CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Condition and Plant Type
2.2. Agrobacterium Mediated Transformation
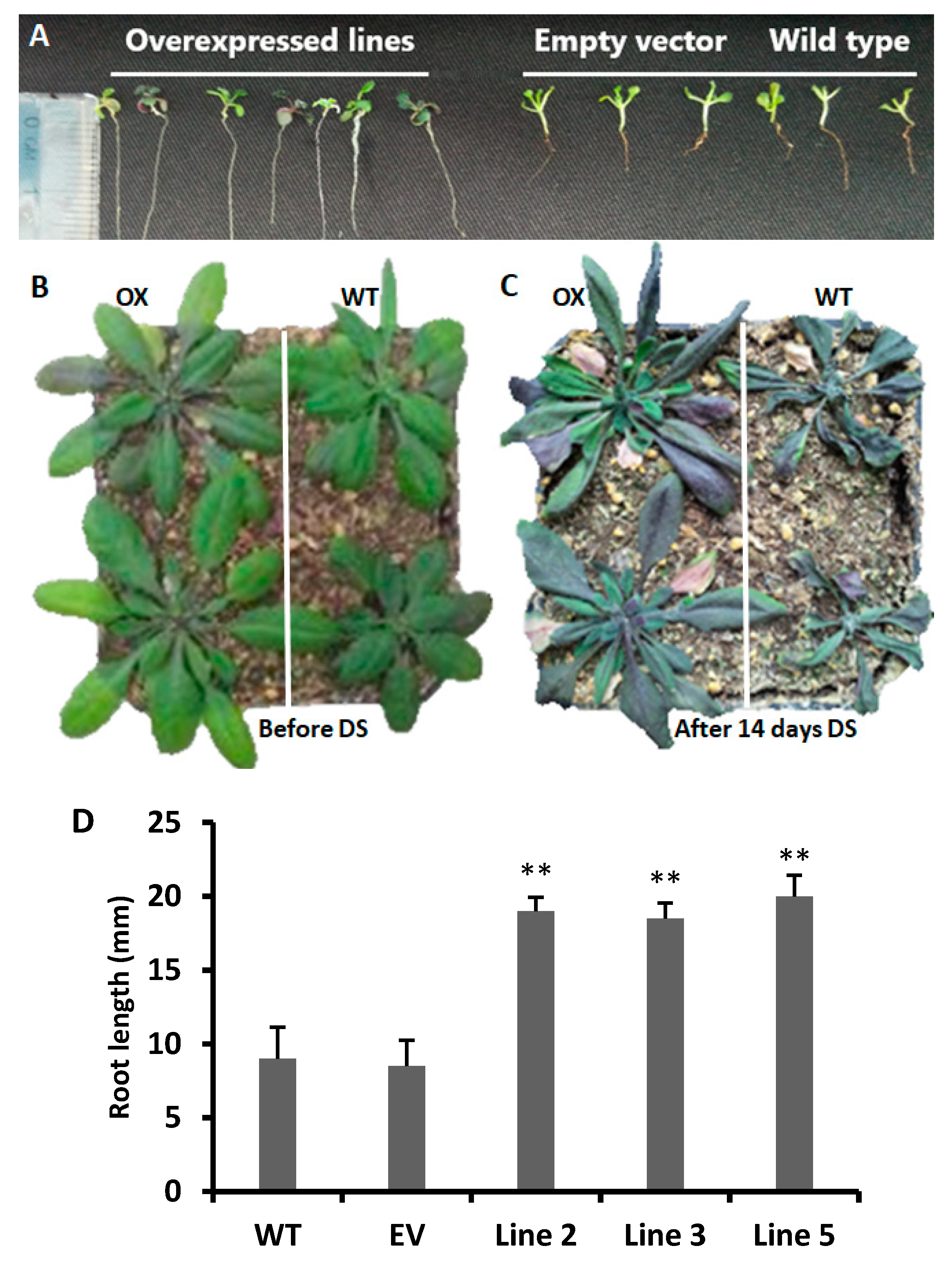
2.3. Transgenic Lines and Drought Stress Conditions
2.4. DNA Extraction and PCR Analysis
2.5. RNA Isolation and Quantitative PCR
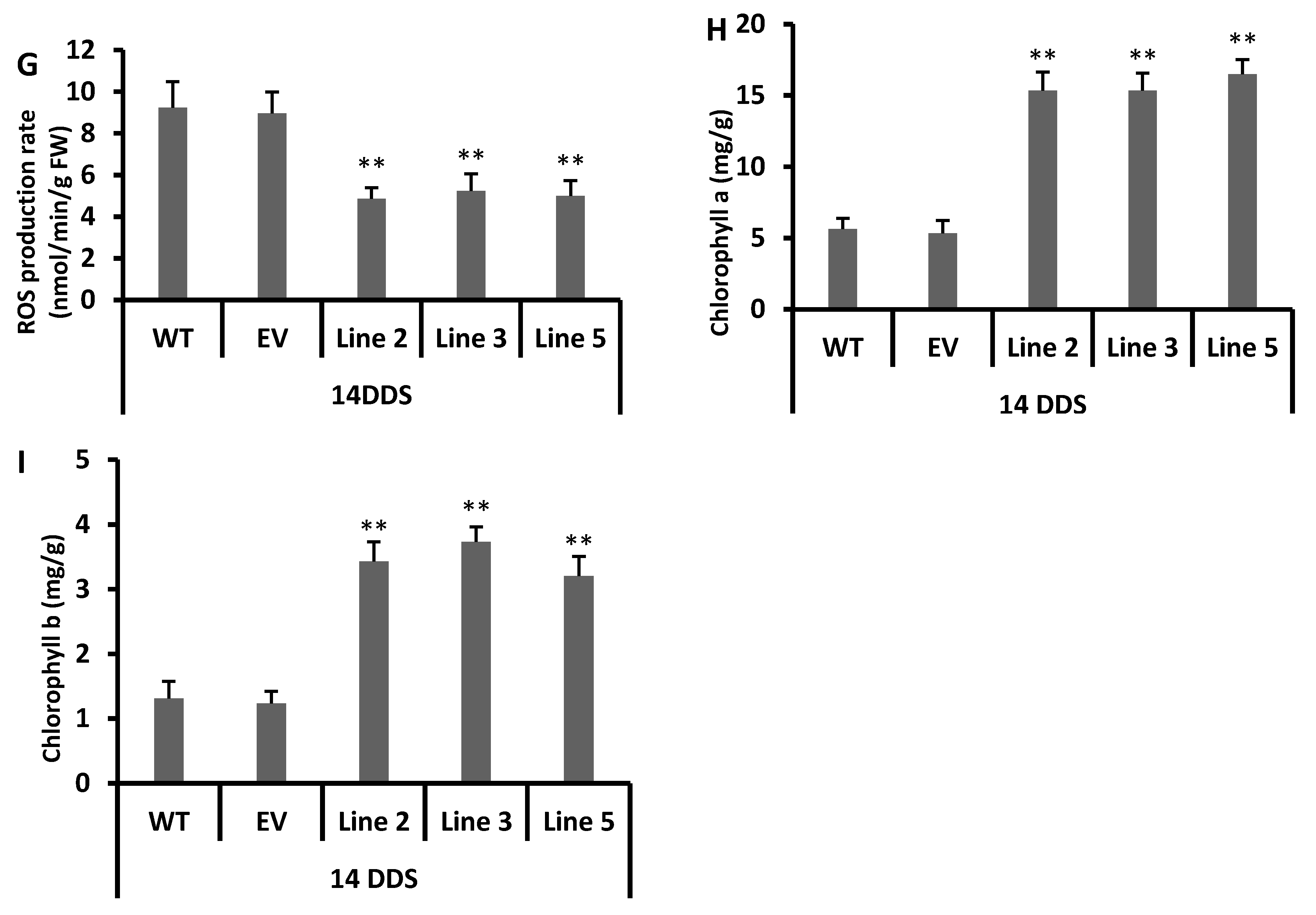
2.6. Chlorophyll a and b Content
2.7. Extraction Procedure of Total Flavonoid and Total Phenolis Contents
2.7.1. Extraction
2.7.2. Estimation of Total Flavonoid Content (TFCs)
2.7.3. Total Phenolic Contents (TPCs)
2.8. Total Anthocyanin Contents (TACs)
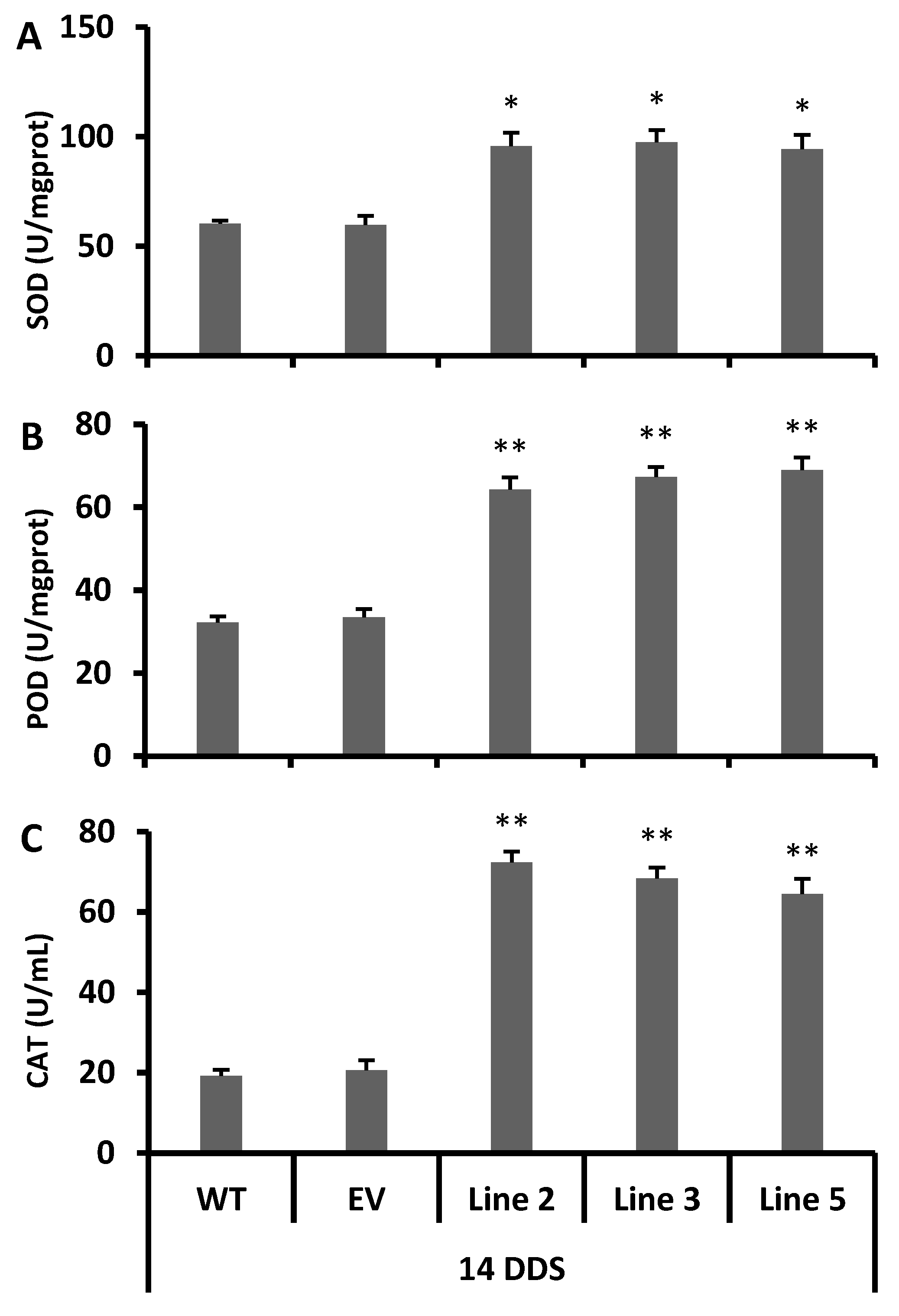
2.9. Antioxidant Enzymatic Activity
2.10. Malondialdehyde and Electrolytic Leakage
2.11. Hydrogen Peroxide (H2O2)
2.12. Reactive Oxygen Species and Superoxide Radicals Determination
2.13. Antioxidant Activity and Capacity (DPPH Free Radical Scavenging Assay)
2.14. Trypan Blue and Nitro-Blue Tetrazolium Staining
2.15. Statistical Analysis
3. Results
3.1. Transcriptome and Gene Expression of CsCYT75B1 Gene
3.2. Phylogenetic Analysis of CsCYT75B1 Gene
3.3. Root Elongation in CYT75B1 Transgenic Lines
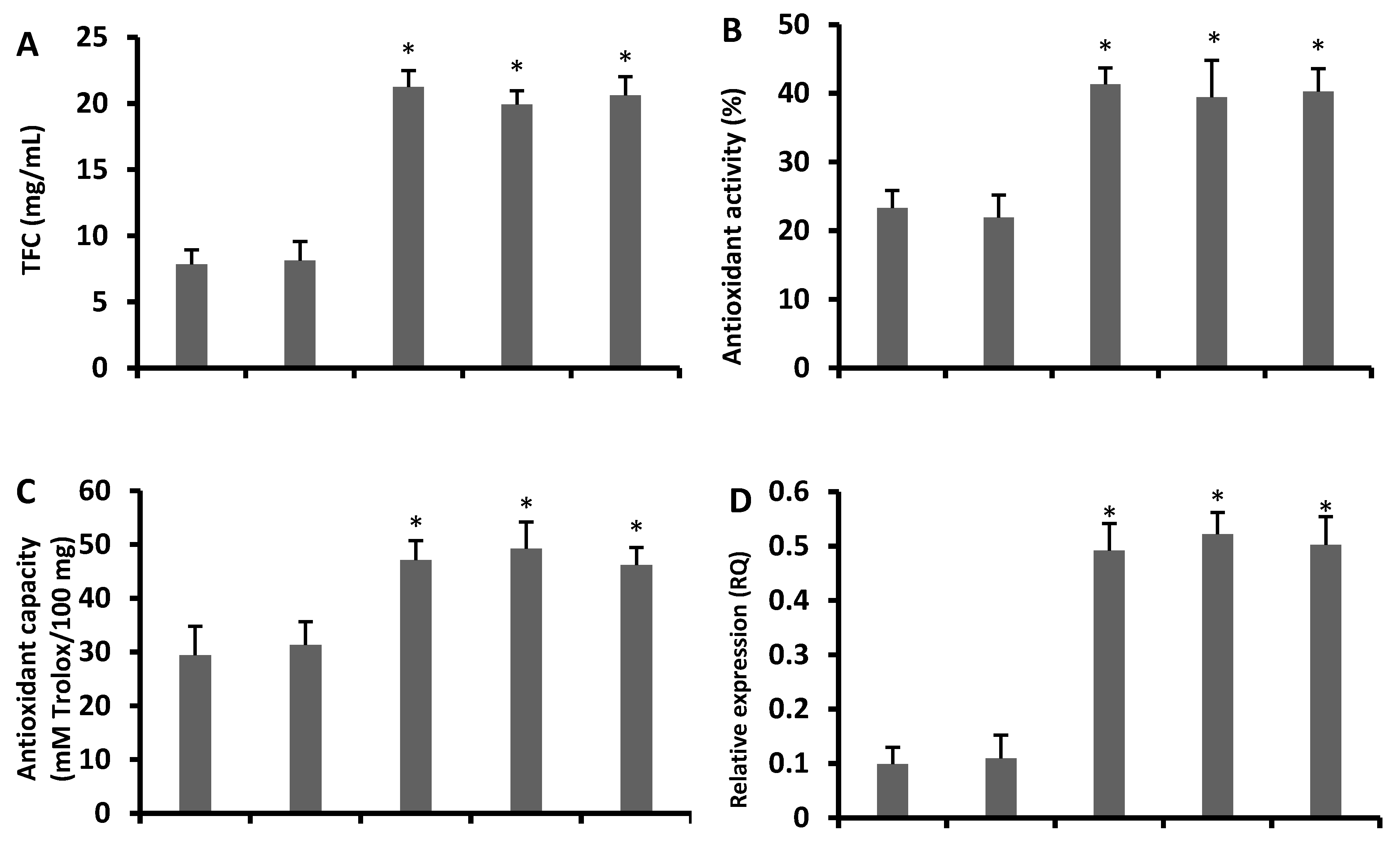
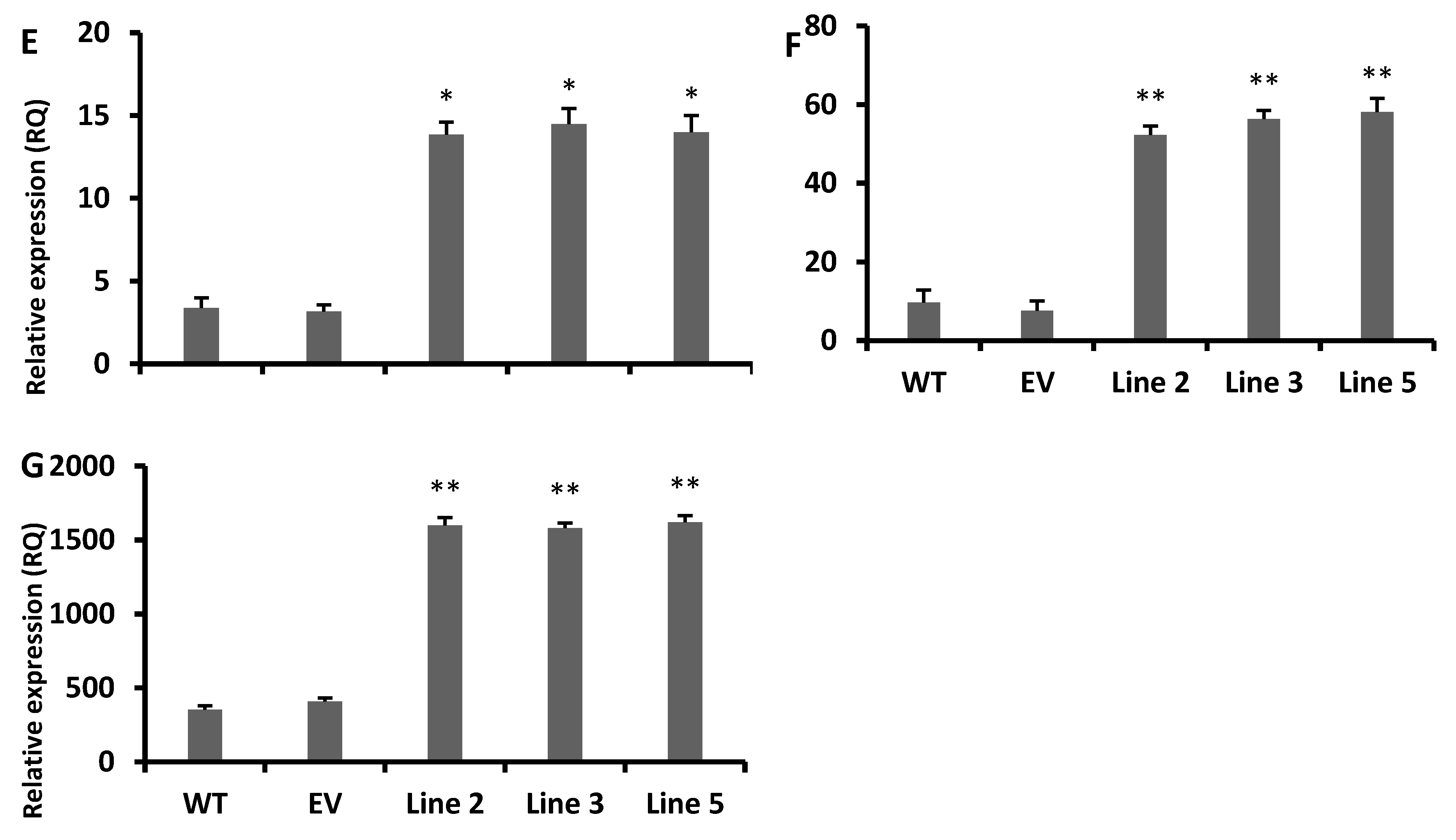
3.4. Increased Antioxidant Flavonoids in Transgenic Arabidopsis Lines
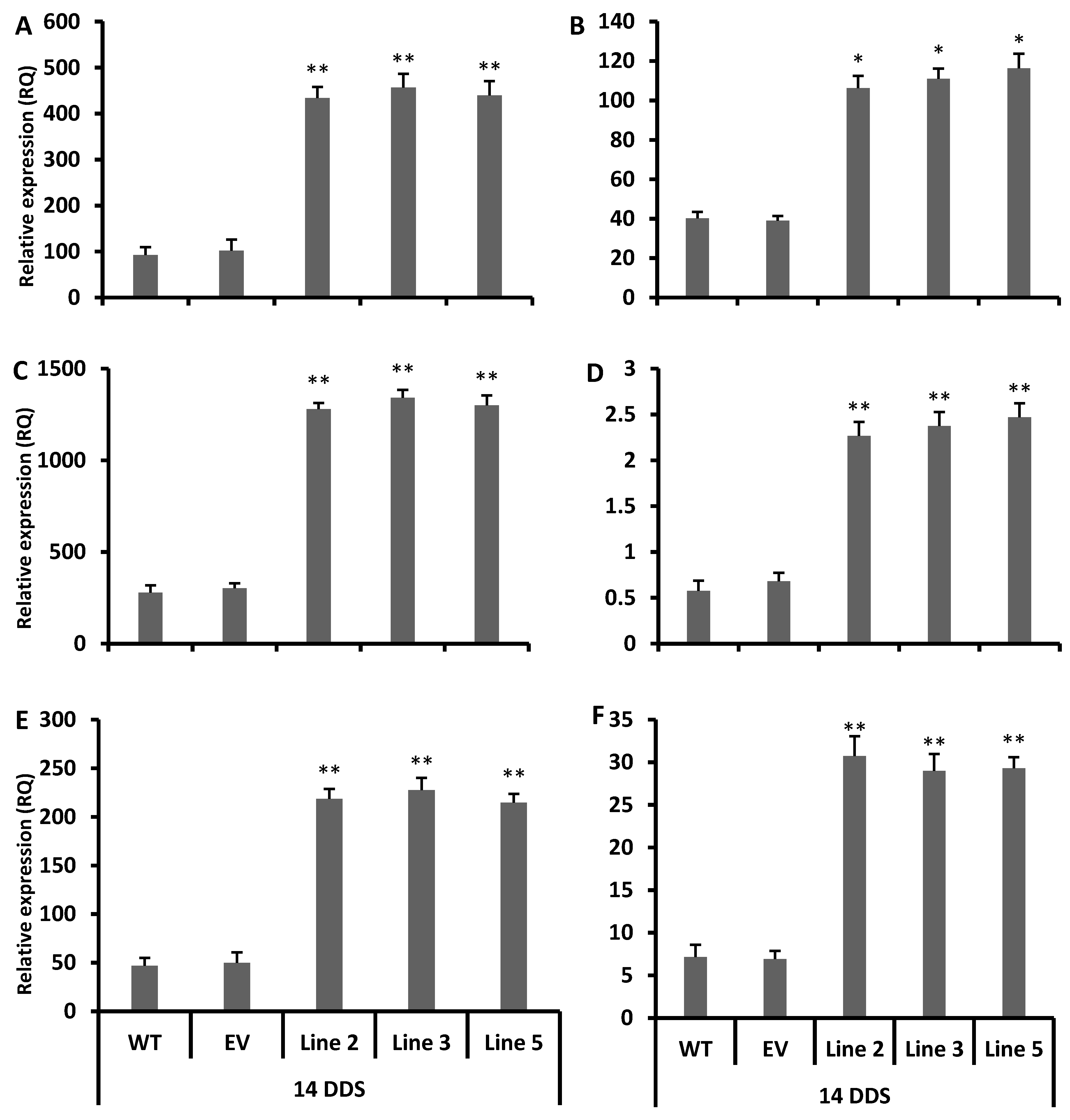
3.5. Accumulation of Antioxidant Flavonoids in Overexpressed Lines after Drought Stress
3.6. Decreased Level of Reactive Oxygen Species in Transgenic Arabidopsis under Drought Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tripoli, E.; La Guardia, M.; Giammanco, S.; Di Majo, D.; Giammanco, M. Citrus flavonoids: Molecular structure, biological activity and nutritional properties: A review. Food Chem. 2007, 104, 466–479. [Google Scholar] [CrossRef]
- Kaur, H.; Heinzel, N.; Schöttner, M.; Baldwin, I.T.; Gális, I. R2R3-NaMYB8 regulates the accumulation of phenylpropanoid-polyamine conjugates, which are essential for local and systemic defense against insect herbivores in Nicotiana attenuata. Plant Physiol. 2010, 152, 1731–1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Fuell, C.; Parr, A.; Hill, L.; Bailey, P.; Elliott, K.; Fairhurst, S.A.; Martin, C.; Michael, A.J. A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell. 2009, 21, 318–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, M.J.; Xu, Y.; Huang, Y.; Tang, X.; Deng, X.; Xu, Q. Ectopic expression of citrus UDP-GLUCOSYL TRANSFERASE gene enhances anthocyanin and proanthocyanidins contents and confers high light tolerance in Arabidopsis. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Wang, X.; Tang, Z.; Yuan, Y.; Xu, Y.; He, J.; Jiang, X.; Peng, S.-A.; Li, L.; Butelli, E. Sub-functionalization of the Ruby2–Ruby1 gene cluster during the domestication of citrus. Nat. Plants 2018, 4, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Heying, E.; Tanumihardjo, S.A. History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 2012, 11, 530–545. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.-P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301. [Google Scholar] [CrossRef]
- Werck-reichhart, D.; Bak, S.; Paquette, S. Cytochromes 450. In The Arabidopsis Book, 1st ed.; American Society of Plant Biologist: New York, NY, USA, 2002; Volume 1, pp. 1–28. [Google Scholar]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [Green Version]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Sales, C.; Beltrán, J.; Gómez-Cadenas, A.; Arbona, V. Activation of secondary metabolism in citrus plants is associated to sensitivity to combined drought and high temperatures. Front. Plant Sci. 2017, 7, 1954. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Reagan, R.L.; Uratsu, S.L.; Phu, M.L.; Albrecht, U.; Zhao, W.; Davis, C.E.; Bowman, K.D.; Dandekar, A.M. Gene regulatory networks elucidating huanglongbing disease mechanisms. PLoS ONE 2013, 8, e74256. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome profiling of Huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Yang, C.; Tu, H.; Zhou, J.; Liu, X.; Cheng, Y.; Luo, J.; Deng, X.; Zhang, H.; Xu, J. Characterization and metabolic diversity of flavonoids in citrus species. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Rao, M.J.; Ding, F.; Wang, N.; Deng, X.; Xu, Q. Metabolic mechanisms of host species against citrus Huanglongbing (Greening Disease). Crit. Rev. Plant Sci. 2019, 37, 496–511. [Google Scholar] [CrossRef]
- Lam, P.Y.; Liu, H.; Lo, C. Completion of tricin biosynthesis pathway in rice: Cytochrome P450 75B4 is a unique chrysoeriol 5′-hydroxylase. Plant Physiol. 2015, 168, 1527–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, B.; Jordán, F.; Osbourn, A. First encounters–deployment of defence-related natural products by plants. New Phytol. 2006, 172, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Hussain, S.; Khalid, M.F.; Saqib, M.; Ahmad, S.; Zafar, W.; Rao, M.J.; Morillon, R.; Anjum, M.A. Drought tolerance in citrus rootstocks is associated with better antioxidant defense mechanism. Acta Physiol. Plant. 2018, 40, 135. [Google Scholar] [CrossRef]
- Arbona, V.; De Ollas, C.J.; Argamasilla, R.; López-Climent, M.F.; Gómez-Cadenas, A. Hormone and metabolite traits related to abiotic stress tolerance in citrus. Acta Hortic. 2015, 1065, 1275–1281. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasajima, I.; Ide, Y.; Ohkama-Ohtsu, N.; Hayashi, H.; Yoneyama, T.; Fujiwara, T. A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 2004, 22, 49–52. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sumanta, N.; Haque, C.I.; Nishika, J.; Suprakash, R. Spectrophotometric analysis of chlorophylls and carotenoids from commonly grown fern species by using various extracting solvents spectrophotometric analysis of chlorophylls and carotenoids from commonly grown fern species by using various extracting solvents. Res. J. Chem. Sci. 2014, 4, 63–69. [Google Scholar]
- Velioglu, Y.S.; Mazza, G.; Gao, L.; Oomah, B.D. Antioxidant activity and total phenolics in selected fruits, vegetables, and grain products. J. Agric. Food Chem. 1998, 46, 4113–4117. [Google Scholar] [CrossRef]
- Dewanto, V.; Wu, X.; Adom, K.K.; Liu, R.H. Thermal processing enhances the nutritional value of tomatoes by increasing total antioxidant activity. J. Agric. Food Chem. 2002, 50, 3010–3014. [Google Scholar] [CrossRef]
- Nakata, M.; Ohme-Takagi, M. Quantification of anthocyanin content. Bio-Protocol 2014, 4, e1098. [Google Scholar] [CrossRef]
- Nakata, M.; Mitsuda, N.; Herde, M.; Koo, A.J.K.; Moreno, J.E.; Suzuki, K.; Howe, G.A.; Ohme-Takagi, M. A bHLH-type transcription factor, ABA-inducible BHLH-type transcription factor/JA-associated MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 2013, 25, 1641–1656. [Google Scholar] [CrossRef] [Green Version]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. In Plant Stress Tolerance; Sunkar, R., Ed.; Springer: New York, NY, USA, 2010; Volume 639, pp. 291–297. [Google Scholar]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, M.; Guo, Q.; Wang, G.; Gong, J.; Xu, Y.; Wang, W. Manipulation of monoubiquitin improves chilling tolerance in transgenic tobacco (Nicotiana tabacum). Plant Physiol. Biochem. 2014, 75, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Özgen, M.; Scheerens, J.C.; Reese, R.N.; Miller, R.A. Total phenolic, anthocyanin contents and antioxidant capacity of selected elderberry (Sambucus canadensis L.) accessions. Pharmacogn. Mag. 2010, 6, 198. [Google Scholar] [CrossRef] [Green Version]
- Daudi, A.; Paudyal, R.; Weizbauer, R. Plant tissue trypan blue staining during phytopathogen infection. Plant J. 2016, 6, 1–7. [Google Scholar]
- Grellet Bournonville, C.F.; Díaz-Ricci, J.C. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. 2011, 22, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Routaboul, J.-M.; Dubos, C.; Beck, G.; Marquis, C.; Bidzinski, P.; Loudet, O.; Lepiniec, L. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 2012, 63, 3749–3764. [Google Scholar] [CrossRef] [Green Version]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.; Song, W. Overexpression of SoCYP85A1, a Spinach Cytochrome p450 gene in transgenic tobacco enhances root development and drought stress tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef] [Green Version]
- De Martino, L.; Mencherini, T.; Mancini, E.; Aquino, R.P.; Fernando, L.; De Almeida, R.; De Feo, V. In vitro phytotoxicity and antioxidant activity of selected flavonoids. Int. J. Mol. Sci. 2012, 13, 5406–5419. [Google Scholar] [CrossRef]
- Borges Bubols, G.; da Rocha Vianna, D.; Medina-Remon, A.; von Poser, G.; Maria Lamuela-Raventos, R.; Lucia Eifler-Lima, V.; Cristina Garcia, S. The antioxidant activity of coumarins and flavonoids. Mini Rev. Med. Chem. 2013, 13, 318–334. [Google Scholar]
- Yu, J.; Wang, L.; Walzem, R.L.; Miller, E.G.; Pike, L.M.; Patil, B.S. Antioxidant activity of citrus limonoids, flavonoids, and coumarins. J. Agric. Food Chem. 2005, 53, 2009–2014. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Rao, M.J.; Anjum, M.A.; Ejaz, S.; Umar, U.U.; Ali, M.A.; Khalid, M.F.; Sohail, M.; Ercisli, S.; Zia-Ul-Haq, M.; et al. Effect of different combinations of antibiotics on fruit quality and antioxidant defense system in Huanglongbing infected Kinnow orchards. AMB Expr. 2019, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Arbona, V.; Hossain, Z.; López-Climent, M.F.; Pérez-Clemente, R.M.; Gómez-Cadenas, A. Antioxidant enzymatic activity is linked to waterlogging stress tolerance in citrus. Physiol. Plant. 2008, 132, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Kuźniak, E.; Urbanek, H. The involvement of hydrogen peroxide in plant responses to stresses. Acta Physiol. Plant. 2000, 22, 195–203. [Google Scholar] [CrossRef]
- Hussain, S.; Rao, M.J.; Anjum, M.A.; Ejaz, S.; Zakir, I.; Ali, M.A.; Ahmad, N.; Ahmad, S. oxidative stress and antioxidant defense in plants under drought conditions. In Plant Abiotic Stress Tolerance; Hasanuzzaman, M., Hakeem, K., Nahar, K., Alharby, H., Eds.; Springer: Cham, Switzerland, 2019; pp. 207–219. [Google Scholar]
- Liu, Z.; Tavares, R.; Forsythe, E.S.; André, F.; Lugan, R.; Jonasson, G.; Boutet-Mercey, S.; Tohge, T.; Beilstein, M.A.; Werck-Reichhart, D. Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat. Commun. 2016, 7, 13026. [Google Scholar] [CrossRef] [Green Version]
- Mittapelli, S.R.; Maryada, S.K. Structural organization, classification and phylogenetic relationship of cytochrome P450 genes in Citrus clementina and Citrus sinensis. Tree Genet Genomes 2014, 10, 399–409. [Google Scholar] [CrossRef]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef]
- Smigocki, A.C.; Wilson, D. Pest and disease resistance enhanced by heterologous suppression of a Nicotiana plumbaginifolia cytochrome P450 gene. Biotechnol. Lett. 2004, 26, 1809–1814. [Google Scholar] [CrossRef]
- Morant, M.; Bak, S.; Møller, B.L.; Werck-Reichhart, D. Plant cytochromes P450: Tools for pharmacology, plant protection and phytoremediation. Curr. Opin. Biotechnol. 2003, 14, 151–162. [Google Scholar] [CrossRef]
- Dos Santos, I.C.; de Almeida, A.-A.F.; Pirovani, C.P.; Costa, M.G.C.; Bellete, B.S.; Freschi, L.; Soares Filho, W.; Coelho Filho, M.A.; da Silva Gesteira, A. Differential accumulation of flavonoids and phytohormones resulting from the canopy/rootstock interaction of citrus plants subjected to dehydration/rehydration. Plant Physiol. Biochem. 2017, 119, 147–158. [Google Scholar] [CrossRef]
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- D’Maris Amick Dempsey, A.C.; Vlot, M.C.W.; Daniel, F.K. Salicylic acid biosynthesis and metabolism. Am. Soc. Plant Biol. 2011, 9, e0156. [Google Scholar]
- Arbona, V.; Manzi, M.; Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Okello, O.P.; Nawiri, M.P.; Musila, W.; Gweyi-Onyango, J.P. Water stress effect on total antioxiant activity and total phenolic content of Solanum scabrum Mill and Solanum scabrum in Kiambu, Kenya. Int. J. Biochem. Res. Rev. 2017, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. In Arabidopsis Book; American Society. Plant Biologists: New York, NY, USA, 2011; Volume 9, p. 0152. [Google Scholar]
- Trojak, M.; Skowron, E. Role of anthocyanins in high-light stress response. World Sci. News 2017, 81, 150–168. [Google Scholar]
- Gonzalez, A.; Brown, M.; Hatlestad, G.; Akhavan, N.; Smith, T.; Hembd, A.; Moore, J.; Montes, D.; Mosley, T.; Resendez, J. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 2016, 419, 54–63. [Google Scholar] [CrossRef]
- Agati, G.; Tattini, M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010, 186, 786–793. [Google Scholar] [CrossRef]
- Rao, M.J.; Hussain, S.; Anjum, M.A.; Saqib, M.; Ahmad, R.; Khalid, M.F.; Sohail, M.; Ejaz, S.; Ali, M.A.; Ahmad, N.; et al. Effect of Seed Priming on Seed Dormancy and Vigor. In Priming and Pretreatment of Seeds and Seedlings; Hasanuzzaman, M., Fotopoulos, V., Eds.; Springer: Singapore, 2019; pp. 135–145. [Google Scholar]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Sarker, U.; Oba, S. Augmentation of leaf color parameters, pigments, vitamins, phenolic acids, flavonoids and antioxidant activity in selected Amaranthus tricolor under salinity stress. Sci. Rep. 2018, 8, 12349. [Google Scholar] [CrossRef] [Green Version]
- Davletova, S.; Rizhsky, L.; Liang, H.; Shengqiang, Z.; Oliver, D.J.; Coutu, J.; Shulaev, V.; Schlauch, K.; Mittler, R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 2005, 17, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Xu, Q. CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis. Antioxidants 2020, 9, 161. https://doi.org/10.3390/antiox9020161
Rao MJ, Xu Y, Tang X, Huang Y, Liu J, Deng X, Xu Q. CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis. Antioxidants. 2020; 9(2):161. https://doi.org/10.3390/antiox9020161
Chicago/Turabian StyleRao, Muhammad Junaid, Yuantao Xu, Xiaomei Tang, Yue Huang, Jihong Liu, Xiuxin Deng, and Qiang Xu. 2020. "CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis" Antioxidants 9, no. 2: 161. https://doi.org/10.3390/antiox9020161
APA StyleRao, M. J., Xu, Y., Tang, X., Huang, Y., Liu, J., Deng, X., & Xu, Q. (2020). CsCYT75B1, a Citrus CYTOCHROME P450 Gene, Is Involved in Accumulation of Antioxidant Flavonoids and Induces Drought Tolerance in Transgenic Arabidopsis. Antioxidants, 9(2), 161. https://doi.org/10.3390/antiox9020161





