Role of Cytochrome P450 Enzymes in Plant Stress Response
Abstract
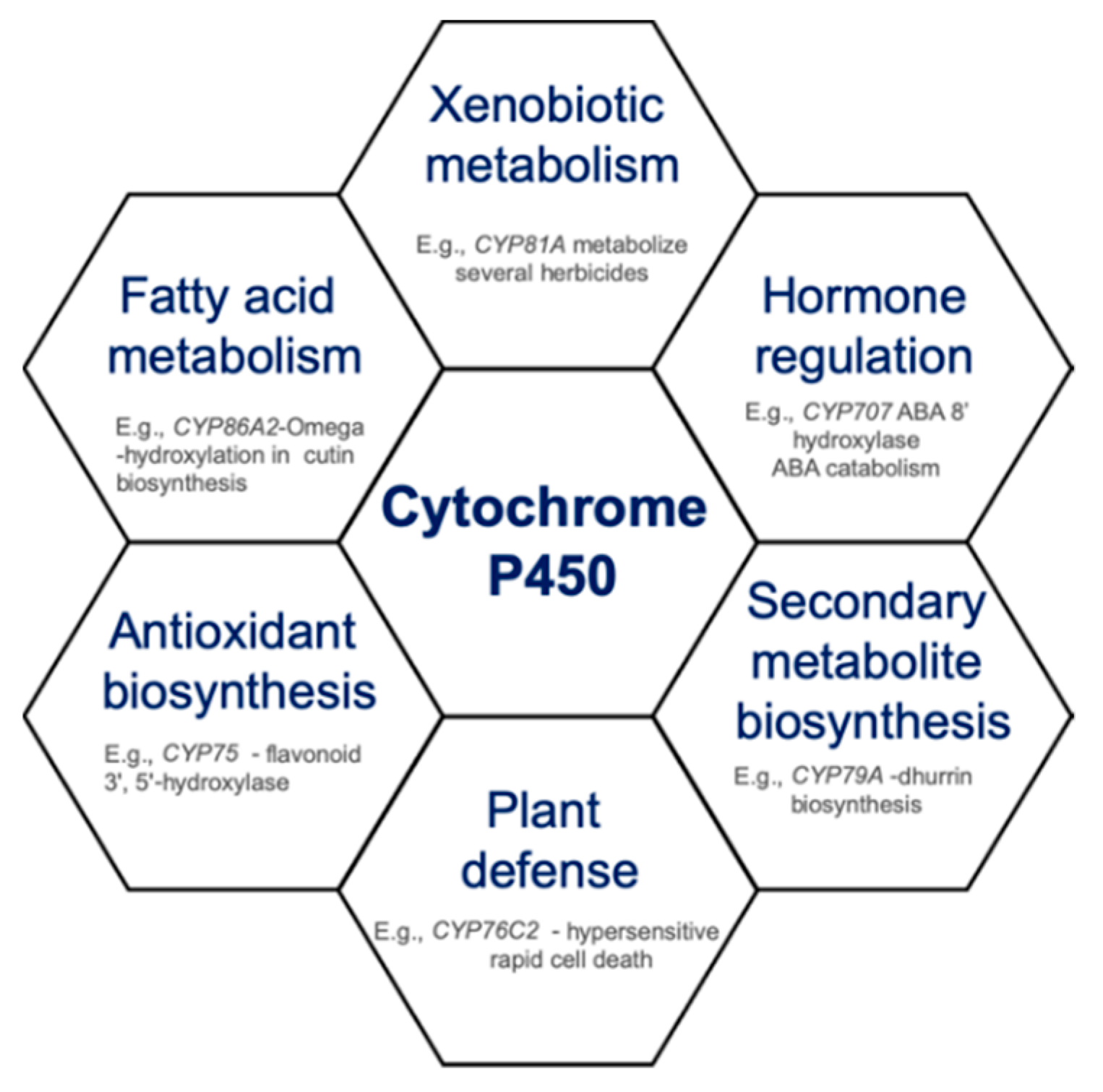
:1. Introduction
2. Classification & Catalysis of CYPs
2.1. Nomenclature and Basic Classification
2.2. Catalysis of CYP Enzymes
3. Role of CYPs in Abiotic and Biotic Stress
3.1. Drought Stress
3.2. Temperature Stress
3.3. Salinity Stress
3.4. Heavy Metal Stress
3.5. Herbicide Stress
3.6. Biotic Stress: Diseases, Insects, and Weeds
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mizutani, M.; Sato, F. Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 2011, 507, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, REVIEWS3003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Cui, X.; Lin, S.; Gan, S.; Xing, H.; Dou, D. GmCYP82A3, a soybean cytochrome p450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE 2016, 11, e0162253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, M.J.; Xu, Y.; Tang, X.; Huang, Y.; Liu, J.; Deng, X.; Xu, Q. CsCYT75B1, a citrus cytochrome P450 gene, is involved in accumulation of antioxidant flavonoids and induces drought tolerance in transgenic Arabidopsis. Antioxidants 2020, 9, 161. [Google Scholar] [CrossRef] [Green Version]
- Chapple, C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 311–343. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef] [Green Version]
- Mafu, S.; Ding, Y.; Murphy, K.M.; Yaacoobi, O.; Addison, J.B.; Wang, Q.; Shen, Z.; Briggs, S.P.; Bohlmann, J.; Castro-Falcon, G.; et al. Discovery, biosynthesis and stress-related accumulation of dolabradiene-derived defenses in maize. Plant Physiol. 2018, 176, 2677–2690. [Google Scholar] [CrossRef]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H.; et al. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Pinot, F.; Beisson, F. Cytochrome P450 metabolizing fatty acids in plants: Characterization and physiological roles. FEBS J. 2011, 278, 195–205. [Google Scholar] [CrossRef]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone Jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [Green Version]
- Qin, D.; Wu, H.; Peng, H.; Yao, Y.; Ni, Z.; Li, Z.; Zhou, C.; Sun, Q. Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat genome array. BMC Genom. 2008, 9, 432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Ma, L.; Li, Y.; Wang, S.; Li, L.; Yang, R. Transcriptome analysis reveals sunflower cytochrome P450 CYP93A1 responses to high salinity treatment at the seedling stage. Genes Genom. 2017, 39, 581–591. [Google Scholar] [CrossRef]
- Rai, A.; Singh, R.; Shirke, P.A.; Tripathi, R.D.; Trivedi, P.K.; Chakrabarty, D. Expression of rice CYP450-like gene (os08g01480) in arabidopsis modulates regulatory network leading to heavy metal and other abiotic stress tolerance. PLoS ONE 2015, 10, e0138574. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A. The role of cytochrome P450 monooxygenases in plant-insect interactions. Plant Physiol. 1996, 112, 1411–1419. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, J.B.; Song, B.; Li, H.P.; Xu, H.Q.; Qu, B.; Dang, F.J.; Liao, Y.C. Resistance to Fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene. Phytopathology 2010, 100, 183–191. [Google Scholar] [CrossRef] [Green Version]
- Paquette, S.M.; Jensen, K.; Bak, S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www.P450.kvl.dk). Phytochemistry 2009, 70, 1940–1947. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef]
- Nelson, D.R. The cytochrome P450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Ortiz de Montellano, P.R. (Ed.) Substrate oxidation by cytochrome P450 Enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2015; pp. 111–176. ISBN 9783319121086. [Google Scholar]
- Črešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2011, 1814, 29–35. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 and the individuality of species. Arch. Biochem. Biophys. 1999, 369, 1–10. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R. Plant cytochrome P450s from moss to poplar. Phytochem. Rev. 2006, 5, 193–204. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Carbone, I.; Dean, R.A. The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol. Biol. 2007, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caspi, R. Available online: https://iubmb.org/wp-content/uploads/sites/2790/2018/12/Classification-of-cytochrome-P-450-enzymes-by-the-Enzyme-Commission.pdf (accessed on 20 May 2020).
- Omura, T.; Sato, R. The carbon monoxide-binding pigment of liver microsomes. i. evidence for its hemoprotein nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar]
- Graham-Lorence, S.; Peterson, J.A.; Amarneh, B.; Simpson, E.R.; White, R.E. A three-dimensional model of aromatase cytochrome P450. Protein Sci. 1995, 4, 1065–1080. [Google Scholar] [CrossRef] [Green Version]
- Peterson, J.A.; Graham, S.E. A close family resemblance: The importance of structure in understanding cytochromes P450. Structure 1998, 6, 1079–1085. [Google Scholar] [CrossRef] [Green Version]
- Werck-Reichhart, D.; Hehn, A.; Didierjean, L. Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci. 2000, 5, 116–123. [Google Scholar] [CrossRef]
- Poulos, T.L. Cytochrome P450. Curr. Opin. Struct. Biol. 1995, 5, 767–774. [Google Scholar] [CrossRef]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2277. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of cytochrome P450-catalyzed oxidations. ACS Catal. 2018, 8, 10964–10976. [Google Scholar] [CrossRef]
- Guengerich, F.P. Cytochromes P450, drugs, and diseases. Mol. Interv. 2003, 3, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Bolwell, G.P.; Bozak, K.; Zimmerlin, A. Plant cytochrome P450. Phytochemistry 1994, 37, 1491–1506. [Google Scholar] [CrossRef]
- Hanukoglu, I. Electron transfer proteins of cytochrome P450 Systems. In Advances in Molecular and Cell Biology; Bittar, E.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1996; Volume 14, pp. 29–56. [Google Scholar]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Seo, M.; Koshiba, T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002, 7, 41–48. [Google Scholar] [CrossRef]
- Cutler, A.J.; Krochko, J.E. Formation and breakdown of ABA. Trends Plant Sci. 1999, 4, 472–478. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Huang, Y.; Xian, W.; Wang, J.; Liao, H. Identification and expression analysis of the Glycine max CYP707A gene family in response to drought and salt stresses. Ann. Bot. 2012, 110, 743–756. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wei, K. Comparative functional genomics analysis of cytochrome P450 gene superfamily in wheat and maize. BMC Plant Biol. 2020, 20, 93. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Lv, Y.; Wan, X.-R.; Li, L.-M.; Hu, B.; Li, L. Cloning and expression analysis of cDNAs encoding ABA 8′-hydroxylase in peanut plants in response to osmotic stress. PLoS ONE 2014, 9, e97025. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Song, Y.; Zhang, H.; Zhang, D. Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol. Biol. Rep. 2013, 31, 946–962. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.-C.; Xu, Y.-Q.; Li, G.-J.; Liao, Y.; Fu, F.-L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Goodwin, S.M.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.-M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.-Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef]
- Nelson, D.R.; Ming, R.; Alam, M.; Schuler, M.A. Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 2008, 1, 216–235. [Google Scholar] [CrossRef]
- Ahmad, P.; Jaleel, C.A.; Salem, M.A.; Nabi, G.; Sharma, S. Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 2010, 30, 161–175. [Google Scholar] [CrossRef]
- Johnson, S.M.; Lim, F.-L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 456. [Google Scholar] [CrossRef] [Green Version]
- Gorantla, M.; Babu, P.R.; Lachagari, V.B.R.; Reddy, A.M.M.; Wusirika, R.; Bennetzen, J.L.; Reddy, A.R. Identification of stress-responsive genes in an indica rice (Oryza sativa L.) using ESTs generated from drought-stressed seedlings. J. Exp. Bot. 2007, 58, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Degenkolbe, T.; Do, P.T.; Zuther, E.; Repsilber, D.; Walther, D.; Hincha, D.K.; Köhl, K.I. Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Mol. Biol. 2009, 69, 133–153. [Google Scholar] [CrossRef] [Green Version]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Hemmati, H.; Gupta, D.; Basu, C. Molecular physiology of heat stress responses in plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Pandey, G.K., Ed.; Springer New York: New York, NY, USA, 2015; Volume 2, pp. 109–142. ISBN 9781493925407. [Google Scholar]
- Zaidi, N.W.; Dar, M.H.; Singh, S.; Singh, U.S. Trichoderma species as abiotic stress relievers in plants. In Biotechnology and Biology of Trichoderma; Gupta, V.K., Schmoll, M., Herrera-Estrella, A., Upadhyay, R.S., Druzhinina, I., Tuohy, M.G., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; Chapter 38; pp. 515–525. ISBN 9780444595768. [Google Scholar]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Wang, M.-X.; Dai, Y.; Wang, Y.; Fan, Y.-F.; Mao, P.; Ma, X.-R. Identification and expression profile of CYPome in perennial ryegrass and tall fescue in response to temperature stress. Front. Plant Sci. 2017, 8, 1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilodeau, P.; Udvardi, M.K.; Peacock, W.J.; Dennis, E.S. A prolonged cold treatment-induced cytochrome P450 gene from Arabidopsis thaliana. Plant Cell Environ. 1999, 22, 791–800. [Google Scholar] [CrossRef]
- Baron, K.N.; Schroeder, D.F.; Stasolla, C. Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci. 2012, 188–189, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Chan, Z. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 2012, 100, 110–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-F.; Wang, Y.; Tang, Y.; Kakani, V.G.; Mahalingam, R. Transcriptome analysis of heat stress response in switchgrass (Panicum virgatum L.). BMC Plant Biol. 2013, 13, 153. [Google Scholar] [CrossRef] [Green Version]
- Obaid, A.Y.; Sabir, J.S.M.; Atef, A.; Liu, X.; Edris, S.; El-Domyati, F.M.; Mutwakil, M.Z.; Gadalla, N.O.; Hajrah, N.H.; Al-Kordy, M.A.; et al. Analysis of transcriptional response to heat stress in Rhazya stricta. BMC Plant Biol. 2016, 16, 252. [Google Scholar] [CrossRef] [Green Version]
- Rahaman, M.; Mamidi, S.; Rahman, M. Genome-wide association study of heat stress-tolerance traits in spring-type Brassica napus L. under controlled conditions. Crop J. 2018, 6, 115–125. [Google Scholar] [CrossRef]
- Wedow, J.M.; Yendrek, C.R.; Mello, T.R.; Creste, S.; Martinez, C.A.; Ainsworth, E.A. Metabolite and transcript profiling of guinea grass (Panicum maximum Jacq) response to elevated [CO2] and temperature. Metabolomics 2019, 15, 51. [Google Scholar] [CrossRef] [Green Version]
- Chopra, R.; Burow, G.; Hayes, C.; Emendack, Y.; Xin, Z.; Burke, J. Transcriptome profiling and validation of gene based single nucleotide polymorphisms (SNPs) in sorghum genotypes with contrasting responses to cold stress. BMC Genom. 2015, 16, 1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sairam, R.K.; Rao, K.V.; Srivastava, G.C. Differential response of wheat genotypes to long term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci. 2002, 163, 1037–1046. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.-Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Yuan, J.; Qin, L.; Shi, W.; Xia, G.; Liu, S. TaCYP81D5, one member in a wheat cytochrome P450 gene cluster, confers salinity tolerance via reactive oxygen species scavenging. Plant Biotechnol. J. 2020, 18, 791–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruah, A.; Simková, K.; Apel, K.; Laloi, C. Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 2009, 70, 547–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javid, M.G.; Sorooshzadeh, A.; Moradi, F.; Modarres Sanavy, S.A.M.; Allahdadi, I. The role of phytohormones in alleviating salt stress in crop plants. Aust. J. Crop Sci. 2011, 5, 726. [Google Scholar]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Peng, M.; Luo, Q.; Jiang, M.; Zhang, X.; Zong, X.; Meng, F.; Li, Y. Isolation and detection of transcript-derived fragments (TDFs) in NaCl-stressed black locust (Robinia pseudoacacia L.) using cDNA-AFLP analysis. Acta Physiol. Plant 2015, 37, 168. [Google Scholar] [CrossRef]
- Wang, X.; Yang, P.; Gao, Q.; Liu, X.; Kuang, T.; Shen, S.; He, Y. Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 2008, 228, 167–177. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals—Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Guo, P.; Bai, G.; Carver, B.; Li, R.; Bernardo, A.; Baum, M. Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum tolerance under aluminum stress. Mol. Genet. Genom. 2007, 277, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yaxley, J.R.; Ross, J.J.; Sherriff, L.J.; Reid, J.B. Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 2001, 125, 627–633. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, S.B.; Sutter, T.R. Microarray analysis of Arabidopsis genome response to aluminum stress. Biol. Plant. 2009, 53, 85–99. [Google Scholar] [CrossRef]
- Ogawa, I.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Time course analysis of gene regulation under cadmium stress in rice. Plant Soil 2009, 325, 97. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, J.; Zhou, Y.; Gong, T.; Wang, J.; Ge, Y. Enhanced phytoremediation of mixed heavy metal (mercury)--organic pollutants (trichloroethylene) with transgenic alfalfa co-expressing glutathione S-transferase and human P450 2E1. J. Hazard. Mater. 2013, 260, 1100–1107. [Google Scholar] [CrossRef]
- Seitz, C.; Eder, C.; Deiml, B.; Kellner, S.; Martens, S.; Forkmann, G. Cloning, functional identification and sequence analysis of flavonoid 3′-hydroxylase and flavonoid 3′, 5′-hydroxylase cDNAs reveals independent evolution of flavonoid 3′, 5′-hydroxylase in the Asteraceae family. Plant Mol. Biol. 2006, 61, 365–381. [Google Scholar] [CrossRef]
- Siminszky, B. Plant cytochrome P450-mediated herbicide metabolism. Phytochem. Rev. 2006, 5, 445–458. [Google Scholar] [CrossRef]
- Liu, X.; Bi, B.; Xu, X.; Li, B.; Tian, S.; Wang, J.; Zhang, H.; Wang, G.; Han, Y.; McElroy, J.S. Rapid identification of a candidate nicosulfuron sensitivity gene (Nss) in maize (Zea mays L.) via combining bulked segregant analysis and RNA-seq. Theor. Appl. Genet. 2019, 132, 1351–1361. [Google Scholar] [CrossRef]
- Pan, G.; Zhang, X.; Liu, K.; Zhang, J.; Wu, X.; Zhu, J.; Tu, J. Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol. Biol. 2006, 61, 933–943. [Google Scholar] [CrossRef]
- Xiang, W.; Wang, X.; Ren, T. Expression of a wheat cytochrome P450 monooxygenase cDNA in yeast catalyzes the metabolism of sulfonylurea herbicides. Pestic. Biochem. Physiol. 2006, 85, 1–6. [Google Scholar] [CrossRef]
- Hamprecht, G.; Witschel, M.; Hawkes, T.R.; Edmunds, A.J.F.; Morris, J.A.; van Almsick, A. Herbicides with Bleaching Properties. In Modern Crop Protection Compounds; ACS Symposium Series; Krämer, W., Schirmer, U., Jeschke, P., Witschel, M., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2011; Volume 58, pp. 197–276. ISBN 9783527644179. [Google Scholar]
- Mitchell, G.; Bartlett, D.W.; Fraser, T.E.; Hawkes, T.R.; Holt, D.C.; Townson, J.K.; Wichert, R.A. Mesotrione: A new selective herbicide for use in maize. Pest Manag. Sci. 2001, 57, 120–128. [Google Scholar] [CrossRef]
- Paporisch, A.; Rubin, B. Isoxadifen safening mechanism in sweet corn genotypes with differential response to P450-metabolized herbicides. Pestic. Biochem. Physiol. 2017, 138, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Nordby, J.N.; Williams, M.M.; Pataky, J.K.; Riechers, D.E.; Lutz, J.D. A Common Genetic Basis in Sweet Corn Inbred Cr1 for Cross Sensitivity to Multiple Cytochrome P450-Metabolized Herbicides. Weed Sci. 2008, 56, 376–382. [Google Scholar] [CrossRef]
- Williams, M.M.; Pataky, J.K. factors affecting differential sensitivity of sweet corn to HPPD-lnhibiting herbicides. Weed Sci. 2010, 58, 289–294. [Google Scholar] [CrossRef]
- Jugulam, M.; Shyam, C. Non-target-site resistance to herbicides: Recent developments. Plants 2019, 8, 417. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [Green Version]
- Blée, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-C.; Kim, S.-Y.; Paek, K.-H.; Choi, D.; Park, J.M. Suppression of CaCYP1, a novel cytochrome P450 gene, compromises the basal pathogen defense response of pepper plants. Biochem. Biophys. Res. Commun. 2006, 345, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Godiard, L.; Sauviac, L.; Dalbin, N.; Liaubet, L.; Callard, D.; Czernic, P.; Marco, Y. CYP76C2, an Arabidopsis thaliana cytochrome P450 gene expressed during hypersensitive and developmental cell death. FEBS Lett. 1998, 438, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Anderson, J.M.; Ohm, H.W. Induction of wheat defense and stress-related genes in response to Fusarium graminearum. Genome 2005, 48, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Rajniak, J.; Barco, B.; Clay, N.K.; Sattely, E.S. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 2015, 525, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Schuhegger, R.; Nafisi, M.; Mansourova, M.; Petersen, B.L.; Olsen, C.E.; Svatos, A.; Halkier, B.A.; Glawischnig, E. CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 2006, 141, 1248–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teutsch, H.G.; Hasenfratz, M.P.; Lesot, A.; Stoltz, C.; Garnier, J.-M.; Jeltsch, J.-M.; Durst, F.; Werck-Reichhart, D. Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate 4-hydroxylase, a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proc. Natl. Acad. Sci. USA 1993, 90, 4102–4106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, M.R.; Deyneka, J.M.; Schuler, M.A. Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol. 1996, 110, 1035–1046. [Google Scholar] [CrossRef] [Green Version]
- Persans, M.W.; Wang, J.; Schuler, M.A. Characterization of maize cytochrome P450 monooxygenases induced in response to safeners and bacterial pathogens. Plant Physiol. 2001, 125, 1126–1138. [Google Scholar] [CrossRef] [Green Version]
- Prince, D.C.; Drurey, C.; Zipfel, C.; Hogenhout, S.A. The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 2014, 164, 2207–2219. [Google Scholar] [CrossRef] [Green Version]
- Irmisch, S.; Zeltner, P.; Handrick, V.; Gershenzon, J.; Köllner, T.G. The maize cytochrome P450 CYP79A61 produces phenylacetaldoxime and indole-3-acetaldoxime in heterologous systems and might contribute to plant defense and auxin formation. BMC Plant Biol. 2015, 15, 128. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Wang, R.; DeParasis, J.; Loughrin, J.H.; Gan, S.; Wagner, G.J. Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat. Biotechnol. 2001, 19, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Spitters, C.J.T.; Van Den Bergh, J.P. Competition between crop and weeds: A system approach. In Biology and Ecology of Weeds; Holzner, W., Numata, M., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 1982; pp. 137–148. ISBN 9789401709163. [Google Scholar]
- Jabran, K.; Mahajan, G.; Sardana, V.; Chauhan, B.S. Allelopathy for weed control in agricultural systems. Crop Prot. 2015, 72, 57–65. [Google Scholar] [CrossRef]
- Netzly, D.H.; Butler, L.G. Roots of sorghum exude hydrophobic droplets containing biologically active components 1. Crop Sci. 1986, 26, 775–778. [Google Scholar] [CrossRef] [Green Version]
- Burgos, N.R.; Talbert, R.E. Weed control and sweet corn (Zea mays var. rugosa) response in a no-till system with cover crops. Weed Sci. 1996, 44, 355–361. [Google Scholar] [CrossRef]
- Czarnota, M.A.; Paul, R.N.; Dayan, F.E.; Nimbal, C.I.; Weston, L.A. Mode of action, localization of production, chemical nature, and activity of sorgoleone: A potent PSII inhibitor in sorghum spp. root exudates. Weed Technol. 2001, 15, 813–825. [Google Scholar] [CrossRef]
- Rehman, A.; Cheema, Z.A.; Khaliq, A.; Arshad, M.; Mohsan, S. Application of sorghum, sunflower and rice water extract combinations helps in reducing herbicide dose for weed management in rice. Int. J. Agric. Biol. 2010, 12, 901–906. [Google Scholar]
- Alsaadawi, I.S.; Dayan, F.E. Potentials and prospects of sorghum allelopathy in agroecosystems. Allelopath. J. 2009, 24, 255. [Google Scholar]
- Pan, Z.; Baerson, S.R.; Wang, M.; Bajsa-Hirschel, J.; Rimando, A.M.; Wang, X.; Nanayakkara, N.P.D.; Noonan, B.P.; Fromm, M.E.; Dayan, F.E.; et al. A cytochrome P450 CYP71 enzyme expressed in Sorghum bicolor root hair cells participates in the biosynthesis of the benzoquinone allelochemical sorgoleone. New Phytol. 2018, 218, 616–629. [Google Scholar] [CrossRef] [Green Version]
- Butrón, A.; Chen, Y.C.; Rottinghaus, G.E.; McMullen, M.D. Genetic variation at bx1 controls DIMBOA content in maize. Theor. Appl. Genet. 2010, 120, 721–734. [Google Scholar] [CrossRef] [Green Version]
- Lam, Y.; Sze, C.W.; Tong, Y.; Ng, T.B.; Tang, S.C.W.; Ho, J.C.M.; Xiang, Q.; Lin, X.; Zhang, Y. Research on the allelopathic potential of wheat. Agric. Sci. 2012, 3, 25455. [Google Scholar] [CrossRef] [Green Version]
- Kato-Noguchi, H.; Sakata, Y.; Takenokuchi, K.; Kosemura, S.; Yamamura, S. Allelopathy in Maize I.: Isolation and identification of allelochemicals in maize seedlings. Plant Prod. Sci. 2000, 3, 43–46. [Google Scholar] [CrossRef]
- Schulz, M.; Marocco, A.; Tabaglio, V.; Macias, F.A.; Molinillo, J.M.G. Benzoxazinoids in rye allelopathy—From discovery to application in sustainable weed control and organic farming. J. Chem. Ecol. 2013, 39, 154–174. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Ishihara, A.; Imaishi, H.; Endo, T.R.; Ohkawa, H.; Iwamura, H. Molecular characterization and chromosomal localization of cytochrome P450 genes involved in the biosynthesis of cyclic hydroxamic acids in hexaploid wheat. Mol. Genet. Genom. 2002, 267, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.; Rattei, T.; Haslbeck, M.; Schwab, W.; Gierl, A.; Frey, M. Comparative analysis of benzoxazinoid biosynthesis in monocots and dicots: Independent recruitment of stabilization and activation functions. Plant Cell 2012, 24, 915–928. [Google Scholar] [CrossRef] [Green Version]
- Jun, X.U.; Wang, X.; Guo, W. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar]
- Feldmann, K.A. Cytochrome P450s as genes for crop improvement. Curr. Opin. Plant Biol. 2001, 4, 162–167. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Tian, L.; Musetti, V.; Kim, J.; Magallanes-Lundback, M.; DellaPenna, D. The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 2004, 101, 402–407. [Google Scholar] [CrossRef] [Green Version]
- Morant, M.; Jørgensen, K.; Schaller, H.; Pinot, F.; Møller, B.L.; Werck-Reichhart, D.; Bak, S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 2007, 19, 1473–1487. [Google Scholar] [CrossRef] [Green Version]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Bak, S.; Olsen, C.E.; Halkier, B.A.; Møller, B.L. Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in dhurrin biosynthesis. Plant Physiol. 2000, 123, 1437–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irmler, S.; Schröder, G.; St-Pierre, B.; Crouch, N.P.; Hotze, M.; Schmidt, J.; Strack, D.; Matern, U.; Schröder, J. Indole alkaloid biosynthesis in Catharanthus roseus: New enzyme activities and identification of cytochrome P450 CYP72A1 as secologanin synthase. Plant J. 2000, 24, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid ω-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| CYP | Identified Species | Biochemical Process | Function | Desirable Trait | Reference |
|---|---|---|---|---|---|
| CYP97C1 | Arabidopsis | Carotenoid ε-ring hydroxylation | Lutein biosynthesis | Abiotic stress resistance | [125] |
| CYP703A2 | Arabidopsis | Hydroxylation of lauric acid | Pollen development | Abiotic stress resistance | [126] |
| CYP83A1 and CYP83B1 | Arabidopsis | Biosynthesis of glucosinolates | Pungency | Insect resistance | [127] |
| CYP79A1 and CYP71E1 | Sorghum | Tyrosine into p-Hydroxymandelonitril | Cyanogenic glucoside (dhurrin) biosynthesis | Insect resistance | [128] |
| CYP72A1 | Catharanthus roseus | Secologanin synthase | Indole alkaloid biosynthesis | Disease resistance | [129] |
| CYP707A | Arabidopsis | ABA 8’-hydroxylases | ABA regulation | Abiotic stress resistance | [38] |
| CYP86A2, A8 | Arabidopsis | Omega-hydroxylation | Cutin biosynthesis | Insect resistance | [46] |
| CYP714A3 | Rice | Gibberellin regulation | Shoot development | Heavy metal stress | [76] |
| CYP88A | Wheat | Gibberellin biosynthesis | |||
| CYP86A1 | Arabidopsis | Omega-hydroxylase | Suberin biosynthesis | Insect resistance | [130] |
| CYP71C | Maize | DIBOA biosynthesis | Allelopathy | Biotic stress resistance | [118] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. https://doi.org/10.3390/antiox9050454
Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PVV, Jugulam M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants. 2020; 9(5):454. https://doi.org/10.3390/antiox9050454
Chicago/Turabian StylePandian, Balaji Aravindhan, Rajendran Sathishraj, Maduraimuthu Djanaguiraman, P.V. Vara Prasad, and Mithila Jugulam. 2020. "Role of Cytochrome P450 Enzymes in Plant Stress Response" Antioxidants 9, no. 5: 454. https://doi.org/10.3390/antiox9050454
APA StylePandian, B. A., Sathishraj, R., Djanaguiraman, M., Prasad, P. V. V., & Jugulam, M. (2020). Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants, 9(5), 454. https://doi.org/10.3390/antiox9050454









