00073-17. [Google Scholar] [CrossRef] [PubMed]
- Beals, E.W. Bray-Curtis ordination—An effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 1984, 14, 1–55. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef] [PubMed]
- Sivandzade, F.; Prasad, S.; Bhalerao, A.; Cucullo, L. Nrf2 and NF-kappaB interplay in cerebrovascular and neurodegenerative disorders: Molecular mechanisms and possible therapeutic approaches. Redox Biol. 2019, 21, 101059. [Google Scholar] [CrossRef] [PubMed]
- Cian, R.E.; Alaiz, M.; Vioque, J.; Drago, S.R. Enzyme proteolysis enhanced extraction of ACE inhibitory and antioxidant compounds (peptides and polyphenols) from Porphyra columbina residual cake. J. Appl. Phycol. 2013, 25, 1197–1206. [Google Scholar] [CrossRef]
- Yu, X.J.; Zhou, C.S.; Yang, H.; Huang, X.Y.; Ma, H.L.; Qin, X.P.; Hu, J.L. Effect of ultrasonic treatment on the degradation and inhibition cancer cell lines of polysaccharides from Porphyra yezoensis. Carbohyd. Polym. 2015, 117, 650–656. [Google Scholar] [CrossRef]
- Venkatraman, K.L.; Mehta, A. Health benefits and pharmacological rffects of Porphyra species. Plant. Foods Hum. Nutr. 2019, 74, 10–17. [Google Scholar] [CrossRef]
- Murray, M.; Dordevic, A.L.; Ryan, L.; Bonham, M.P. The impact of a single dose of a polyphenol-rich seaweed extract on postprandial glycaemic control in healthy adults: A randomised cross-over trial. Nutrients 2018, 10, 270. [Google Scholar] [CrossRef]
- Corsetto, P.A.; Montorfano, G.; Zava, S.; Colombo, I.; Ingadottir, B.; Jonsdottir, R.; Sveinsdottir, K.; Rizzo, A.M. Characterization of antioxidant potential of seaweed extracts for enrichment of convenience food. Antioxidants 2020, 9, 249. [Google Scholar] [CrossRef]
- Gacesa, R.; Lawrence, K.P.; Georgakopoulos, N.D.; Yabe, K.; Dunlap, W.C.; Barlow, D.J.; Wells, G.; Young, A.R.; Long, P.F. The mycosporine-like amino acids porphyra-334 and shinorine are antioxidants and direct antagonists of Keap1-Nrf2 binding. Biochimie 2018, 154, 35–44. [Google Scholar] [CrossRef]
- Ryu, J.; Kwon, M.J.; Nam, T.J. Nrf2 and NF-kappaB signaling pathways contribute to porphyra-334-mediated inhibition of UVA-induced inflammation in skin fibroblasts. Mar. Drugs 2015, 13, 4721–4732. [Google Scholar] [CrossRef]
- Chen, Y.; Chou, K.; Fuchs, E.; Havran, W.L.; Boismenu, R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14338–14343. [Google Scholar] [CrossRef]
- Iliev, I.D.; Mileti, E.; Matteoli, G.; Chieppa, M.; Rescigno, M. Intestinal epithelial cells promote colitis-protective regulatory T-cell differentiation through dendritic cell conditioning. Mucosal Immunol. 2009, 2, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Annacker, O.; Coombes, J.L.; Malmstrom, V.; Uhlig, H.H.; Bourne, T.; Johansson-Lindbom, B.; Agace, W.W.; Parker, C.M.; Powrie, F. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J. Exp. Med. 2005, 202, 1051–1061. [Google Scholar] [CrossRef] [PubMed]
- Nyborg, A.C.; Zacco, A.; Ettinger, R.; Jack Borrok, M.; Zhu, J.; Martin, T.; Woods, R.; Kiefer, C.; Bowen, M.A.; Suzanne Cohen, E.; et al. Development of an antibody that neutralizes soluble IgE and eliminates IgE expressing B cells. Cell. Mol. Immunol. 2016, 13, 391–400. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, X.; Chen, J.; Chen, T.; Shi, Z.; Lei, M.; Zhang, Y.; Bai, P.; Li, Y.; Fei, X. The pentacyclic triterpene Lupeol switches M1 macrophages to M2 and ameliorates experimental inflammatory bowel disease. Int. Immunopharmacol. 2016, 30, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wu, C.; Zhang, Z.; Liu, M.; Maruthi Prasad, E.; Chen, Y.; Wang, K. Pinocembrin protects against dextran sulfate sodium-Induced rats colitis by ameliorating inflammation, improving barrier function and modulating gut microbiota. Front. Physiol. 2019, 10, 908. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, M.; Nusrat, S.; Alam, P.; Malik, S.; Chaturvedi, S.K.; Ajmal, M.R.; Abdelhameed, A.S.; Khan, R.H. Investigating the site selective binding of busulfan to human serum albumin: Biophysical and molecular docking approaches. Int. J. Biol. Macromol. 2018, 107, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Dieleman, L.A.; Ridwan, B.U.; Tennyson, G.S.; Beagley, K.W.; Bucy, R.P.; Elson, C.O. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology 1994, 107, 1643–1652. [Google Scholar] [CrossRef]
- Shin, E.S.; Hwang, H.J.; Kim, I.H.; Nam, T.J. A glycoprotein from Porphyra yezoensis produces anti-inflammatory effects in liposaccharide-stimulated macrophages via the TLR4 signaling pathway. Int. J. Mol. Med. 2011, 28, 809–815. [Google Scholar] [CrossRef]
- Zenewicz, L.A.; Antov, A.; Flavell, R.A. CD4 T-cell differentiation and inflammatory bowel disease. Trends Mol. Med. 2009, 15, 199–207. [Google Scholar] [CrossRef]
- Galvez, J. Role of Th17 cells in the pathogenesis of human IBD. ISRN Inflamm. 2014, 2014, 928461. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Van Immerseel, F.; Croubels, S.; De Baere, S.; Haesebrouck, F.; Ducatelle, R.; Louis, P.; Vandamme, P. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb. Biotechnol. 2011, 4, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Jin, L.; Xia, D.; Zhang, Q.; Ma, L.; Zheng, H.; Xu, T.; Chang, S.; Li, X.; Xun, Z.; et al. Nitrate ameliorates dextran sodium sulfate-induced colitis by regulating the homeostasis of the intestinal microbiota. Free Radic. Biol. Med. 2020, 152, 609–621. [Google Scholar] [CrossRef]
- Vaziri, N.D.; Liu, S.M.; Lau, W.L.; Khazaeli, M.; Nazertehrani, S.; Farzaneh, S.H.; Kieffer, D.A.; Adams, S.H.; Martin, R.J. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS ONE 2014, 9, e114881. [Google Scholar] [CrossRef]
- Kim, D.H.; Sung, B.; Kang, Y.J.; Jang, J.Y.; Hwang, S.Y.; Lee, Y.; Kim, M.; Im, E.; Yoon, J.H.; Kim, C.M.; et al. Anti-inflammatory effects of betaine on AOM/DSSinduced colon tumorigenesis in ICR male mice. Int. J. Oncol. 2014, 45, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. mBio 2014, 5, e00889. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Tatiya-aphiradee, N.; Chatuphonprasert, W.; Jarukamjorn, K. Oxidative stress exacerbates dextran sulfate sodium-induced ulcerative colitis in ICR mice. Biologia 2020. [Google Scholar] [CrossRef]
- Crapo, J.D. Oxidative stress as an initiator of cytokine release and cell damage. Eur. Respir. J. 2003, 22, 4s–6s. [Google Scholar] [CrossRef]
- Guan, G.; Lan, S. Implications of antioxidant systems in inflammatory bowel disease. BioMed Res. Int. 2018, 2018, 1290179. [Google Scholar] [CrossRef]
- Toumpanakis, D.; Karatza, M.H.; Katsaounou, P.; Roussos, C.; Zakynthinos, S.; Papapetropoulos, A.; Vassilakopoulos, T. Antioxidant supplementation alters cytokine production from monocytes. J. Interferon. Cytokine Res. 2009, 29, 741–748. [Google Scholar] [CrossRef]
- Chae, H.S.; Park, H.J.; Hwang, H.R.; Kwon, A.; Lim, W.H.; Yi, W.J.; Han, D.H.; Kim, Y.H.; Baek, J.H. The effect of antioxidants on the production of pro-inflammatory cytokines and orthodontic tooth movement. Mol. Cells 2011, 32, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Reimund, J.M.; Allison, A.C.; Muller, C.D.; Dumont, S.; Kenney, J.S.; Baumann, R.; Duclos, B.; Poindron, P. Antioxidants inhibit the in vitro production of inflammatory cytokines in Crohn’s disease and ulcerative colitis. Eur. J. Clin. Investig. 1998, 28, 145–150. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |



| Gene | Forward Sequences | Reverse Sequences | NCBI Number |
|---|---|---|---|
| TNF-α | AGCCCCCAGTCTGTATCCTT | CATTCGAGGCTCCAGTGAAT | NM_013693.3 |
| IL-6 | AGTTGCCTTCTTGGGACTGA | CAGAATTGCCATTGCACAAC | NM_031168.2 |
| IL-1β | GGGCCTCAAAGGAAAGAATC | TACCAGTTGGGGAACTCTGC | NM_008361.4 |
| COX-2 | AGAAGGAAATGGCTGCAGAA | GCTCGGCTTCCAGTATTGAG | NM_011198.4 |
| β-actin | GGTGGGAATGGGTCAGAAGG | CAGCACAGGGTGCTCCTC | NM_007393.5 |
| Extracts | Total Polyphenol Content (mg GAE/g) | DPPH Radical Scavenging (IC50 µg/mL) |
|---|---|---|
| PT | 32.3 ± 0.8 | 798.4 ± 80 |
| Group | Live Cell | B Cell (CD45R/B220) | T Cell (CD3) | |
|---|---|---|---|---|
| Tc Cell (CD8) | Th/Treg Cell (CD4) | |||
| Control n = 10 | 10,377 ± 55 | 5056 ± 179 (48.74%) | 4595 ± 172 (44.27%) | |
| 1344 ± 52 (29.43%) | 3151 ± 154 (68.41%) | |||
| PT extracts-L n = 10 | 9687 ± 16 | 2528 ± 91 (26.10%) | 6424 ± 106 * (66.31%) | |
| 1804 ± 47 * (28.16%) | 4499 ± 134 * (69.93%) | |||
| PT extracts-H n = 10 | 9635 ± 32 | 2788 ± 105 (28.93%) | 6240 ± 156 * (64.75%) | |
| 1707 ± 44 * (27.44%) | 4435 ± 158 * (70.97%) | |||
| Group | D0 | D7 | Body Weight Loss |
|---|---|---|---|
| Control (n = 10) | 21.6 ± 0.51 | 20.9 ± 0.32 | 0.8 ± 0.57 |
| DSS (n = 10) | 21.4 ± 0.73 | 19.0 ± 0.58 | 2.3 ± 0.15 # |
| PTE (n = 10) | 21.6 ± 0.61 | 20.3 ± 0.66 | 0.9 ± 0.34 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Choi, J.H.; Ko, G.; Jo, H.; Oh, T.; Ahn, B.; Unno, T. Anti-Inflammatory Properties and Gut Microbiota Modulation of Porphyra tenera Extracts in Dextran Sodium Sulfate-Induced Colitis in Mice. Antioxidants 2020, 9, 988. https://doi.org/10.3390/antiox9100988
Kim J, Choi JH, Ko G, Jo H, Oh T, Ahn B, Unno T. Anti-Inflammatory Properties and Gut Microbiota Modulation of Porphyra tenera Extracts in Dextran Sodium Sulfate-Induced Colitis in Mice. Antioxidants. 2020; 9(10):988. https://doi.org/10.3390/antiox9100988
Chicago/Turabian StyleKim, Jungman, Jae Ho Choi, Gwangpyo Ko, Hyejun Jo, Taehwan Oh, Byungjae Ahn, and Tatsuya Unno. 2020. "Anti-Inflammatory Properties and Gut Microbiota Modulation of Porphyra tenera Extracts in Dextran Sodium Sulfate-Induced Colitis in Mice" Antioxidants 9, no. 10: 988. https://doi.org/10.3390/antiox9100988
APA StyleKim, J., Choi, J. H., Ko, G., Jo, H., Oh, T., Ahn, B., & Unno, T. (2020). Anti-Inflammatory Properties and Gut Microbiota Modulation of Porphyra tenera Extracts in Dextran Sodium Sulfate-Induced Colitis in Mice. Antioxidants, 9(10), 988. https://doi.org/10.3390/antiox9100988






