Development of an Analytical Assay for Electrochemical Detection and Quantification of Protein-Bound 3-Nitrotyrosine in Biological Samples and Comparison with Classical, Antibody-Based Methods
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Animals Handling and Euthanasia
2.3. Nitration of Bovine Serum Albumin (BSA) or Biological Samples
2.4. Dot Blot Analysis for Protein-Bound 3-Nitrotyrosine
2.5. ELISA Quantification of Protein-Bound 3-Nitrotyrosine
2.6. HPLC/ECD and UV/Vis Detection of 3-Nitrotyrosine
2.7. Detection of Mitochondrial Superoxide Formation by mitoSOX HPLC Method and Plate Reader Assay
2.8. LC-MS/MS Analysis
2.9. Statistical Analysis
3. Results
3.1. Comparison of the Detection and Quantification of 3-NT Standards as Well as Nitrated BSA Standards by HPLC/ECD, Dot Blot and ELISA
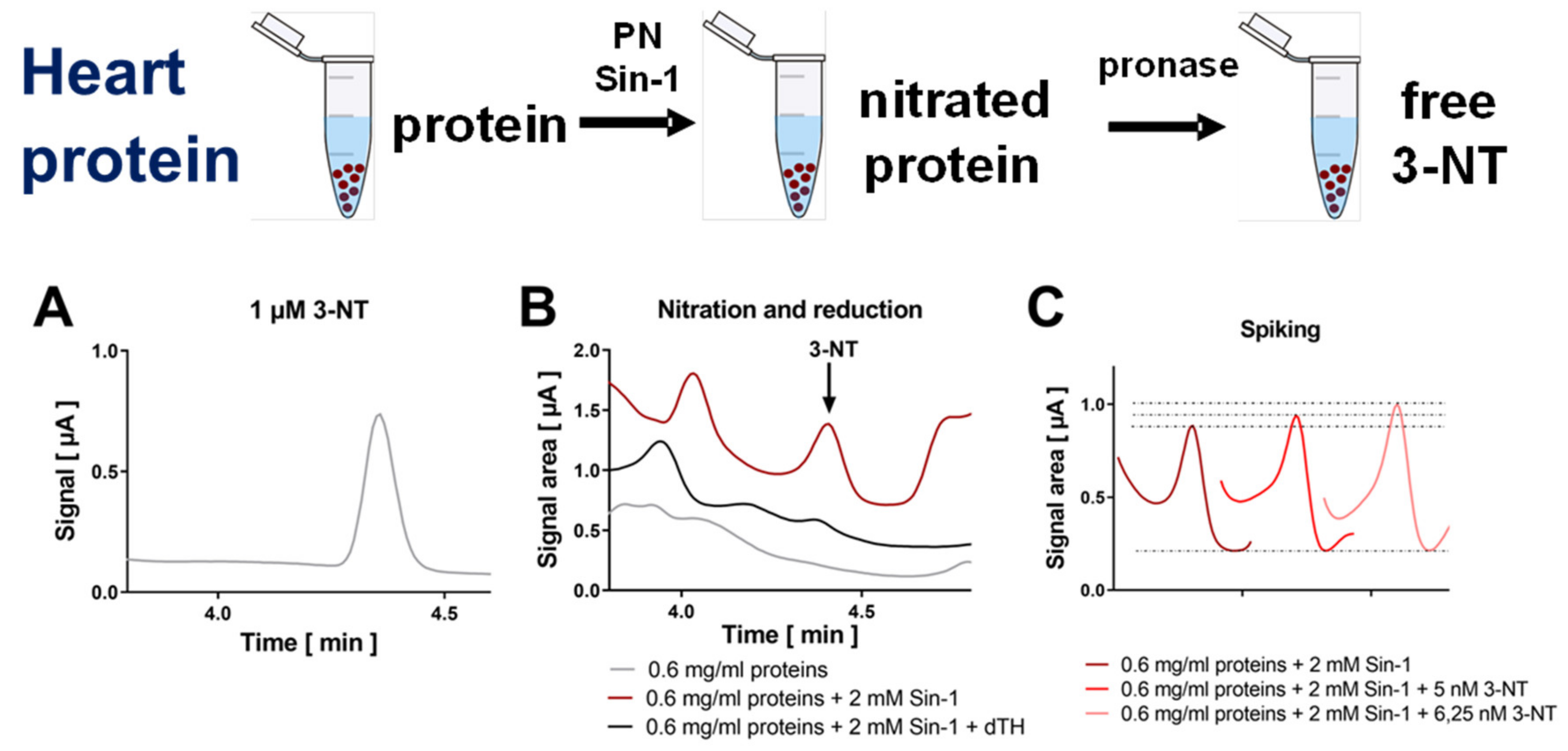
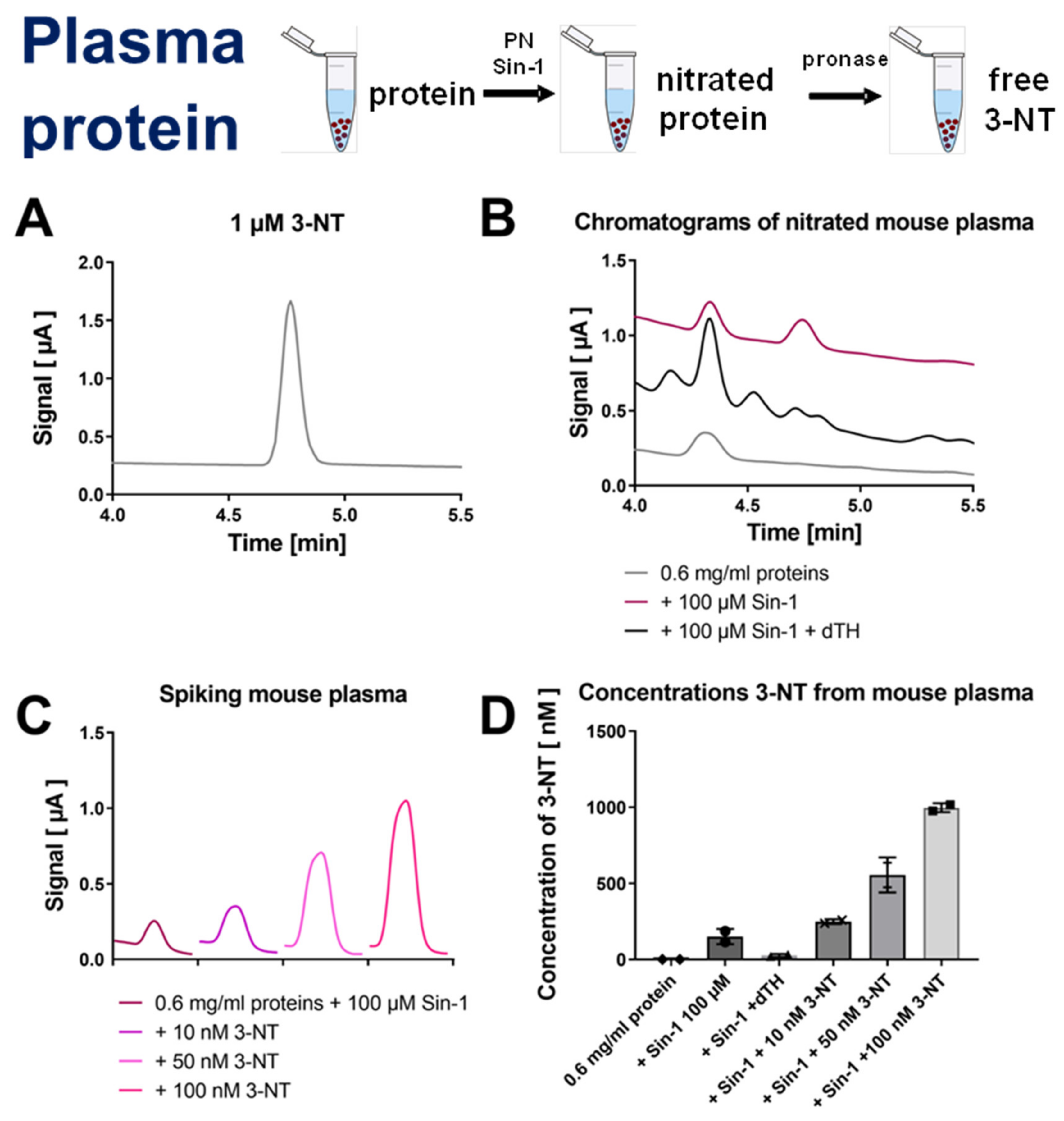
3.2. Detection and Quantification of Free 3-NT from Nitrated Tissue Homogenates and Plasma Samples by HPLC/ECD as Well as Comparison with HPLC/UV or ELISA
3.3. Detection and Quantification of Free 3-NT from Tissue Samples of Diabetic Rats with or without Combined Sin-1 Nitration by HPLC/ECD as Well as Comparison with LC-MS
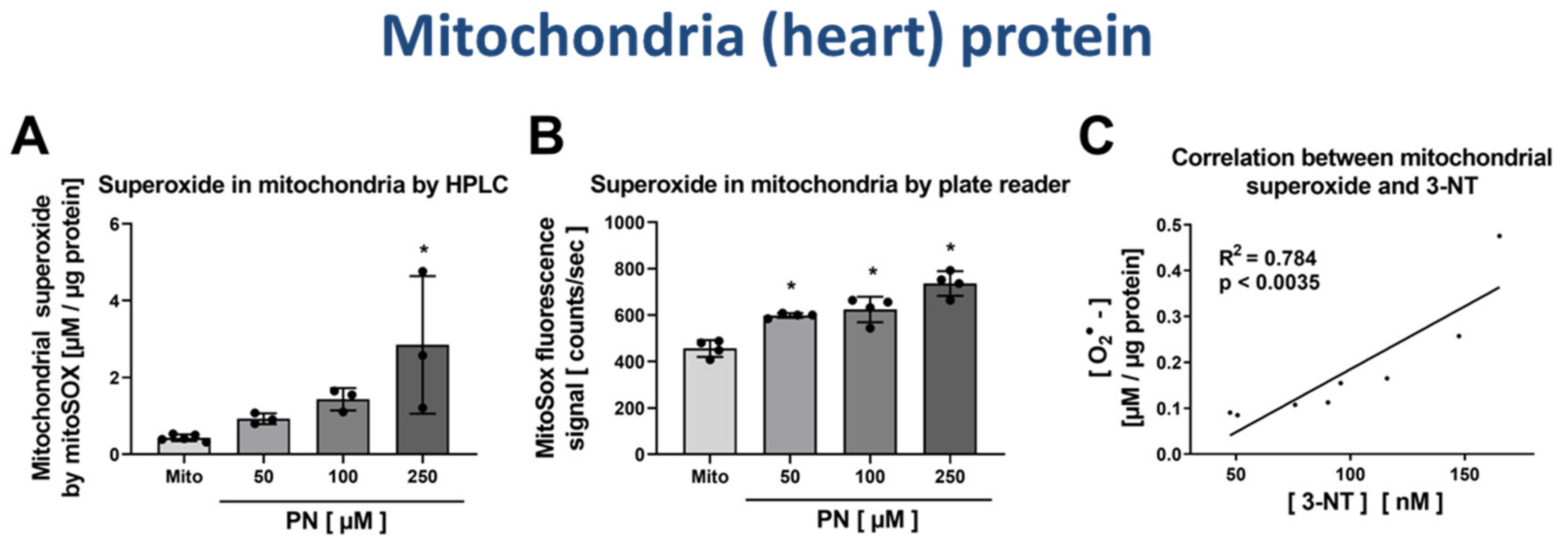
3.4. Correlation of Mitochondrial Nitration and Superoxide Formation in Response to PN Treatment
4. Discussion
4.1. Importance of the Quantification of Oxidative Stress in General and 3-Nitrotyrosine in Particular
4.2. Biological Consequences of Protein Tyrosine Nitration
4.3. Comparison of Previous Reports on Detection and Quantification of Protein Tyrosine Nitration
4.4. Comparison of Theory and Our Empiric Data
4.5. Strengths and Limitations of the Present Study
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Griendling, K.K.; FitzGerald, G.A. Oxidative stress and cardiovascular injury: Part I: Basic mechanisms and in vivo monitoring of ROS. Circulation 2003, 108, 1912–1916. [Google Scholar] [CrossRef] [PubMed]
- Griendling, K.K.; FitzGerald, G.A. Oxidative stress and cardiovascular injury: Part II: Animal and human studies. Circulation 2003, 108, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Ischiropoulos, H.; Beckman, J.S. Oxidative stress and nitration in neurodegeneration: Cause, effect, or association? J. Clin. Investig. 2003, 111, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, S.; Rupprecht, H.J.; Bickel, C.; Torzewski, M.; Hafner, G.; Tiret, L.; Smieja, M.; Cambien, F.; Meyer, J.; Lackner, K.J. Glutathione peroxidase 1 activity and cardiovascular events in patients with coronary artery disease. N. Engl. J. Med. 2003, 349, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Schottker, B.; Brenner, H.; Jansen, E.H.; Gardiner, J.; Peasey, A.; Kubinova, R.; Pajak, A.; Topor-Madry, R.; Tamosiunas, A.; Saum, K.U.; et al. Evidence for the free radical/oxidative stress theory of ageing from the CHANCES consortium: A meta-analysis of individual participant data. BMC Med. 2015, 13, 300. [Google Scholar] [CrossRef]
- Sies, H. Oxidative Stress: Oxidants and Antioxidants; Academic Press: London, UK, 1991. [Google Scholar]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Daiber, A.; Xia, N.; Steven, S.; Oelze, M.; Hanf, A.; Kroller-Schon, S.; Munzel, T.; Li, H. New therapeutic implications of endothelial nitric oxide synthase (eNOS) function/dysfunction in cardiovascular disease. Int. J. Mol. Sci. 2019, 20, 187. [Google Scholar] [CrossRef]
- Steven, S.; Frenis, K.; Oelze, M.; Kalinovic, S.; Kuntic, M.; Bayo Jimenez, M.T.; Vujacic-Mirski, K.; Helmstadter, J.; Kroller-Schon, S.; Munzel, T.; et al. Vascular inflammation and oxidative stress: Major triggers for cardiovascular disease. Oxid. Med. Cell. Longev. 2019, 2019, 7092151. [Google Scholar] [CrossRef]
- Quijano, C.; Hernandez-Saavedra, D.; Castro, L.; McCord, J.M.; Freeman, B.A.; Radi, R. Reaction of peroxynitrite with Mn-superoxide dismutase. Role of the metal center in decomposition kinetics and nitration. J. Biol. Chem. 2001, 276, 11631–11638. [Google Scholar] [CrossRef]
- MacMillan-Crow, L.A.; Crow, J.P.; Kerby, J.D.; Beckman, J.S.; Thompson, J.A. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc. Natl. Acad. Sci. USA 1996, 93, 11853–11858. [Google Scholar] [CrossRef]
- MacMillan-Crow, L.A.; Crow, J.P.; Thompson, J.A. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry 1998, 37, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Abello, N.; Kerstjens, H.A.; Postma, D.S.; Bischoff, R. Protein tyrosine nitration: Selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteom. Res. 2009, 8, 3222–3238. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Taylor, S.W.; Zhang, B.; Ghosh, S.S.; Capaldi, R.A. Oxidative damage to mitochondrial complex I due to peroxynitrite: Identification of reactive tyrosines by mass spectrometry. J. Biol. Chem. 2003, 278, 37223–37230. [Google Scholar] [CrossRef] [PubMed]
- Radi, R.; Cassina, A.; Hodara, R.; Quijano, C.; Castro, L. Peroxynitrite reactions and formation in mitochondria. Free Radic. Biol. Med. 2002, 33, 1451–1464. [Google Scholar] [CrossRef]
- Sies, H.; Dafre, A.L.; Ji, Y.; Akerboom, T.P. Protein S-thiolation and redox regulation of membrane-bound glutathione transferase. Chem. Biol. Interact. 1998, 111, 177–185. [Google Scholar] [CrossRef]
- Lassegue, B.; Sorescu, D.; Szocs, K.; Yin, Q.; Akers, M.; Zhang, Y.; Grant, S.L.; Lambeth, J.D.; Griendling, K.K. Novel gp91(phox) homologues in vascular smooth muscle cells: Nox1 mediates angiotensin II-induced superoxide formation and redox-sensitive signaling pathways. Circ. Res. 2001, 88, 888–894. [Google Scholar] [CrossRef]
- Li, M.; Absher, P.M.; Liang, P.; Russell, J.C.; Sobel, B.E.; Fukagawa, N.K. High glucose concentrations induce oxidative damage to mitochondrial DNA in explanted vascular smooth muscle cells. Exp. Biol. Med. 2001, 226, 450–457. [Google Scholar] [CrossRef]
- Bachschmid, M.; Schildknecht, S.; Ullrich, V. Redox regulation of vascular prostanoid synthesis by the nitric oxide-superoxide system. Biochem. Biophys. Res. Commun. 2005, 338, 536–542. [Google Scholar] [CrossRef]
- Daiber, A.; Bachschmid, M. Enzyme inhibition by peroxynitrite-mediated tyrosine nitration and thiol oxidation. Curr. Enzym. Inhib. 2007, 3, 103–117. [Google Scholar] [CrossRef]
- Turko, I.V.; Murad, F. Protein nitration in cardiovascular diseases. Pharm. Rev. 2002, 54, 619–634. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of peroxynitrite and protein tyrosine nitration. Chem. Rev. 2018, 118, 1338–1408. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Martin, C.; Ullrich, V. Tyrosine nitration as a mechanism of selective inactivation of prostacyclin synthase by peroxynitrite. Biol. Chem. 1997, 378, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.H.; Daiber, A.; Peterson, J.A.; Shoun, H.; Ullrich, V. Rapid reactions of peroxynitrite with heme-thiolate proteins as the basis for protection of prostacyclin synthase from inactivation by nitration. Arch. Biochem. Biophys. 2000, 376, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Daiber, A.; Bachschmid, M.; Beckman, J.S.; Munzel, T.; Ullrich, V. The impact of metal catalysis on protein tyrosine nitration by peroxynitrite. Biochem. Biophys. Res. Commun. 2004, 317, 873–881. [Google Scholar] [CrossRef]
- Daiber, A.; Herold, S.; Schoneich, C.; Namgaladze, D.; Peterson, J.A.; Ullrich, V. Nitration and inactivation of cytochrome P450BM-3 by peroxynitrite. Stopped-flow measurements prove ferryl intermediates. Eur. J. Biochem. FEBS 2000, 267, 6729–6739. [Google Scholar] [CrossRef]
- Steven, S.; Oelze, M.; Hanf, A.; Kroller-Schon, S.; Kashani, F.; Roohani, S.; Welschof, P.; Kopp, M.; Godtel-Armbrust, U.; Xia, N.; et al. The SGLT2 inhibitor empagliflozin improves the primary diabetic complications in ZDF rats. Redox Biol. 2017, 13, 370–385. [Google Scholar] [CrossRef]
- Daiber, A.; Oelze, M.; Coldewey, M.; Bachschmid, M.; Wenzel, P.; Sydow, K.; Wendt, M.; Kleschyov, A.L.; Stalleicken, D.; Ullrich, V.; et al. Oxidative stress and mitochondrial aldehyde dehydrogenase activity: A comparison of pentaerythritol tetranitrate with other organic nitrates. Mol. Pharm. 2004, 66, 1372–1382. [Google Scholar] [CrossRef]
- Wenzel, P.; Schulz, E.; Gori, T.; Ostad, M.A.; Mathner, F.; Schildknecht, S.; Gobel, S.; Oelze, M.; Stalleicken, D.; Warnholtz, A.; et al. Monitoring white blood cell mitochondrial aldehyde dehydrogenase activity: Implications for nitrate therapy in humans. J. Pharm. Exp. 2009, 330, 63–71. [Google Scholar] [CrossRef]
- Oelze, M.; Kroller-Schon, S.; Steven, S.; Lubos, E.; Doppler, C.; Hausding, M.; Tobias, S.; Brochhausen, C.; Li, H.; Torzewski, M.; et al. Glutathione peroxidase-1 deficiency potentiates dysregulatory modifications of endothelial nitric oxide synthase and vascular dysfunction in aging. Hypertension 2014, 63, 390–396. [Google Scholar] [CrossRef]
- Oelze, M.; Knorr, M.; Schell, R.; Kamuf, J.; Pautz, A.; Art, J.; Wenzel, P.; Munzel, T.; Kleinert, H.; Daiber, A. Regulation of human mitochondrial aldehyde dehydrogenase (ALDH-2) activity by electrophiles in vitro. J. Biol. Chem. 2011, 286, 8893–8900. [Google Scholar] [CrossRef]
- Daiber, A.; Schoneich, C.; Schmidt, P.; Jung, C.; Ullrich, V. Autocatalytic nitration of P450CAM by peroxynitrite. J. Inorg. Biochem. 2000, 81, 213–220. [Google Scholar] [CrossRef]
- Daiber, A.; Daub, S.; Bachschmid, M.; Schildknecht, S.; Oelze, M.; Steven, S.; Schmidt, P.; Megner, A.; Wada, M.; Tanabe, T.; et al. Protein tyrosine nitration and thiol oxidation by peroxynitrite-strategies to prevent these oxidative modifications. Int. J. Mol. Sci. 2013, 14, 7542–7570. [Google Scholar] [CrossRef]
- Link, A.J.; LaBaer, J. Trichloroacetic acid (TCA) precipitation of proteins. Cold Spring Harb. Protoc. 2011, 2011, 993–994. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Srinivasan, S.; Hardy, M.; Ouari, O.; Lopez, M.; Vasquez-Vivar, J.; Avadhani, N.G.; Kalyanaraman, B. Cytochrome c-mediated oxidation of hydroethidine and mito-hydroethidine in mitochondria: Identification of homo- and heterodimers. Free Radic. Biol. Med. 2008, 44, 835–846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kalinovic, S.; Oelze, M.; Kroller-Schon, S.; Steven, S.; Vujacic-Mirski, K.; Kvandova, M.; Schmal, I.; Al Zuabi, A.; Munzel, T.; Daiber, A. Comparison of mitochondrial superoxide detection ex vivo/in vivo by mitoSOX HPLC method with classical assays in three different animal models of oxidative stress. Antioxidants 2019, 8, 11. [Google Scholar] [CrossRef]
- Daiber, A. Redox signaling (cross-talk) from and to mitochondria involves mitochondrial pores and reactive oxygen species. Biochim. Biophys. Acta 2010, 1797, 897–906. [Google Scholar] [CrossRef]
- Daiber, A.; Di Lisa, F.; Oelze, M.; Kroller-Schon, S.; Steven, S.; Schulz, E.; Munzel, T. Crosstalk of mitochondria with NADPH oxidase via reactive oxygen and nitrogen species signalling and its role for vascular function. Br. J. Pharm. 2017, 174, 1670–1689. [Google Scholar] [CrossRef]
- Kroller-Schon, S.; Steven, S.; Kossmann, S.; Scholz, A.; Daub, S.; Oelze, M.; Xia, N.; Hausding, M.; Mikhed, Y.; Zinssius, E.; et al. Molecular mechanisms of the crosstalk between mitochondria and NADPH oxidase through reactive oxygen species-studies in white blood cells and in animal models. Antioxid. Redox Signal. 2014, 20, 247–266. [Google Scholar] [CrossRef]
- Wenzel, P.; Mollnau, H.; Oelze, M.; Schulz, E.; Wickramanayake, J.M.; Muller, J.; Schuhmacher, S.; Hortmann, M.; Baldus, S.; Gori, T.; et al. First evidence for a crosstalk between mitochondrial and NADPH oxidase-derived reactive oxygen species in nitroglycerin-triggered vascular dysfunction. Antioxid. Redox Signal. 2008, 10, 1435–1447. [Google Scholar] [CrossRef]
- Zinkevich, N.S.; Gutterman, D.D. ROS-induced ROS release in vascular biology: Redox-redox signaling. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H647–H653. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial ROS-induced ROS release: An update and review. Biochim. Biophys. Acta 2006, 1757, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Heitzer, T.; Schlinzig, T.; Krohn, K.; Meinertz, T.; Munzel, T. Endothelial dysfunction, oxidative stress, and risk of cardiovascular events in patients with coronary artery disease. Circulation 2001, 104, 2673–2678. [Google Scholar] [CrossRef] [PubMed]
- Fratta Pasini, A.; Albiero, A.; Stranieri, C.; Cominacini, M.; Pasini, A.; Mozzini, C.; Vallerio, P.; Cominacini, L.; Garbin, U. Serum oxidative stress-induced repression of Nrf2 and GSH depletion: A mechanism potentially involved in endothelial dysfunction of young smokers. PLoS ONE 2012, 7, e30291. [Google Scholar] [CrossRef]
- Yilmaz, M.I.; Saglam, M.; Caglar, K.; Cakir, E.; Sonmez, A.; Ozgurtas, T.; Aydin, A.; Eyileten, T.; Ozcan, O.; Acikel, C.; et al. The determinants of endothelial dysfunction in CKD: Oxidative stress and asymmetric dimethylarginine. Am. J. Kidney Dis. J. Natl. Kidney Found. 2006, 47, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Gamez, B.; Fernandez-Marin, M.C.; Gomez-Chaparro, J.L.; Munoz-Cabrera, L.; Lopez-Barea, J.; Perez-Jimenez, F.; Lopez-Miranda, J. Relationship of oxidative stress and endothelial dysfunction in sleep apnoea. Eur. Respir. J. 2011, 37, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.R. Inhibitors of the leukocyte superoxide generating oxidase: Mechanisms of action and methods for their elucidation. Free Radic Biol Med. 1990, 8, 71–93. [Google Scholar] [CrossRef]
- Flaherty, J.T.; Weisfeldt, M.L. Reperfusion injury. Free Radic Biol Med. 1988, 5, 409–419. [Google Scholar] [CrossRef]
- Linnane, A.W.; Eastwood, H. Cellular redox regulation and prooxidant signaling systems: A new perspective on the free radical theory of aging. Ann. N. Y. Acad. Sci. 2006, 1067, 47–55. [Google Scholar] [CrossRef]
- Afanas’Ev, I.B. Mechanism of superoxide-mediated damage relevance to mitochondrial aging. Ann. N. Y. Acad. Sci. 2004, 1019, 34–35. [Google Scholar] [CrossRef]
- Beckman, J.S.; Beckman, T.W.; Chen, J.; Marshall, P.A.; Freeman, B.A. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc. Natl. Acad. Sci. USA 1990, 87, 1620–1624. [Google Scholar] [CrossRef] [PubMed]
- Munzel, T.; Daiber, A.; Ullrich, V.; Mulsch, A. Vascular consequences of endothelial nitric oxide synthase uncoupling for the activity and expression of the soluble guanylyl cyclase and the cGMP-dependent protein kinase. Arter. Thromb. Vasc. Biol. 2005, 25, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Bassenge, E.; Schneider, H.T.; Daiber, A. Oxidative stress and cardiovascular diseases. Deutsche Medizinische Wochenschrift 2005, 130, 2904–2909. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, V.; Namgaladze, D.; Frein, D. Superoxide as inhibitor of calcineurin and mediator of redox regulation. Toxicol. Lett. 2003, 139, 107–110. [Google Scholar] [CrossRef]
- Rahman, I.; Biswas, S.K.; Kode, A. Oxidant and antioxidant balance in the airways and airway diseases. Eur J. Pharm. 2006, 533, 222–239. [Google Scholar] [CrossRef]
- Gutteridge, J.M.; Halliwell, B. Free radicals and antioxidants in the year 2000. A historical look to the future. Ann. N. Y. Acad. Sci. 2000, 899, 136–147. [Google Scholar] [CrossRef]
- Gryglewski, R.J.; Palmer, R.M.; Moncada, S. Superoxide anion is involved in the breakdown of endothelium-derived vascular relaxing factor. Nature 1986, 320, 454–456. [Google Scholar] [CrossRef]
- Denicola, A.; Freeman, B.A.; Trujillo, M.; Radi, R. Peroxynitrite reaction with carbon dioxide/bicarbonate: Kinetics and influence on peroxynitrite-mediated oxidations. Arch. Biochem. Biophys. 1996, 333, 49–58. [Google Scholar] [CrossRef]
- Bonini, M.G.; Radi, R.; Ferrer-Sueta, G.; Ferreira, A.M.; Augusto, O. Direct EPR detection of the carbonate radical anion produced from peroxynitrite and carbon dioxide. J. Biol. Chem. 1999, 274, 10802–10806. [Google Scholar] [CrossRef]
- Ferrer-Sueta, G.; Radi, R. Chemical biology of peroxynitrite: Kinetics, diffusion, and radicals. ACS Chem. Biol. 2009, 4, 161–177. [Google Scholar] [CrossRef]
- Souza, J.M.; Daikhin, E.; Yudkoff, M.; Raman, C.S.; Ischiropoulos, H. Factors determining the selectivity of protein tyrosine nitration. Arch. Biochem. Biophys. 1999, 371, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Greenacre, S.A.; Ischiropoulos, H. Tyrosine nitration: Localisation, quantification, consequences for protein function and signal transduction. Free Radic. Res. 2001, 34, 541–581. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.; Youhnovski, N.; Daiber, A.; Balan, A.; Arsic, M.; Bachschmid, M.; Przybylski, M.; Ullrich, V. Specific nitration at tyrosine 430 revealed by high resolution mass spectrometry as basis for redox regulation of bovine prostacyclin synthase. J. Biol. Chem. 2003, 278, 12813–12819. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Jendral, M.; Ullrich, V. Prostaglandin endoperoxide-dependent vasospasm in bovine coronary arteries after nitration of prostacyclin synthase. Br. J. Pharm. 1999, 126, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Cassina, A.M.; Hodara, R.; Souza, J.M.; Thomson, L.; Castro, L.; Ischiropoulos, H.; Freeman, B.A.; Radi, R. Cytochrome c nitration by peroxynitrite. J. Biol. Chem. 2000, 275, 21409–21415. [Google Scholar] [CrossRef]
- Vadseth, C.; Souza, J.M.; Thomson, L.; Seagraves, A.; Nagaswami, C.; Scheiner, T.; Torbet, J.; Vilaire, G.; Bennett, J.S.; Murciano, J.C.; et al. Pro-thrombotic state induced by post-translational modification of fibrinogen by reactive nitrogen species. J. Biol. Chem. 2004, 279, 8820–8826. [Google Scholar] [CrossRef]
- Nowak, P.; Kolodziejczyk, J.; Wachowicz, B. Peroxynitrite and fibrinolytic system: The effect of peroxynitrite on plasmin activity. Mol. Cell Biochem. 2004, 267, 141–146. [Google Scholar] [CrossRef]
- Heffron, S.P.; Parastatidis, I.; Cuchel, M.; Wolfe, M.L.; Tadesse, M.G.; Mohler, E.R., III; Ischiropoulos, H.; Rader, D.J.; Reilly, M.P. Inflammation induces fibrinogen nitration in experimental human endotoxemia. Free Radic. Biol. Med. 2009, 47, 1140–1146. [Google Scholar] [CrossRef]
- Parastatidis, I.; Thomson, L.; Burke, A.; Chernysh, I.; Nagaswami, C.; Visser, J.; Stamer, S.; Liebler, D.C.; Koliakos, G.; Heijnen, H.F.; et al. Fibrinogen beta-chain tyrosine nitration is a prothrombotic risk factor. J. Biol. Chem. 2008, 283, 33846–33853. [Google Scholar] [CrossRef]
- Brennan, M.L.; Wu, W.; Fu, X.; Shen, Z.; Song, W.; Frost, H.; Vadseth, C.; Narine, L.; Lenkiewicz, E.; Borchers, M.T.; et al. A tale of two controversies: Defining both the role of peroxidases in nitrotyrosine formation in vivo using eosinophil peroxidase and myeloperoxidase-deficient mice, and the nature of peroxidase-generated reactive nitrogen species. J. Biol. Chem. 2002, 277, 17415–17427. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Lass, A.; Schmidt, K.; Mayer, B. Protein tyrosine nitration in cytokine-activated murine macrophages. Involvement of a peroxidase/nitrite pathway rather than peroxynitrite. J. Biol. Chem. 2001, 276, 34051–34058. [Google Scholar] [CrossRef] [PubMed]
- Schildknecht, S.; Heinz, K.; Daiber, A.; Hamacher, J.; Kavakli, C.; Ullrich, V.; Bachschmid, M. Autocatalytic tyrosine nitration of prostaglandin endoperoxide synthase-2 in LPS-stimulated RAW 264.7 macrophages. Biochem. Biophys. Res. Commun. 2006, 340, 318–325. [Google Scholar] [CrossRef]
- Thomson, L.; Christie, J.; Vadseth, C.; Lanken, P.N.; Fu, X.; Hazen, S.L.; Ischiropoulos, H. Identification of immunoglobulins that recognize 3-nitrotyrosine in patients with acute lung injury after major trauma. Am. J. Respir. Cell Mol. Biol. 2007, 36, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Thomson, L.; Tenopoulou, M.; Lightfoot, R.; Tsika, E.; Parastatidis, I.; Martinez, M.; Greco, T.M.; Doulias, P.T.; Wu, Y.; Tang, W.H.; et al. Immunoglobulins against tyrosine-nitrated epitopes in coronary artery disease. Circulation 2012, 126, 2392–2401. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Hasegawa, T.; Mukai, K.; Watanabe, M.; Nishino, H. Determination of nitrotyrosine by HPLC-ECD and its application. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2002, 64, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Shigenaga, M.K.; Lee, H.H.; Blount, B.C.; Christen, S.; Shigeno, E.T.; Yip, H.; Ames, B.N. Inflammation and NO(X)-induced nitration: Assay for 3-nitrotyrosine by HPLC with electrochemical detection. Proc. Natl. Acad. Sci. USA 1997, 94, 3211–3216. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, H.; Celan, I.; Chazotte, L.; Pignatelli, B.; Mower, H.F. Analysis of 3-nitrotyrosine in biological fluids and protein hydrolyzates by high-performance liquid chromatography using a postseparation, on-line reduction column and electrochemical detection: Results with various nitrating agents. Nitric Oxide 1999, 3, 132–141. [Google Scholar] [CrossRef]
- Hensley, K.; Maidt, M.L.; Pye, Q.N.; Stewart, C.A.; Wack, M.; Tabatabaie, T.; Floyd, R.A. Quantitation of protein-bound 3-nitrotyrosine and 3,4-dihydroxyphenylalanine by high-performance liquid chromatography with electrochemical array detection. Anal. Biochem. 1997, 251, 187–195. [Google Scholar] [CrossRef]
- Sodum, R.S.; Akerkar, S.A.; Fiala, E.S. Determination of 3-nitrotyrosine by high-pressure liquid chromatography with a dual-mode electrochemical detector. Anal. Biochem. 2000, 280, 278–285. [Google Scholar] [CrossRef]
- Nuriel, T.; Deeb, R.S.; Hajjar, D.P.; Gross, S.S. Protein 3-nitrotyrosine in complex biological samples: Quantification by high-pressure liquid chromatography/electrochemical detection and emergence of proteomic approaches for unbiased identification of modification sites. Methods Enzym. 2008, 441, 1–17. [Google Scholar]
- Maruyama, W.; Hashizume, Y.; Matsubara, K.; Naoi, M. Identification of 3-nitro-L-tyrosine, a product of nitric oxide and superoxide, as an indicator of oxidative stress in the human brain. J. Chromatogr. B Biomed. Appl. 1996, 676, 153–158. [Google Scholar] [CrossRef]
- Skinner, K.A.; Crow, J.P.; Skinner, H.B.; Chandler, R.T.; Thompson, J.A.; Parks, D.A. Free and protein-associated nitrotyrosine formation following rat liver preservation and transplantation. Arch. Biochem Biophys 1997, 342, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, M.; Hao, G.; Gross, S.S. Detection of cysteine S-nitrosylation and tyrosine 3-nitration in kidney proteins. Methods Mol. Med. 2003, 86, 373–384. [Google Scholar] [PubMed]
- Kumarathasan, P.; Vincent, R. New approach to the simultaneous analysis of catecholamines and tyrosines in biological fluids. J. Chromatogr. A 2003, 987, 349–358. [Google Scholar] [CrossRef]
- Frijhoff, J.; Winyard, P.G.; Zarkovic, N.; Davies, S.S.; Stocker, R.; Cheng, D.; Knight, A.R.; Taylor, E.L.; Oettrich, J.; Ruskovska, T.; et al. Clinical relevance of biomarkers of oxidative stress. Antioxid Redox Signal. 2015, 23, 1144–1170. [Google Scholar] [CrossRef]
- Daiber, A.; Bachschmid, M.; Frein, D.; Ullrich, V. Reply to “Trouble with the analysis of nitrite, nitrate, S-nitrosothiols, and 3-nitrotyrosine: Freezing-induced artefacts”. Nitric Oxide 2004, 11, 214–215. [Google Scholar] [CrossRef]
- Daiber, A.; Bachschmid, M.; Kavakli, C.; Frein, D.; Wendt, M.; Ullrich, V.; Munzel, T. A new pitfall in detecting biological end products of nitric oxide-nitration, nitros(yl)ation and nitrite/nitrate artefacts during freezing. Nitric Oxide 2003, 9, 44–52. [Google Scholar] [CrossRef]
- Girault, I.; Karu, A.E.; Schaper, M.; Barcellos-Hoff, M.H.; Hagen, T.; Vogel, D.S.; Ames, B.N.; Christen, S.; Shigenaga, M.K. Immunodetection of 3-nitrotyrosine in the liver of zymosan-treated rats with a new monoclonal antibody: Comparison to analysis by HPLC. Free Radic. Biol. Med. 2001, 31, 1375–1387. [Google Scholar] [CrossRef]
- Knight, A.R.; Taylor, E.L.; Lukaszewski, R.; Jensen, K.T.; Jones, H.E.; Carre, J.E.; Isupov, M.N.; Littlechild, J.A.; Bailey, S.J.; Brewer, E.; et al. A high-sensitivity electrochemiluminescence-based ELISA for the measurement of the oxidative stress biomarker, 3-nitrotyrosine, in human blood serum and cells. Free Radic. Biol. Med. 2018, 120, 246–254. [Google Scholar] [CrossRef]
- Degendorfer, G.; Chuang, C.Y.; Kawasaki, H.; Hammer, A.; Malle, E.; Yamakura, F.; Davies, M.J. Peroxynitrite-mediated oxidation of plasma fibronectin. Free Radic. Biol. Med. 2016, 97, 602–615. [Google Scholar] [CrossRef]
- Degendorfer, G.; Chuang, C.Y.; Mariotti, M.; Hammer, A.; Hoefler, G.; Hagglund, P.; Malle, E.; Wise, S.G.; Davies, M.J. Exposure of tropoelastin to peroxynitrous acid gives high yields of nitrated tyrosine residues, di-tyrosine cross-links and altered protein structure and function. Free Radic Biol Med. 2018, 115, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Gerding, H.R.; Karreman, C.; Daiber, A.; Delp, J.; Hammler, D.; Mex, M.; Schildknecht, S.; Leist, M. Reductive modification of genetically encoded 3-nitrotyrosine sites in alpha synuclein expressed in E. coli. Redox Biol. 2019, 26, 101251. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Filburn, C.R.; Klotz, L.O.; Zweier, J.L.; Sollott, S.J. Reactive oxygen species (ROS)-induced ROS release: A new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J. Exp. Med. 2000, 192, 1001–1014. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vujacic-Mirski, K.; Bruns, K.; Kalinovic, S.; Oelze, M.; Kröller-Schön, S.; Steven, S.; Mojovic, M.; Korac, B.; Münzel, T.; Daiber, A. Development of an Analytical Assay for Electrochemical Detection and Quantification of Protein-Bound 3-Nitrotyrosine in Biological Samples and Comparison with Classical, Antibody-Based Methods. Antioxidants 2020, 9, 388. https://doi.org/10.3390/antiox9050388
Vujacic-Mirski K, Bruns K, Kalinovic S, Oelze M, Kröller-Schön S, Steven S, Mojovic M, Korac B, Münzel T, Daiber A. Development of an Analytical Assay for Electrochemical Detection and Quantification of Protein-Bound 3-Nitrotyrosine in Biological Samples and Comparison with Classical, Antibody-Based Methods. Antioxidants. 2020; 9(5):388. https://doi.org/10.3390/antiox9050388
Chicago/Turabian StyleVujacic-Mirski, Ksenija, Kai Bruns, Sanela Kalinovic, Matthias Oelze, Swenja Kröller-Schön, Sebastian Steven, Milos Mojovic, Bato Korac, Thomas Münzel, and Andreas Daiber. 2020. "Development of an Analytical Assay for Electrochemical Detection and Quantification of Protein-Bound 3-Nitrotyrosine in Biological Samples and Comparison with Classical, Antibody-Based Methods" Antioxidants 9, no. 5: 388. https://doi.org/10.3390/antiox9050388
APA StyleVujacic-Mirski, K., Bruns, K., Kalinovic, S., Oelze, M., Kröller-Schön, S., Steven, S., Mojovic, M., Korac, B., Münzel, T., & Daiber, A. (2020). Development of an Analytical Assay for Electrochemical Detection and Quantification of Protein-Bound 3-Nitrotyrosine in Biological Samples and Comparison with Classical, Antibody-Based Methods. Antioxidants, 9(5), 388. https://doi.org/10.3390/antiox9050388









