The KEAP1/NRF2 Signaling Pathway in Keratinization
Abstract
1. Introduction and Overview
Keratinization as an Environmental Response: Beneficial or Detrimental
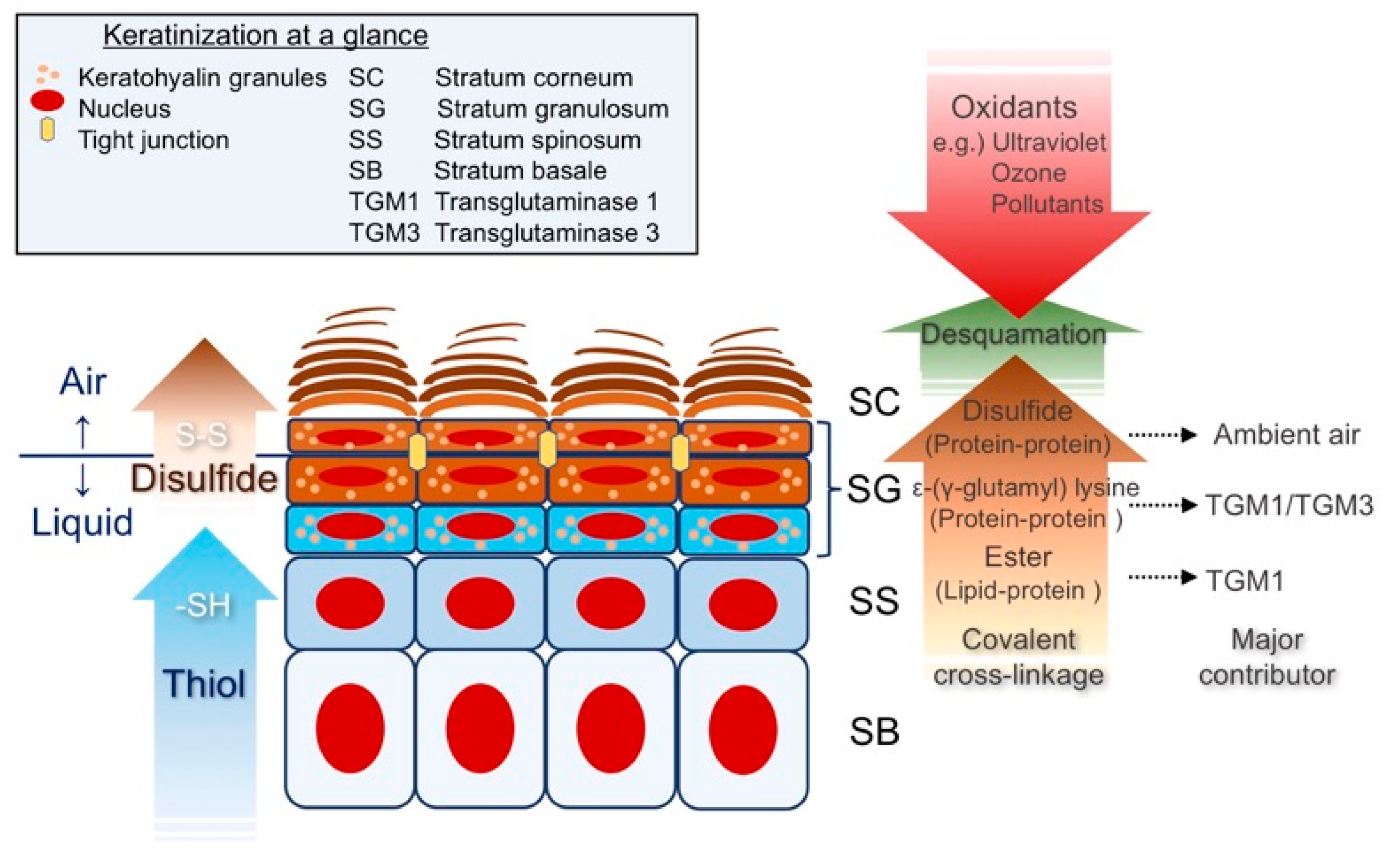
2. Keratinization at a Glance
Epidermal Differentiation; a Search for the Thiol-Rich Protein
3. The KEAP1/NRF2 Signaling Pathway and the EDC
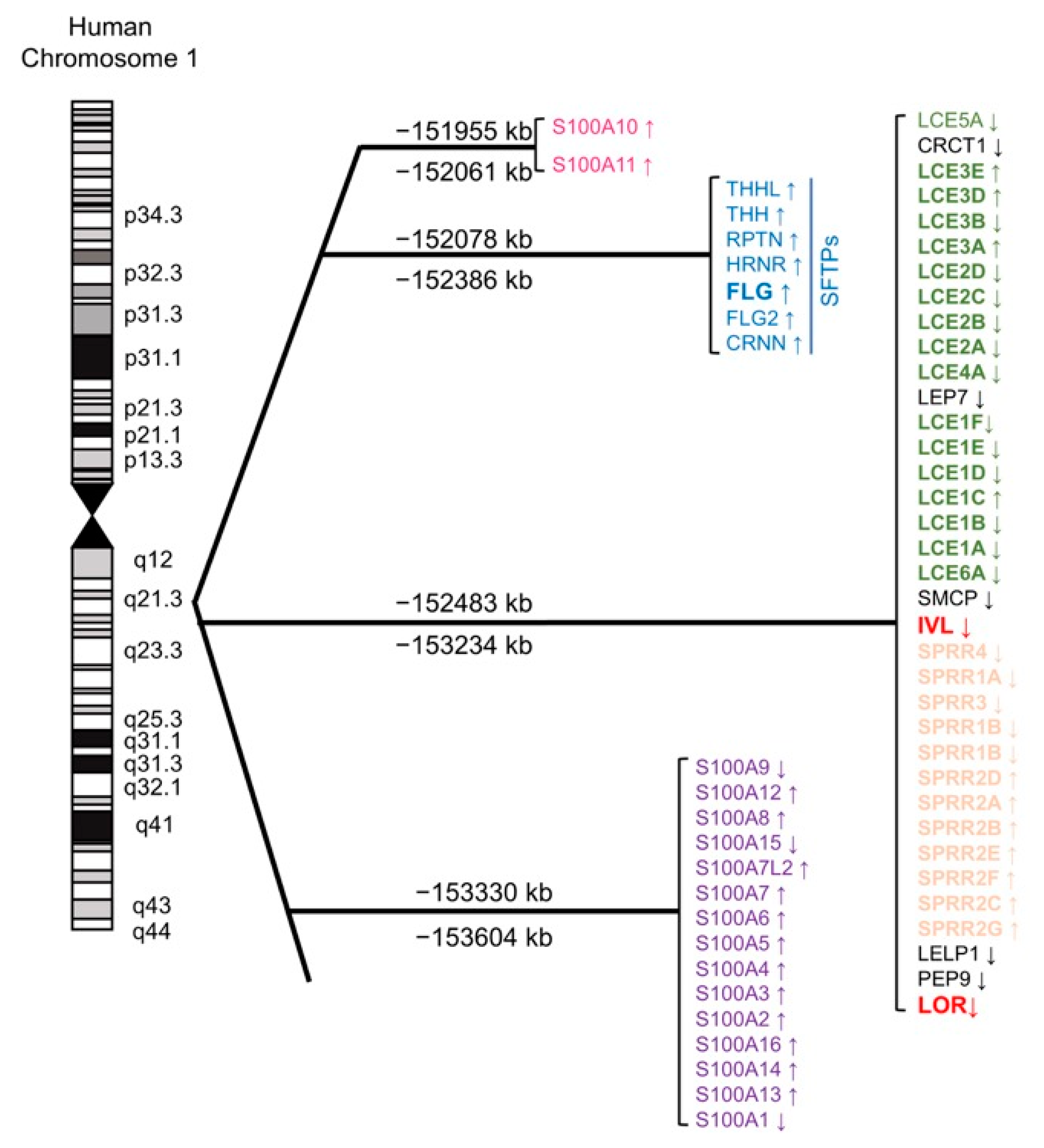
3.1. The Evolution of the EDC Gene Cluster; Analogy to the β-globin Gene Cluster
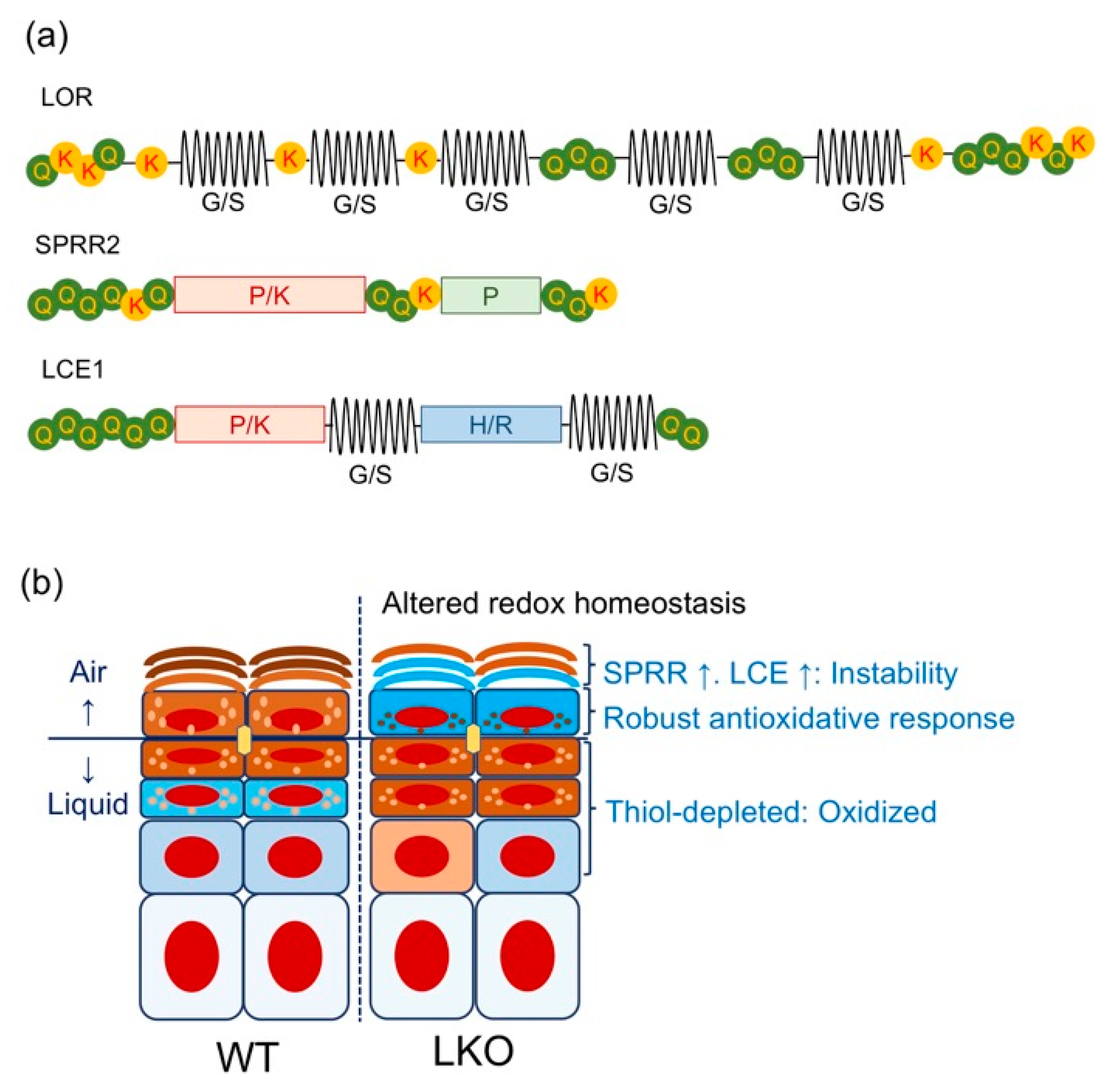
3.2. Lessons from Lor-Deficient Mice: Quasi-Normalcy is not Synonymous with Normalcy
4. The KEAP1/NRF2 Signaling Pathway and Epidermal Homeostasis
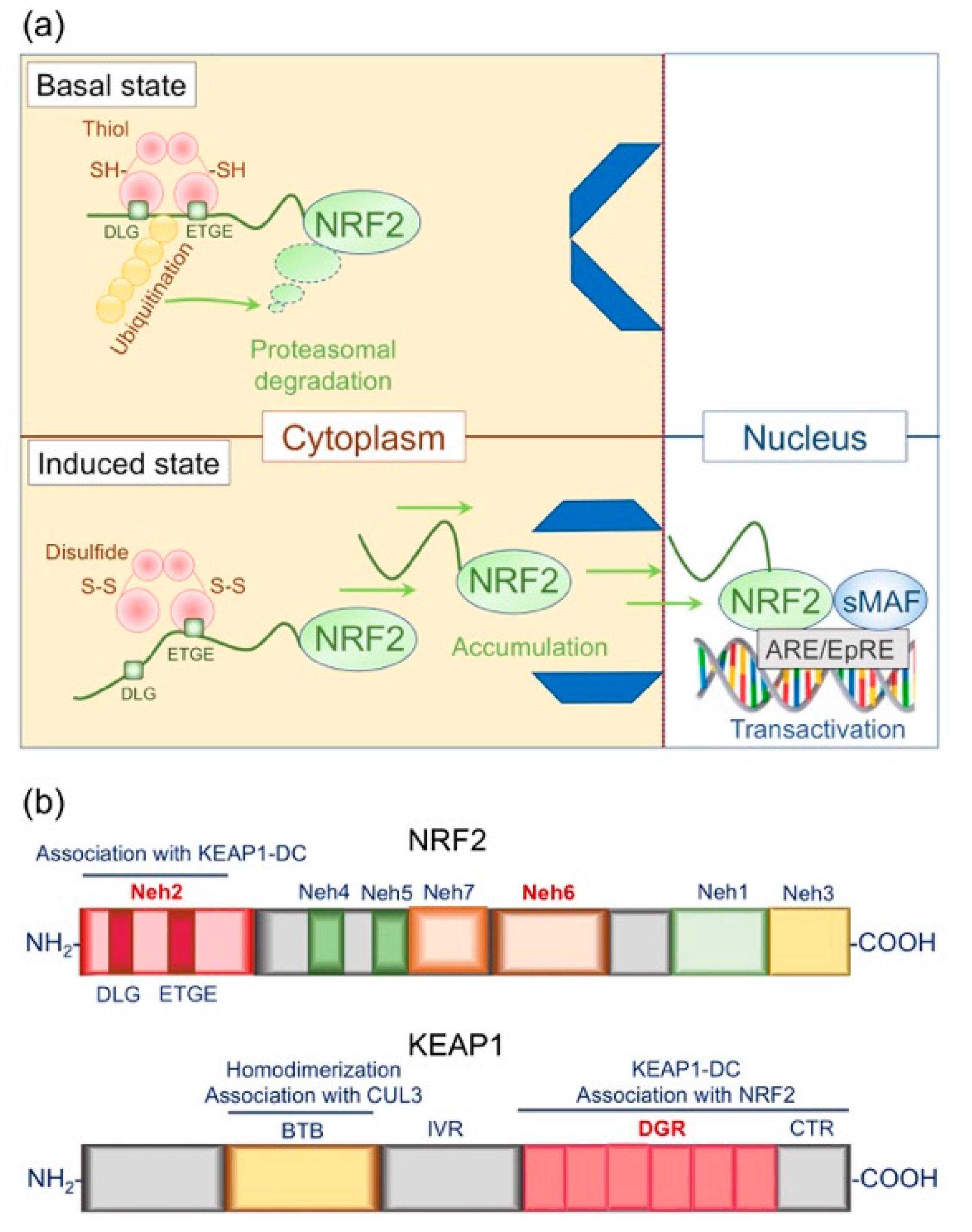
4.1. Structural and Functional Basis of the KEAP1/NRF2 System: Form Follows Function
4.1.1. Structural Basis of NRF2
4.1.2. Structural Basis of KEAP1
4.1.3. Functional Basis of the KEAP1/NRF2 Signaling Pathway: Principles of Action
4.1.4. What Makes the KEAP1/NRF2 Signaling Pathway Sensitive and Efficient?
4.2. Nrf2 and Epidermal Barrier Function: Maintaining Epidermal Redox Balance
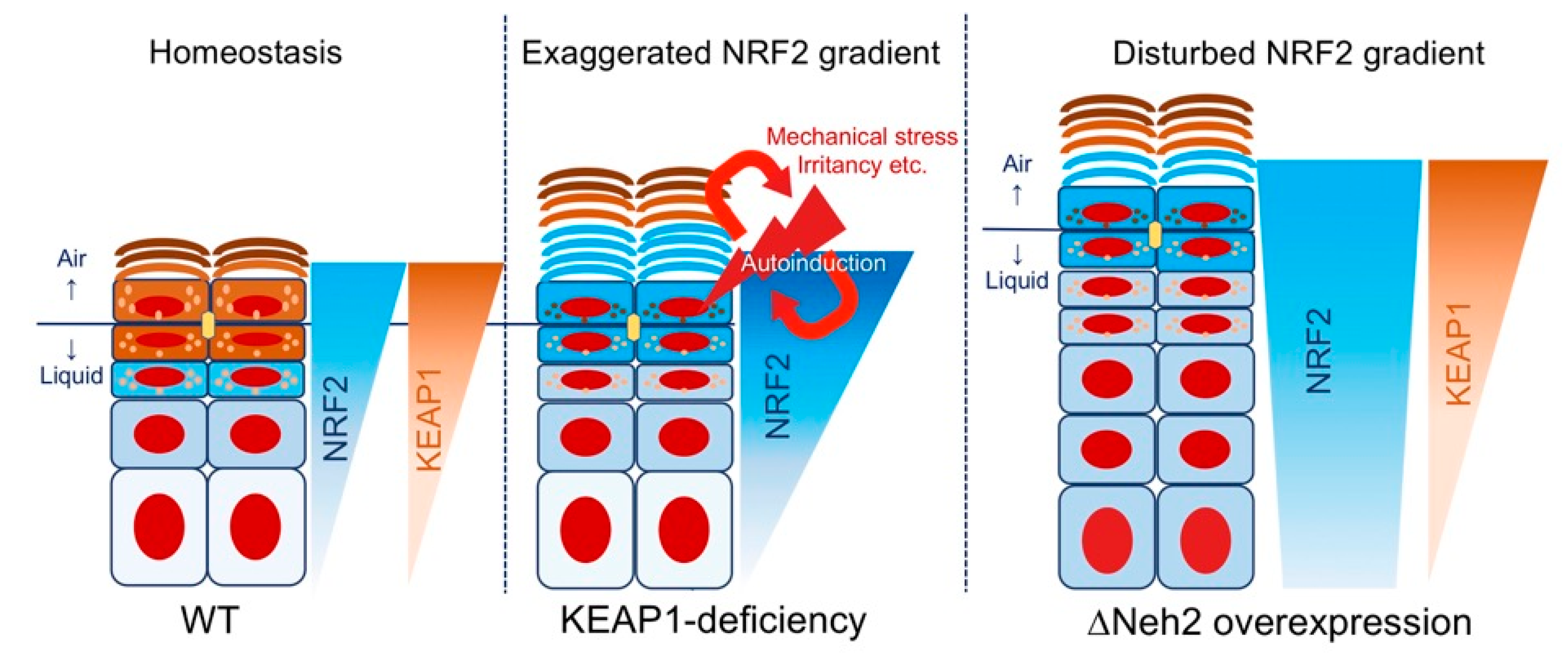
4.2.1. Keap1-Deficient Mice: the Epidermal “Keaper” and Striker
4.2.2. Neh2-Nrf2 Mice: A Model of Gene-Environment Mismatch?
4.2.3. Summary: Why are Keap1 and Neh2 Deficiencies Phenotypically Different?
4.3. Epidermal Barrier Repair in LKO Mice
4.4. NRF2 Activation and Epidermal Carcinogenesis: Friend or Foe?
5. Concluding Remarks
Funding
Conflicts of Interest
Abbreviations
| AD | atopic dermatitis |
| ARCI | autosomal recessive congenital ichthyosis |
| ARE | antioxidant response element |
| ARE | antioxidant responsive element |
| BTB | broad-complex, tramtrack, and bric-à-brac |
| bZIP | basic leucine zipper |
| CE | cornified cell envelope |
| CMV | cytomegalovirus |
| CNC | cap ´n´ collar |
| CTR | C-terminal region |
| CUL3 | cullin 3 |
| CYP1A1 | cytochrome P450 family 1 subfamily A member 1 |
| DGR | double glycine repeat |
| DMBA | 7,12- dimethylbenz(a)anthracene |
| DMBADE | DMBA-3,4-diol-1,2-epoxide |
| ΔNeh2-Nrf2 | Nrf2 that lacks KEAP1-interacting domain |
| dnNrf2 | dominant-negative Nrf2 |
| E | embryonic day |
| EDC | epidermal differentiation complex |
| EpRE | electrophile response element |
| FLG | filaggrin |
| GATA1 | GATA protein binding protein 1 |
| HIF-1α | hypoxia inducible factor 1 alpha |
| IκBα | NF-κB inhibitor alpha |
| IVL | involucrin |
| KEAP1 | Kelch-like erythroid cell-derived protein with cap´n´collar homology-associated protein 1 |
| KEAP1-DC | KEAP1 double glycine repeat and the C-terminal region |
| LCE | late cornified envelope |
| LG | lamellar granules |
| LKO | Lor-knockout (LKO) |
| LOR | loricrin |
| MADISH | metabolizing acquired dioxin-induced skin hamartomas |
| Neh | Nrf2-erythroid cell-derived protein with CNC homology |
| NF-κB | nuclear factor κB |
| NFE2 | nuclear factor erythroid 2 |
| NQO1 | NAD(P)H quinone dehydrogenase 1 |
| NRF2 | NFE2-related factor 2 |
| PRP | proline-rich proteins |
| S100 | S100 calcium-binding proteins |
| SC | stratum corneum |
| SCC | squamous cell carcinoma |
| SCF | complex S-phase kinase associated protein 1-cullin 1-ring-box 1 |
| SG | stratum granulosum |
| SPRR | small proline-rich protein |
| SQSTM1 | sequestosome 1 |
| TCDD | 2,3,7,8-tetrachlorodibenzo-p-dioxin |
| TF | transcription factor |
| TP53 | tumor protein p53 |
| UV | ultraviolet |
References
- Marks, R.; Plewig, G. Stratum Corneum; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Nemes, Z.; Steinert, P.M. Bricks and mortar of the epidermal barrier. Exp. Mol. Med. 1999, 31, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, T.; Akiyama, M. Inherited ichthyosis: Non-syndromic forms. J. Dermatol. 2016, 43, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Zhuang, D.Z.; Han, R.; Isaac, G.; Tobin, J.J.; McKee, M.; Welti, R.; Brissette, J.L.; Fitzgerald, M.L.; Freeman, M.W. ABCA12 maintains the epidermal lipid permeability barrier by facilitating formation of ceramide linoleic esters. J. Biol. Chem. 2008, 283, 36624–36635. [Google Scholar] [CrossRef]
- Yanagi, T.; Akiyama, M.; Nishihara, H.; Ishikawa, J.; Sakai, K.; Miyamura, Y.; Naoe, A.; Kitahara, T.; Tanaka, S.; Shimizu, H. Self-improvement of keratinocyte differentiation defects during skin maturation in ABCA12-deficient harlequin ichthyosis model mice. Am. J. Pathol. 2010, 177, 106–118. [Google Scholar] [CrossRef]
- Kuramoto, N.; Takizawa, T.; Takizawa, T.; Matsuki, M.; Morioka, H.; Robinson, J.M.; Yamanishi, K. Development of ichthyosiform skin compensates for defective permeability barrier function in mice lacking transglutaminase 1. J. Clin. Investig. 2002, 109, 243–250. [Google Scholar] [CrossRef]
- Palmer, C.N.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef]
- Paller, A.S.; Spergel, J.M.; Mina-Osorio, P.; Irvine, A.D. The atopic march and atopic multimorbidity: Many trajectories, many pathways. J. Allergy Clin. Immunol. 2019, 143, 46–55. [Google Scholar] [CrossRef]
- Lieberman, D. The Story of the Human Body: Evolution, Health, and Disease; Vintage: Dewey, WA, USA, 2014. [Google Scholar]
- Jones, C.L.; Reiners, J.J., Jr. Differentiation status of cultured murine keratinocytes modulates induction of genes responsive to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Arch. Biochem. Biophys. 1997, 347, 163–173. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Itoh, K.; Wakabayashi, J.; Motohashi, H.; Noda, S.; Takahashi, S.; Imakado, S.; Kotsuji, T.; Otsuka, F.; Roop, D.R.; et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003, 35, 238–245. [Google Scholar] [CrossRef]
- Magin, T.M. A keaper and a striker maintain epidermal homeostasis. Nat. Genet. 2003, 35, 202–204. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, Y.; Huebner, A.J.; Rice, R.H.; Koch, P.J.; Speransky, V.V.; Steven, A.C.; Roop, D.R. Lce1 Family Members Are Nrf2-Target Genes that Are Induced to Compensate for the Loss of Loricrin. J. Investig. Dermatol. 2016, 136, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Huebner, A.J.; Dai, D.; Morasso, M.; Schmidt, E.E.; Schafer, M.; Werner, S.; Roop, D.R. Amniotic fluid activates the nrf2/keap1 pathway to repair an epidermal barrier defect in utero. Dev. Cell 2012, 23, 1238–1246. [Google Scholar] [CrossRef] [PubMed]
- Rolfs, F.; Huber, M.; Kuehne, A.; Kramer, S.; Haertel, E.; Muzumdar, S.; Wagner, J.; Tanner, Y.; Bohm, F.; Smola, S.; et al. Nrf2 Activation Promotes Keratinocyte Survival during Early Skin Carcinogenesis via Metabolic Alterations. Cancer Res. 2015, 75, 4817–4829. [Google Scholar] [CrossRef] [PubMed]
- Schafer, M.; Farwanah, H.; Willrodt, A.H.; Huebner, A.J.; Sandhoff, K.; Roop, D.; Hohl, D.; Bloch, W.; Werner, S. Nrf2 links epidermal barrier function with antioxidant defense. EMBO Mol. Med. 2012, 4, 364–379. [Google Scholar] [CrossRef] [PubMed]
- Schafer, M.; Willrodt, A.H.; Kurinna, S.; Link, A.S.; Farwanah, H.; Geusau, A.; Gruber, F.; Sorg, O.; Huebner, A.J.; Roop, D.R.; et al. Activation of Nrf2 in keratinocytes causes chloracne (MADISH)-like skin disease in mice. EMBO Mol. Med. 2014, 6, 442–457. [Google Scholar] [CrossRef]
- Van Scott, E.; Flesch, P. Sulfhydryl and disulfide in keratinization. Science 1954, 119, 70–71. [Google Scholar] [CrossRef]
- Ishitsuka, Y.; Roop, D.R. Loricrin: Past, Present, and Future. Int. J. Mol. Sci. 2020, 21, 2271. [Google Scholar] [CrossRef]
- Rice, R.H.; Green, H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell 1977, 11, 417–422. [Google Scholar] [CrossRef]
- Rice, R.H.; Green, H. Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: Activation of the cross-linking by calcium ions. Cell 1979, 18, 681–694. [Google Scholar] [CrossRef]
- Bern, H.A.; Harkness, D.R.; Blair, S.M. Radioautographic studies of keratin formation. Proc. Natl. Acad. Sci. USA 1955, 41, 55. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, K.; Epstein, W.L. Sulfur-containing proteins and epidermal keratinization. J. Cell Biol. 1969, 40, 830. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, T.; Hirai, R. The synthesis of the cystine-rich proteins in rat epidermis: I. Analysis by [35S]cystine incorporation. Curr. Probl. Dermatol. 1980, 10, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, T.; Takahashi, M. The cystine-rich envelope protein from human epidermal stratum corneum cells. J. Investig. Dermatol. 1987, 88, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Mehrel, T.; Hohl, D.; Rothnagel, J.A.; Longley, M.A.; Bundman, D.; Cheng, C.; Lichti, U.; Bisher, M.E.; Steven, A.C.; Steinert, P.M.; et al. Identification of a major keratinocyte cell envelope protein, loricrin. Cell 1990, 61, 1103–1112. [Google Scholar] [CrossRef]
- Matsui, T.; Amagai, M. Dissecting the formation, structure and barrier function of the stratum corneum. Int. Immunol. 2015, 27, 269–280. [Google Scholar] [CrossRef]
- Lavker, R.M.; Matoltsy, A.G. Formation of horny cells: The fate of cell organelles and differentiation products in ruminal epithelium. J. Cell Biol. 1970, 44, 501–512. [Google Scholar] [CrossRef]
- Nemes, Z.; Marekov, L.N.; Fesus, L.; Steinert, P.M. A novel function for transglutaminase 1: Attachment of long-chain omega-hydroxyceramides to involucrin by ester bond formation. Proc. Natl. Acad. Sci. USA 1999, 96, 8402–8407. [Google Scholar] [CrossRef]
- Meyer, A.; Zardoya, R. Recent advances in the (molecular) phylogeny of vertebrates. Ann. Rev. Ecol. Evol. Syst. 2003, 34, 311–338. [Google Scholar] [CrossRef]
- Strasser, B.; Mlitz, V.; Hermann, M.; Rice, R.H.; Eigenheer, R.A.; Alibardi, L.; Tschachler, E.; Eckhart, L. Evolutionary origin and diversification of epidermal barrier proteins in amniotes. Mol. Biol. Evol. 2014, 31, 3194–3205. [Google Scholar] [CrossRef]
- Steven, A.C.; Bisher, M.E.; Roop, D.R.; Steinert, P.M. Biosynthetic pathways of filaggrin and loricrin--two major proteins expressed by terminally differentiated epidermal keratinocytes. J. Struct. Biol. 1990, 104, 150–162. [Google Scholar] [CrossRef]
- Steinert, P.M.; Cantieri, J.S.; Teller, D.C.; Lonsdale-Eccles, J.D.; Dale, B.A. Characterization of a class of cationic proteins that specifically interact with intermediate filaments. Proc. Natl. Acad. Sci. USA 1981, 78, 4097–4101. [Google Scholar] [CrossRef] [PubMed]
- Steinert, P.M.; Marekov, L.N. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J. Biol. Chem. 1995, 270, 17702–17711. [Google Scholar] [CrossRef] [PubMed]
- Rice, R.H.; Durbin-Johnson, B.P.; Ishitsuka, Y.; Salemi, M.; Phinney, B.S.; Rocke, D.M.; Roop, D.R. Proteomic Analysis of Loricrin Knockout Mouse Epidermis. J. Proteome Res. 2016, 15, 2560–2566. [Google Scholar] [CrossRef] [PubMed]
- Hohl, D.; Mehrel, T.; Lichti, U.; Turner, M.L.; Roop, D.R.; Steinert, P.M. Characterization of human loricrin. Structure and function of a new class of epidermal cell envelope proteins. J. Biol. Chem. 1991, 266, 6626–6636. [Google Scholar] [PubMed]
- Fay, J.C.; Wu, C.I. Hitchhiking under positive Darwinian selection. Genetics 2000, 155, 1405–1413. [Google Scholar]
- Dawkins, R. The Selfish Gene; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Choi, O.R.; Engel, J.D. Developmental regulation of beta-globin gene switching. Cell 1988, 55, 17–26. [Google Scholar] [CrossRef]
- Natarajan, C.; Hoffmann, F.G.; Weber, R.E.; Fago, A.; Witt, C.C.; Storz, J.F. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science 2016, 354, 336–339. [Google Scholar] [CrossRef]
- Taylor, S.M.; Parobek, C.M.; Fairhurst, R.M. Haemoglobinopathies and the clinical epidemiology of malaria: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 457–468. [Google Scholar] [CrossRef]
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef]
- Itoh, K.; Igarashi, K.; Hayashi, N.; Nishizawa, M.; Yamamoto, M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 1995, 15, 4184–4193. [Google Scholar] [CrossRef] [PubMed]
- Pevny, L.; Simon, M.C.; Robertson, E.; Klein, W.H.; Tsai, S.F.; D’Agati, V.; Orkin, S.H.; Costantini, F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 1991, 349, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Shivdasani, R.A.; Orkin, S.H. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc. Natl. Acad. Sci. USA 1995, 92, 8690–8694. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Lu, R.; Chang, J.C.; Kan, Y.W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 1996, 93, 13943–13948. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef]
- Kitamura, Y.; Umemura, T.; Kanki, K.; Kodama, Y.; Kitamoto, S.; Saito, K.; Itoh, K.; Yamamoto, M.; Masegi, T.; Nishikawa, A.; et al. Increased susceptibility to hepatocarcinogenicity of Nrf2-deficient mice exposed to 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Sci. 2007, 98, 19–24. [Google Scholar] [CrossRef]
- Ramos-Gomez, M.; Kwak, M.K.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Talalay, P.; Kensler, T.W. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA 2001, 98, 3410–3415. [Google Scholar] [CrossRef]
- Xu, C.; Huang, M.T.; Shen, G.; Yuan, X.; Lin, W.; Khor, T.O.; Conney, A.H.; Kong, A.N. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006, 66, 8293–8296. [Google Scholar] [CrossRef]
- Schafer, M.; Dutsch, S.; auf dem Keller, U.; Navid, F.; Schwarz, A.; Johnson, D.A.; Johnson, J.A.; Werner, S. Nrf2 establishes a glutathione-mediated gradient of UVB cytoprotection in the epidermis. Genes Dev. 2010, 24, 1045–1058. [Google Scholar] [CrossRef]
- Kim, T.H.; Shivdasani, R.A. Stomach development, stem cells and disease. Development 2016, 143, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Jarnik, M.; de Viragh, P.A.; Scharer, E.; Bundman, D.; Simon, M.N.; Roop, D.R.; Steven, A.C. Quasi-normal cornified cell envelopes in loricrin knockout mice imply the existence of a loricrin backup system. J. Investig. Dermatol. 2002, 118, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Ishitsuka, Y.; Inoue, S.; Nakamura, Y.; Saito, A.; Okiyama, N.; Fujisawa, Y.; Furuta, J.; Watanabe, R.; Fujimoto, M. Nuclear Factor Erythroid 2-Related Factor 2 (Nrf2) Regulates Epidermal Keratinization under Psoriatic Skin Inflammation. Am. J. Pathol. 2020, 190, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Redondo, P.; Bauza, A. Topical N-acetylcysteine for lamellar ichthyosis. Lancet 1999, 354, 1880. [Google Scholar] [CrossRef]
- Li, J.; Ichikawa, T.; Villacorta, L.; Janicki, J.S.; Brower, G.L.; Yamamoto, M.; Cui, T. Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Mischke, D.; Korge, B.P.; Marenholz, I.; Volz, A.; Ziegler, A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J. Investig. Dermatol 1996, 106, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Kypriotou, M.; Huber, M.; Hohl, D. The human epidermal differentiation complex: Cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012, 21, 643–649. [Google Scholar] [CrossRef]
- Efstratiadis, A.; Posakony, J.W.; Maniatis, T.; Lawn, R.M.; O’Connell, C.; Spritz, R.A.; DeRiel, J.K.; Forget, B.G.; Weissman, S.M.; Slightom, J.L.; et al. The structure and evolution of the human beta-globin gene family. Cell 1980, 21, 653–668. [Google Scholar] [CrossRef]
- Backendorf, C.; Hohl, D. A common origin for cornified envelope proteins? Nat. Genet. 1992, 2, 91. [Google Scholar] [CrossRef]
- Eckert, R.L.; Green, H. Structure and evolution of the human involucrin gene. Cell 1986, 46, 583–589. [Google Scholar] [CrossRef]
- Teumer, J.; Green, H. Divergent evolution of part of the involucrin gene in the hominoids: Unique intragenic duplications in the gorilla and human. Proc. Natl. Acad. Sci. USA 1989, 86, 1283–1286. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.W.; Chung, O.; Cho, Y.S.; MacGregor, G.R.; Plikus, M.V. Gene loss in keratinization programs accompanies adaptation of cetacean skin to aquatic lifestyle. Exp. Dermatol. 2015, 24, 572–573. [Google Scholar] [CrossRef] [PubMed]
- Strasser, B.; Mlitz, V.; Fischer, H.; Tschachler, E.; Eckhart, L. Comparative genomics reveals conservation of filaggrin and loss of caspase-14 in dolphins. Exp. Dermatol 2015, 24, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.R.; Aho, S.; Bosko, C.A. Filaggrin-revisited. Int. J. Cosmet. Sci. 2013, 35, 412–423. [Google Scholar] [CrossRef]
- Goodwin, Z.A.; de Guzman Strong, C. Recent Positive Selection in Genes of the Mammalian Epidermal Differentiation Complex Locus. Front. Genet. 2016, 7, 227. [Google Scholar] [CrossRef]
- Bickenbach, J.R.; Greer, J.M.; Bundman, D.S.; Rothnagel, J.A.; Roop, D.R. Loricrin expression is coordinated with other epidermal proteins and the appearance of lipid lamellar granules in development. J. Investig. Dermatol. 1995, 104, 405–410. [Google Scholar] [CrossRef]
- Hardman, M.J.; Sisi, P.; Banbury, D.N.; Byrne, C. Patterned acquisition of skin barrier function during development. Development 1998, 125, 1541–1552. [Google Scholar]
- Koch, P.J.; de Viragh, P.A.; Scharer, E.; Bundman, D.; Longley, M.A.; Bickenbach, J.; Kawachi, Y.; Suga, Y.; Zhou, Z.; Huber, M.; et al. Lessons from loricrin-deficient mice: Compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 2000, 151, 389–400. [Google Scholar] [CrossRef]
- Steinert, P.M. The complexity and redundancy of epithelial barrier function. J. Cell Biol. 2000, 151, F5–F8. [Google Scholar] [CrossRef]
- Ishitsuka, Y.; Roop, D.R. Loricrin Confers Photoprotective Function against UVB in Corneocytes. J. Investig. Dermatol. 2018, 138, 2684–2687. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Akiyama, M.; Sugiyama-Nakagiri, Y.; Sakai, K.; Goto, M.; McMillan, J.R.; Ota, M.; Sawamura, D.; Shimizu, H. Expression of the keratinocyte lipid transporter ABCA12 in developing and reconstituted human epidermis. Am. J. Pathol. 2007, 171, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, S.; Fijneman, R.; Wiegant, J.; van Kessel, A.G.; van De Putte, P.; Backendorf, C. Molecular characterization and evolution of the SPRR family of keratinocyte differentiation markers encoding small proline-rich proteins. Genomics 1993, 16, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.; Tilli, C.M.; Hardman, M.J.; Avilion, A.A.; MacLeod, M.C.; Ashcroft, G.S.; Byrne, C. Late cornified envelope family in differentiating epithelia--response to calcium and ultraviolet irradiation. J. Investig. Dermatol. 2005, 124, 1062–1070. [Google Scholar] [CrossRef] [PubMed]
- Vermeij, W.P.; Backendorf, C. Skin cornification proteins provide global link between ROS detoxification and cell migration during wound healing. PLoS ONE 2010, 5, e11957. [Google Scholar] [CrossRef]
- de Cid, R.; Riveira-Munoz, E.; Zeeuwen, P.L.; Robarge, J.; Liao, W.; Dannhauser, E.N.; Giardina, E.; Stuart, P.E.; Nair, R.; Helms, C.; et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 2009, 41, 211–215. [Google Scholar] [CrossRef]
- Sevilla, L.M.; Nachat, R.; Groot, K.R.; Klement, J.F.; Uitto, J.; Djian, P.; Maatta, A.; Watt, F.M. Mice deficient in involucrin, envoplakin, and periplakin have a defective epidermal barrier. J. Cell Biol. 2007, 179, 1599–1612. [Google Scholar] [CrossRef]
- Huppke, P.; Weissbach, S.; Church, J.A.; Schnur, R.; Krusen, M.; Dreha-Kulaczewski, S.; Kuhn-Velten, W.N.; Wolf, A.; Huppke, B.; Millan, F.; et al. Activating de novo mutations in NFE2L2 encoding NRF2 cause a multisystem disorder. Nat. Commun. 2017, 8, 818. [Google Scholar] [CrossRef]
- Menegon, S.; Columbano, A.; Giordano, S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef]
- Taguchi, K.; Maher, J.M.; Suzuki, T.; Kawatani, Y.; Motohashi, H.; Yamamoto, M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 2010, 30, 3016–3026. [Google Scholar] [CrossRef]
- Suzuki, T.; Murakami, S.; Biswal, S.S.; Sakaguchi, S.; Harigae, H.; Yamamoto, M.; Motohashi, H. Systemic activation of NRF2 alleviates lethal autoimmune inflammation in scurfy mice. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef]
- Rushmore, T.H.; Morton, M.R.; Pickett, C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991, 266, 11632–11639. [Google Scholar] [PubMed]
- Wasserman, W.W.; Fahl, W.E. Functional antioxidant responsive elements. Proc. Natl. Acad. Sci. USA 1997, 94, 5361–5366. [Google Scholar] [CrossRef] [PubMed]
- Nioi, P.; Nguyen, T.; Sherratt, P.J.; Pickett, C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 2005, 25, 10895–10906. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Seng, S.; Avraham, H.K.; Jiang, S.; Yang, S.; Sekine, M.; Kimelman, N.; Li, H.; Avraham, S. The nuclear matrix protein, NRP/B, enhances Nrf2-mediated oxidative stress responses in breast cancer cells. Cancer Res. 2007, 67, 8596–8604. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRalpha inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef]
- Prestera, T.; Zhang, Y.; Spencer, S.R.; Wilczak, C.A.; Talalay, P. The electrophile counterattack response: Protection against neoplasia and toxicity. Adv. Enzym. Regul. 1993, 33, 281–296. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Cole, R.N.; Itoh, K.; Wakabayashi, N.; Katoh, Y.; Yamamoto, M.; Talalay, P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA 2002, 99, 11908–11913. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 2006, 21, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Cullinan, S.B.; Gordan, J.D.; Jin, J.; Harper, J.W.; Diehl, J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 2004, 24, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef]
- Tong, K.I.; Padmanabhan, B.; Kobayashi, A.; Shang, C.; Hirotsu, Y.; Yokoyama, S.; Yamamoto, M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007, 27, 7511–7521. [Google Scholar] [CrossRef]
- Ogura, T.; Tong, K.I.; Mio, K.; Maruyama, Y.; Kurokawa, H.; Sato, C.; Yamamoto, M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. USA 2010, 107, 2842–2847. [Google Scholar] [CrossRef]
- Kang, M.I.; Kobayashi, A.; Wakabayashi, N.; Kim, S.G.; Yamamoto, M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2046–2051. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- Matzinger, P. The danger model: A renewed sense of self. Science 2002, 296, 301–305. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell 1988, 53, 211–217. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Baltimore, D. I kappa B: A specific inhibitor of the NF-kappa B transcription factor. Science 1988, 242, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.K.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: Role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002, 22, 2883–2892. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.A.; Jaiswal, A.K. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003, 278, 44675–44682. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 12475–12480. [Google Scholar] [CrossRef]
- Ciechanover, A. The ubiquitin-proteasome proteolytic pathway. Cell 1994, 79, 13–21. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE 2007, 2007, cm8. [Google Scholar] [CrossRef]
- Michael, D.; Oren, M. The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 2003, 13, 49–58. [Google Scholar] [CrossRef]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef]
- Nguyen, T.; Sherratt, P.J.; Huang, H.C.; Yang, C.S.; Pickett, C.B. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 2003, 278, 4536–4541. [Google Scholar] [CrossRef]
- Kobayashi, M.; Itoh, K.; Suzuki, T.; Osanai, H.; Nishikawa, K.; Katoh, Y.; Takagi, Y.; Yamamoto, M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 2002, 7, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Katoh, Y.; Iida, K.; Kang, M.I.; Kobayashi, A.; Mizukami, M.; Tong, K.I.; McMahon, M.; Hayes, J.D.; Itoh, K.; Yamamoto, M. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 2005, 433, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Tong, K.I.; Kobayashi, A.; Katsuoka, F.; Yamamoto, M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 2006, 387, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.; Killeen, E.; Naquin, R.; Alam, S.; Alam, J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 2003, 278, 2396–2402. [Google Scholar] [CrossRef] [PubMed]
- Zandi, E.; Rothwarf, D.M.; Delhase, M.; Hayakawa, M.; Karin, M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 1997, 91, 243–252. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef]
- Ogawa, T.; Ishitsuka, Y.; Roop, D.; Fujimoto, M. 314 Loricrin protects against chemical carcinogenesis but affects cancer immunoediting. J. Investig. Dermatol. 2019, 139, S54. [Google Scholar] [CrossRef][Green Version]
- Saurat, J.H.; Kaya, G.; Saxer-Sekulic, N.; Pardo, B.; Becker, M.; Fontao, L.; Mottu, F.; Carraux, P.; Pham, X.C.; Barde, C.; et al. The cutaneous lesions of dioxin exposure: Lessons from the poisoning of Victor Yushchenko. Toxicol. Sci. 2012, 125, 310–317. [Google Scholar] [CrossRef]
- Klaassen, C.D.; Reisman, S.A. Nrf2 the rescue: Effects of the antioxidative/electrophilic response on the liver. Toxicol. Appl. Pharmacol. 2010, 244, 57–65. [Google Scholar] [CrossRef]
- Common, J.E.A.; Barker, J.N.; van Steensel, M.A.M. What does acne genetics teach us about disease pathogenesis? Br. J. Dermatol. 2019, 181, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.C.; Dellavalle, R.P.; Garner, S. Acne vulgaris. Lancet 2012, 379, 361–372. [Google Scholar] [CrossRef]
- Woo, T.E.; Sibley, C.D. The emerging utility of the cutaneous microbiome in the treatment of acne and atopic dermatitis. J. Am. Acad. Dermatol. 2020, 82, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Vodermaier, H.C. APC/C and SCF: Controlling each other and the cell cycle. Curr. Biol. 2004, 14, R787–R796. [Google Scholar] [CrossRef] [PubMed]
- Steinert, P.M.; Idler, W.W. Postsynthetic modifications of mammalian epidermal alpha-keratin. Biochemistry 1979, 18, 5664–5669. [Google Scholar] [CrossRef] [PubMed]
- Thiele, J.J.; Hsieh, S.N.; Briviba, K.; Sies, H. Protein oxidation in human stratum corneum: Susceptibility of keratins to oxidation in vitro and presence of a keratin oxidation gradient in vivo. J. Investig. Dermatol. 1999, 113, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, K.; Steinert, P.M. Overexpression of human loricrin in transgenic mice produces a normal phenotype. Proc. Natl. Acad. Sci. USA 1993, 90, 10754–10758. [Google Scholar] [CrossRef]
- de Koning, H.D.; van den Bogaard, E.H.; Bergboer, J.G.; Kamsteeg, M.; van Vlijmen-Willems, I.M.; Hitomi, K.; Henry, J.; Simon, M.; Takashita, N.; Ishida-Yamamoto, A.; et al. Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair. Br. J. Dermatol. 2012, 166, 1245–1254. [Google Scholar] [CrossRef]
- Ekanayake-Mudiyanselage, S.; Aschauer, H.; Schmook, F.P.; Jensen, J.M.; Meingassner, J.G.; Proksch, E. Expression of epidermal keratins and the cornified envelope protein involucrin is influenced by permeability barrier disruption. J. Investig. Dermatol. 1998, 111, 517–523. [Google Scholar] [CrossRef]
- Vermeij, W.P.; Alia, A.; Backendorf, C. ROS quenching potential of the epidermal cornified cell envelope. J. Investig. Dermatol. 2011, 131, 1435–1441. [Google Scholar] [CrossRef]
- Steinert, P.M.; Candi, E.; Kartasova, T.; Marekov, L. Small proline-rich proteins are cross-bridging proteins in the cornified cell envelopes of stratified squamous epithelia. J. Struct. Biol. 1998, 122, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Steinert, P.M.; Kartasova, T.; Marekov, L.N. Biochemical evidence that small proline-rich proteins and trichohyalin function in epithelia by modulation of the biomechanical properties of their cornified cell envelopes. J. Biol. Chem. 1998, 273, 11758–11769. [Google Scholar] [CrossRef] [PubMed]
- Hagerman, A.E.; Butler, L.G. The specificity of proanthocyanidin-protein interactions. J. Biol. Chem. 1981, 256, 4494–4497. [Google Scholar] [PubMed]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar] [CrossRef] [PubMed]
- auf dem Keller, U.; Huber, M.; Beyer, T.A.; Kumin, A.; Siemes, C.; Braun, S.; Bugnon, P.; Mitropoulos, V.; Johnson, D.A.; Johnson, J.A.; et al. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol. Cell. Biol. 2006, 26, 3773–3784. [Google Scholar] [CrossRef] [PubMed]
- Braun, S.; Hanselmann, C.; Gassmann, M.G.; auf dem Keller, U.; Born-Berclaz, C.; Chan, K.; Kan, Y.W.; Werner, S. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 2002, 22, 5492–5505. [Google Scholar] [CrossRef]
- Fan, X.; Wang, D.; Burgmaier, J.E.; Teng, Y.; Romano, R.A.; Sinha, S.; Yi, R. Single Cell and Open Chromatin Analysis Reveals Molecular Origin of Epidermal Cells of the Skin. Dev. Cell 2018, 47, 21–37. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Shibata, T.; Ohta, T.; Tong, K.I.; Kokubu, A.; Odogawa, R.; Tsuta, K.; Asamura, H.; Yamamoto, M.; Hirohashi, S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA 2008, 105, 13568–13573. [Google Scholar] [CrossRef]
- Miyata, M.; Kudo, G.; Lee, Y.H.; Yang, T.J.; Gelboin, H.V.; Fernandez-Salguero, P.; Kimura, S.; Gonzalez, F.J. Targeted disruption of the microsomal epoxide hydrolase gene. Microsomal epoxide hydrolase is required for the carcinogenic activity of 7,12-dimethylbenz[a]anthracene. J. Biol. Chem. 1999, 274, 23963–23968. [Google Scholar] [CrossRef]
- Schaper, I.D.; Marcuzzi, G.P.; Weissenborn, S.J.; Kasper, H.U.; Dries, V.; Smyth, N.; Fuchs, P.; Pfister, H. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res. 2005, 65, 1394–1400. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Motohashi, H. NRF2 addiction in cancer cells. Cancer Sci. 2018, 109, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Terzian, T.; Suh, Y.A.; Iwakuma, T.; Post, S.M.; Neumann, M.; Lang, G.A.; Van Pelt, C.S.; Lozano, G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008, 22, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019, 26, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Ishitsuka, Y.; Nakamura, Y.; Kubota, N.; Saito, A.; Fujisawa, Y.; Watanabe, R.; Okiyama, N.; Suga, Y.; Roop, D.R.; et al. NRF2 Augments Epidermal Antioxidant Defenses and Promotes Atopy. J. Immunol. 2020, 205, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef]
- Akinduro, O.; Sully, K.; Patel, A.; Robinson, D.J.; Chikh, A.; McPhail, G.; Braun, K.M.; Philpott, M.P.; Harwood, C.A.; Byrne, C.; et al. Constitutive Autophagy and Nucleophagy during Epidermal Differentiation. J. Investig. Dermatol. 2016, 136, 1460–1470. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kumanomidou, T.; Sou, Y.S.; Mizushima, T.; Ezaki, J.; Ueno, T.; Kominami, E.; Yamane, T.; Tanaka, K.; Komatsu, M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008, 283, 22847–22857. [Google Scholar] [CrossRef]
- Ong, P.Y.; Leung, D.Y. Bacterial and Viral Infections in Atopic Dermatitis: A Comprehensive Review. Clin. Rev. Allergy Immunol. 2016, 51, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, K.; Dykes, P.J.; Marks, R. Inflammation due to intra-cutaneous implantation of stratum corneum. Br. J. Exp. Pathol. 1984, 65, 107–115. [Google Scholar] [PubMed]
- Gahring, L.C.; Buckley, A.; Daynes, R.A. Presence of epidermal-derived thymocyte activating factor/interleukin 1 in normal human stratum corneum. J. Clin. Investig. 1985, 76, 1585–1591. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishitsuka, Y.; Ogawa, T.; Roop, D. The KEAP1/NRF2 Signaling Pathway in Keratinization. Antioxidants 2020, 9, 751. https://doi.org/10.3390/antiox9080751
Ishitsuka Y, Ogawa T, Roop D. The KEAP1/NRF2 Signaling Pathway in Keratinization. Antioxidants. 2020; 9(8):751. https://doi.org/10.3390/antiox9080751
Chicago/Turabian StyleIshitsuka, Yosuke, Tatsuya Ogawa, and Dennis Roop. 2020. "The KEAP1/NRF2 Signaling Pathway in Keratinization" Antioxidants 9, no. 8: 751. https://doi.org/10.3390/antiox9080751
APA StyleIshitsuka, Y., Ogawa, T., & Roop, D. (2020). The KEAP1/NRF2 Signaling Pathway in Keratinization. Antioxidants, 9(8), 751. https://doi.org/10.3390/antiox9080751




