Performance of Layer-by-Layer-Modified Multibore® Ultrafiltration Capillary Membranes for Salt Retention and Removal of Antibiotic Resistance Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Membrane and Chemicals
2.2. Semi-Automated LbL-Coating of Ultrafiltration Membranes
Coating Procedure
2.3. Coating of PES Microparticles and Zeta-Potential Determination
2.4. Evaluation of Performance of the LbL-Modified Membranes
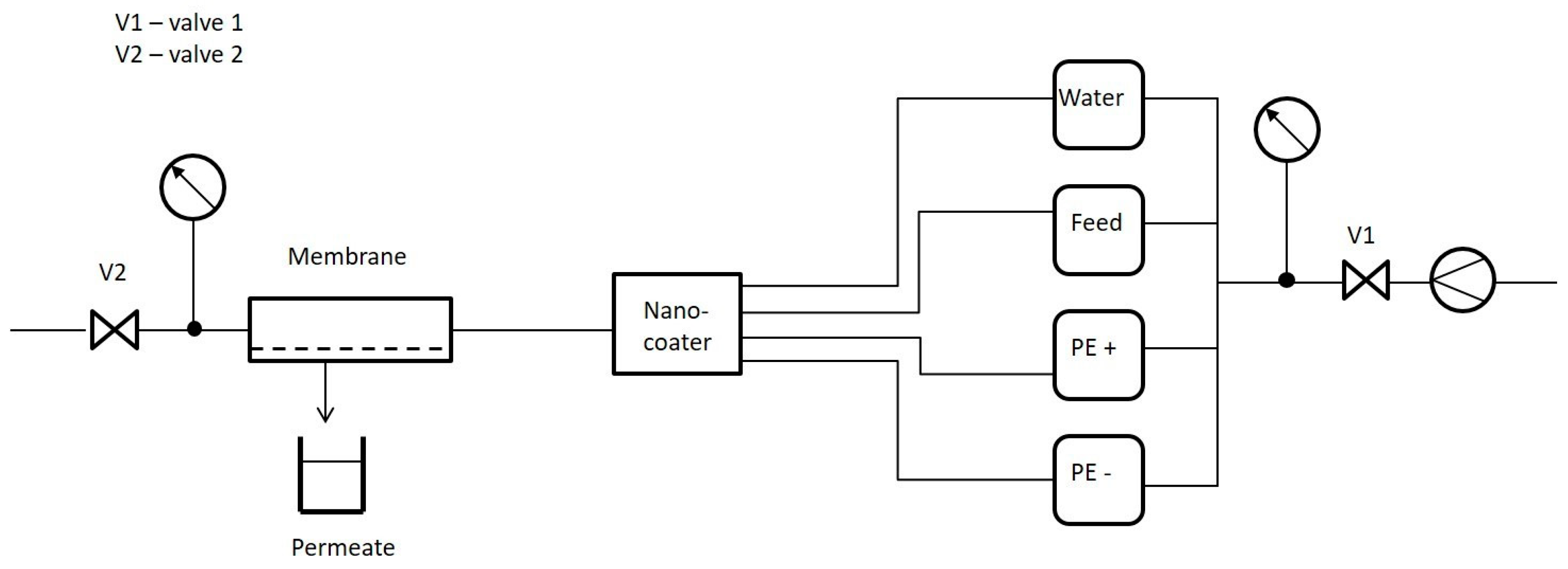
2.4.1. Membrane Filtration Unit
2.4.2. Membrane Pretreatment Prior to Filtration Experiments
2.4.3. Measurement of Pure Water Permeability and Salt Retention
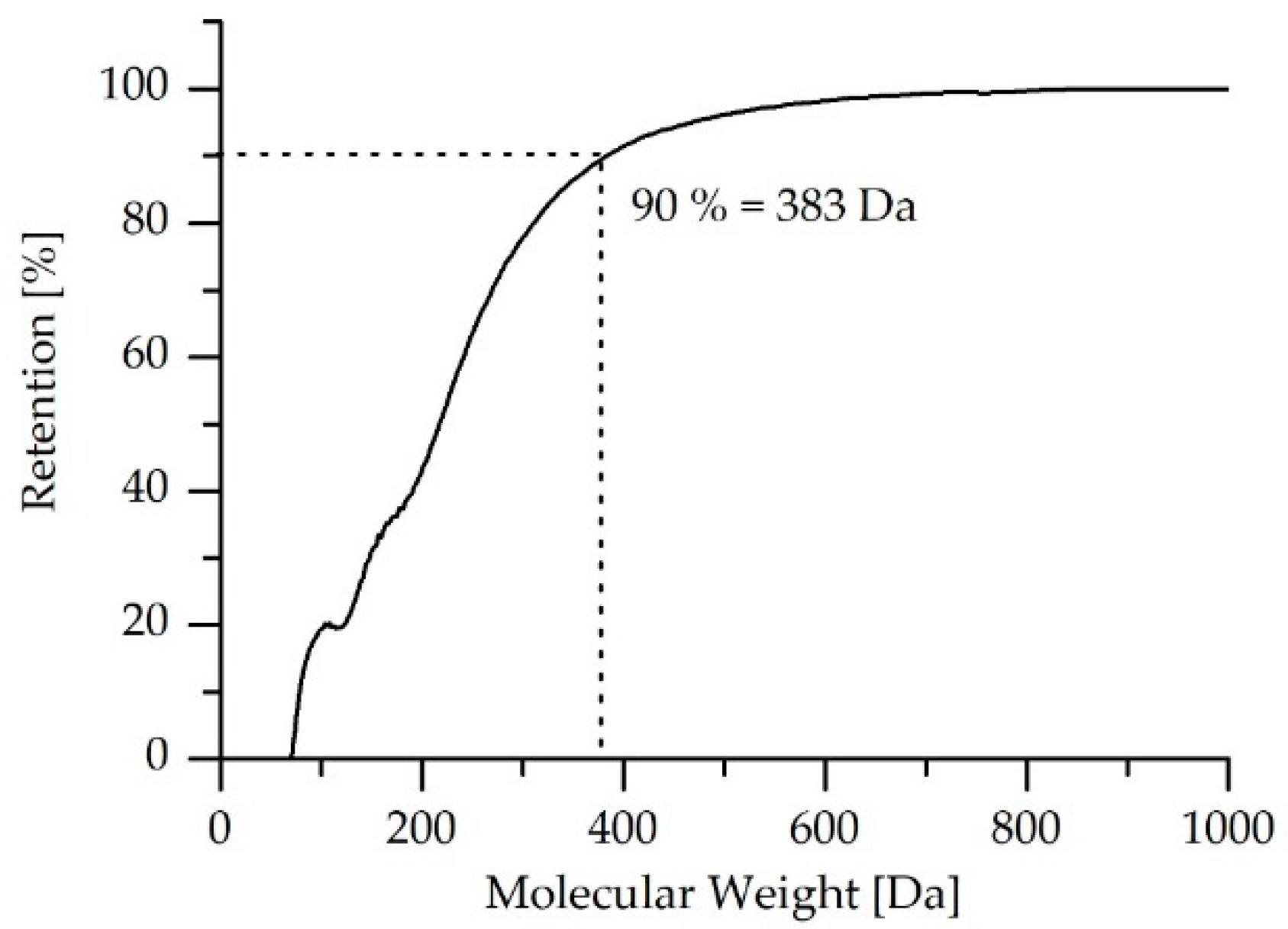
2.4.4. Measurement of Molecular Weight Cut-Off
2.4.5. Investigation of Backwashing Flux Influence on the Stability of LbL-Modified Membranes
2.5. Laboratory Filtration Experiments of ARGs
2.6. Determination of Interactions Between the ARGs and Membrane
2.7. qPCR Analysis
3. Results and Discussion
3.1. Characterization of LbL-Modifed Membranes and the Polyelectrolyte Coating
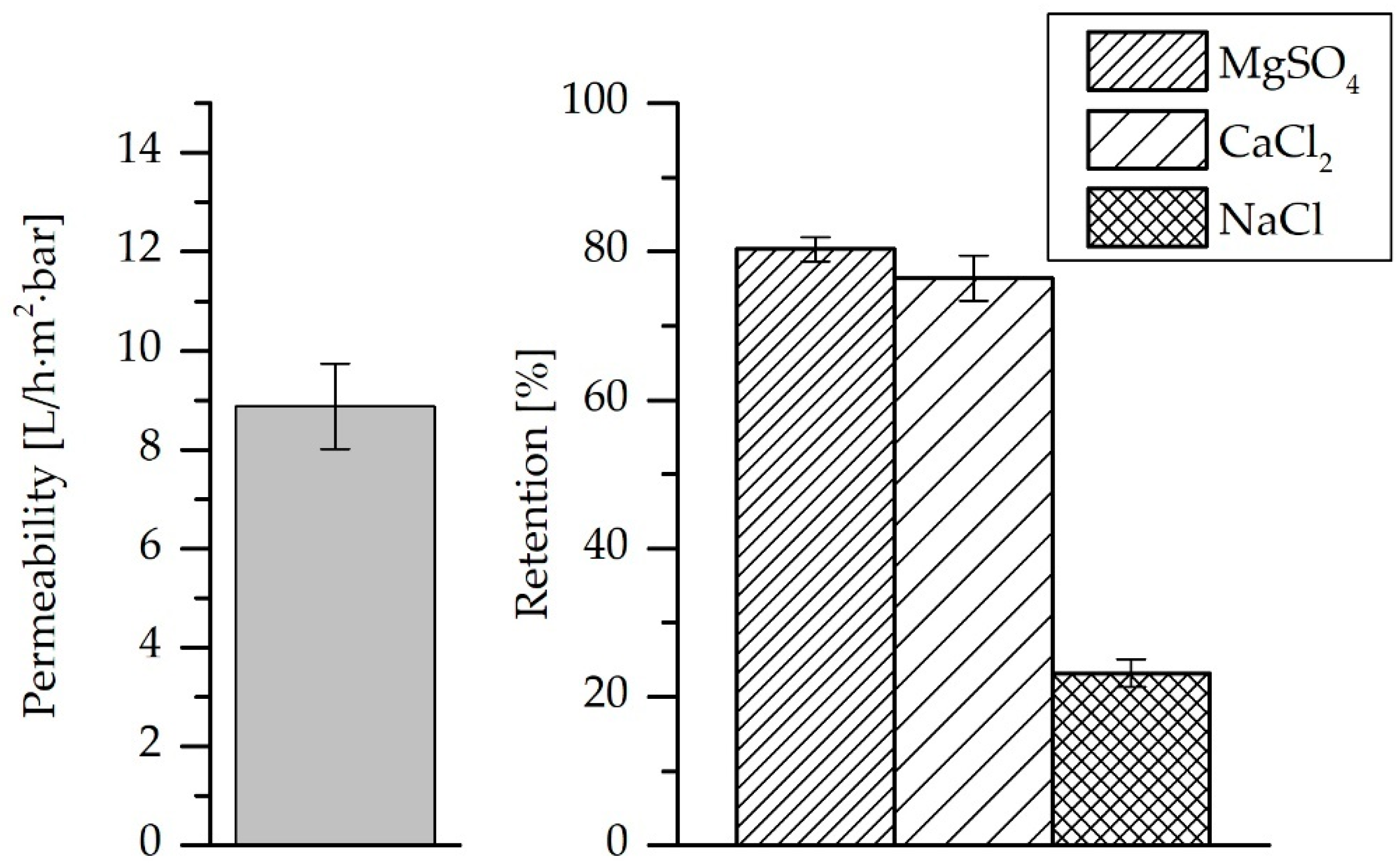
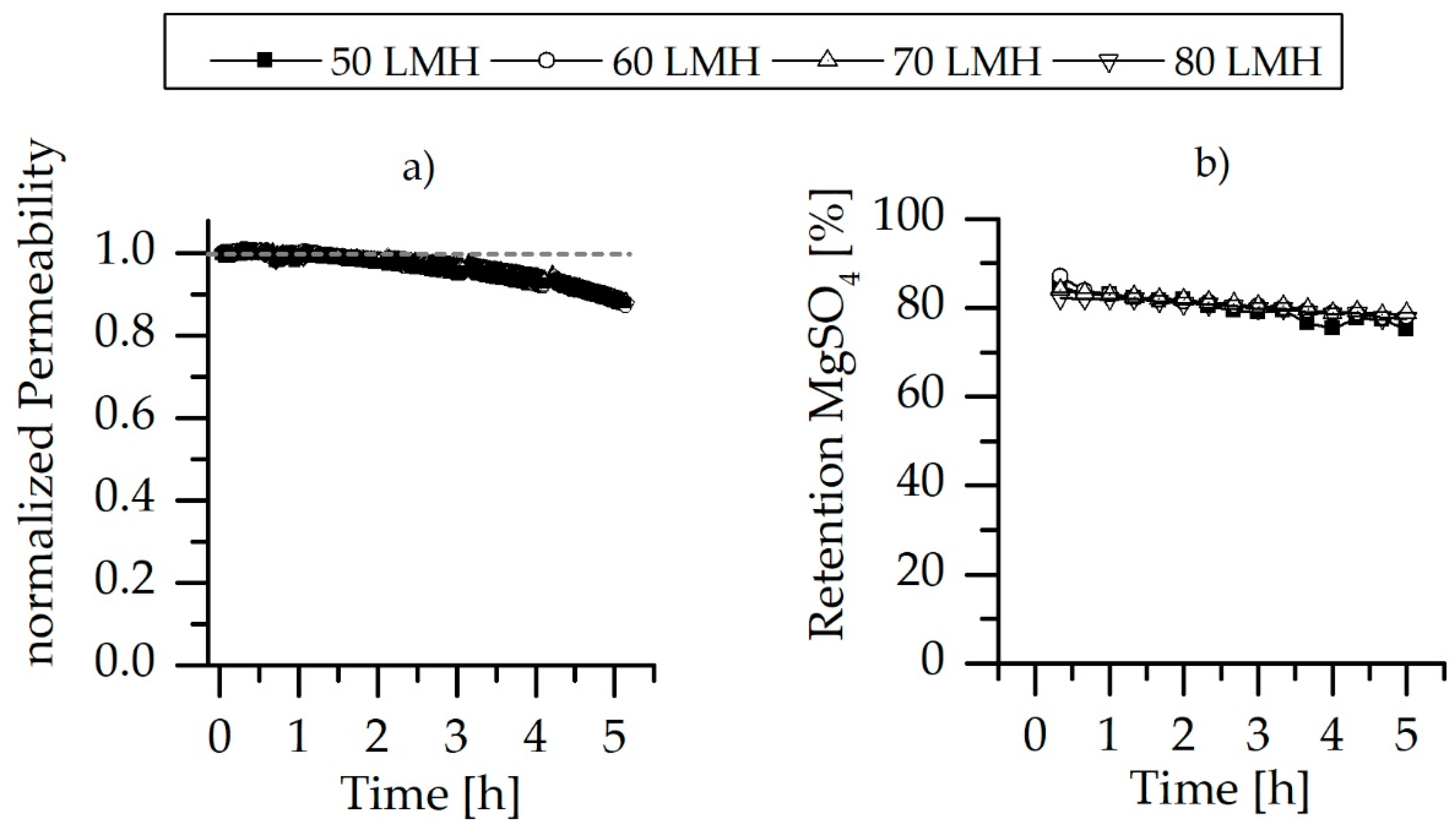
3.2. Pure Water Permeability and Salt Retention
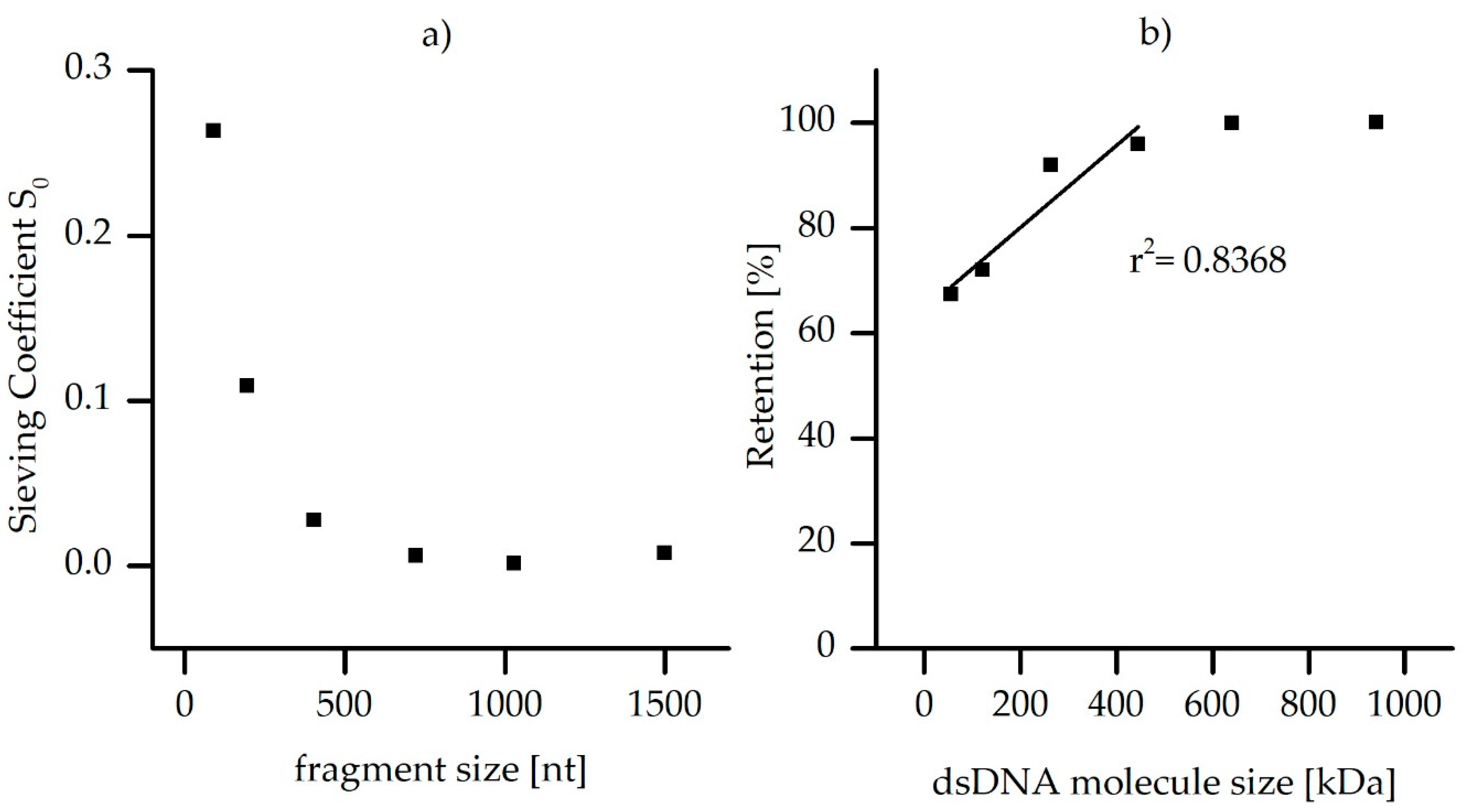
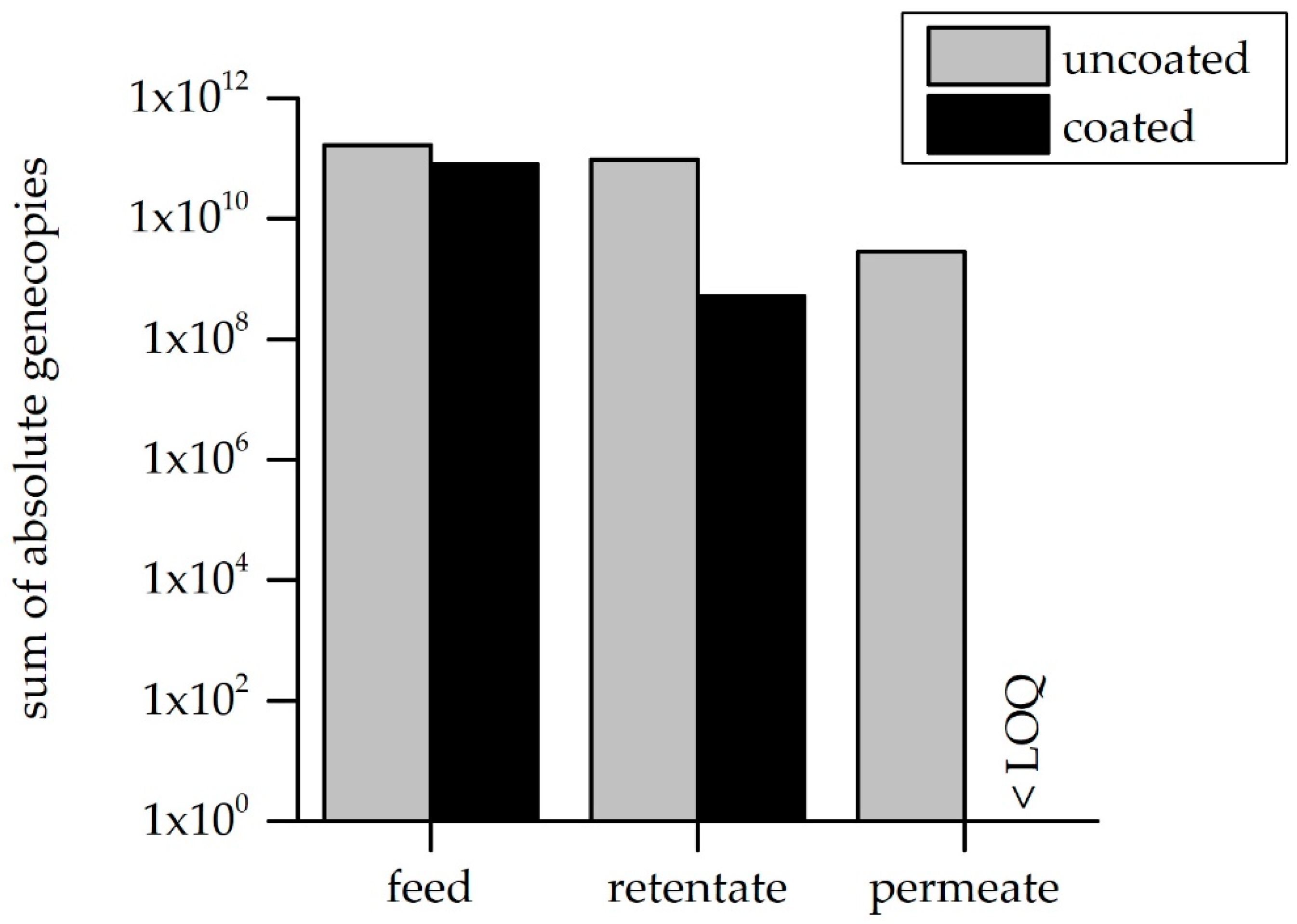
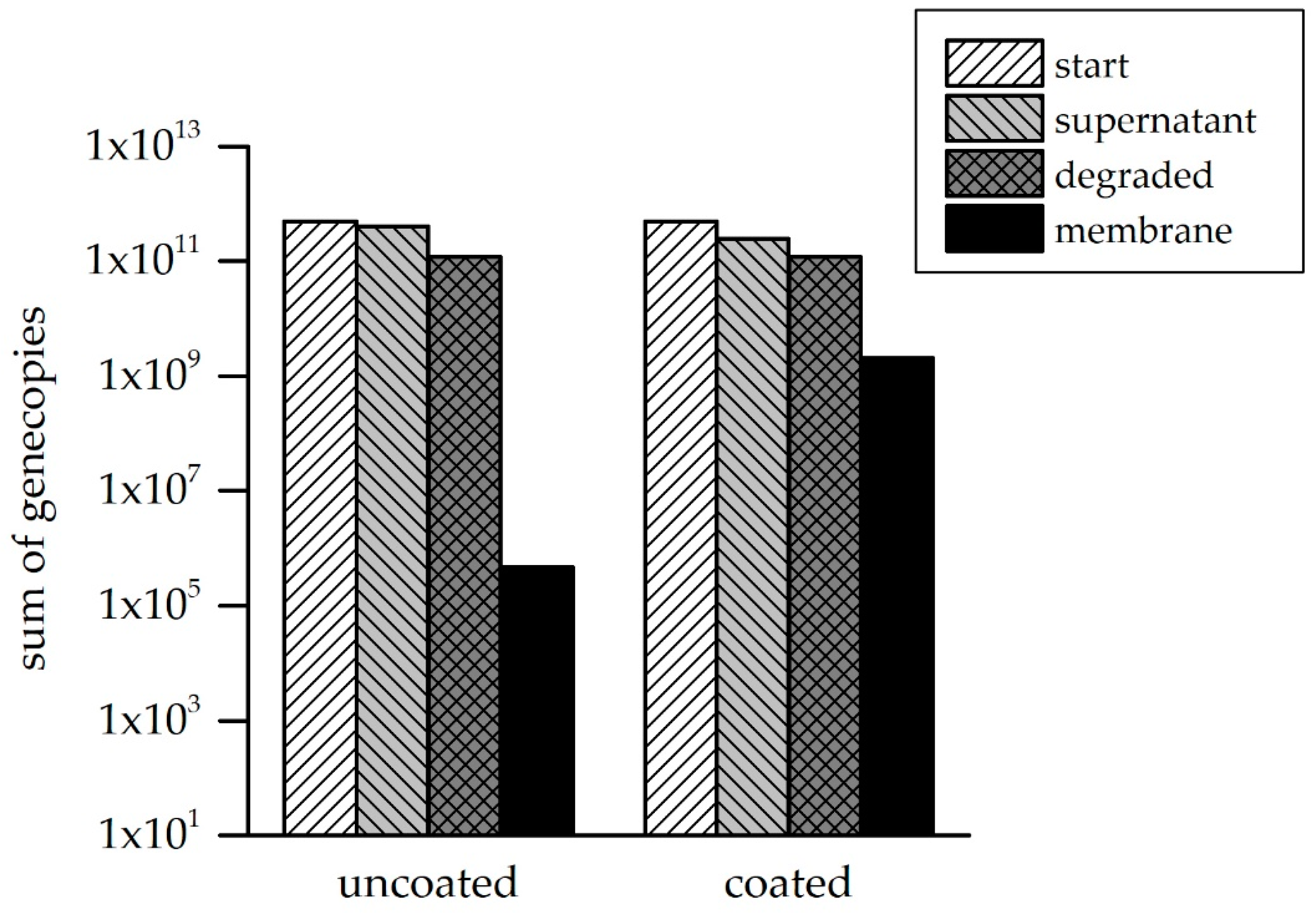
3.3. ARG Retention
3.4. Mechnical Stability of the LbL Modification
3.5. Chemical Stability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Paltrinieri, L.; Remmen, K.; Müller, B.; Chu, L.; Köser, J.; Wintgens, T.; Wessling, M.; de Smet, L.C.; Sudhölter, E.J. Improved phosphoric acid recovery from sewage sludge ash using layer-by-layer modified membranes. J. Membr. Sci. 2019, 587, 117162. [Google Scholar] [CrossRef]
- Remmen, K.; Müller, B.; Köser, J.; Wessling, M.; Wintgens, T. Phosphorus recovery in an acidic environment using layer-by-layer modified membranes. J. Membr. Sci. 2019, 582, 254–263. [Google Scholar] [CrossRef]
- Ishigami, T.; Amano, K.; Fujii, A.; Ohmukai, Y.; Kamio, E.; Maruyama, T.; Matsuyama, H. Fouling reduction of reverse osmosis membrane by surface modification via layer-by-layer assembly. Sep. Purif. Technol. 2012, 99, 1–7. [Google Scholar] [CrossRef]
- Fadhillah, F.; Zaidi, S.; Khan, Z.; Khaled, M.M.; Rahman, F.; Hammond, P.T. Development of polyelectrolyte multilayer thin film composite membrane for water desalination application. Desalination 2013, 318, 19–24. [Google Scholar] [CrossRef]
- Menne, D.; Üzüm, C.; Koppelmann, A.; Wong, J.E.; van Foeken, C.; Borre, F.; Dähne, L.; Laakso, T.; Pihlajamäki, A.; Wessling, M. Regenerable polymer/ceramic hybrid nanofiltration membrane based on polyelectrolyte assembly by layer-by-layer technique. J. Membr. Sci. 2016, 520, 924–932. [Google Scholar] [CrossRef]
- de Grooth, J.; Haakmeester, B.; Wever, C.; Potreck, J.; de Vos, W.M.; Nijmeijer, K. Long term physical and chemical stability of polyelectrolyte multilayer membranes. J. Membr. Sci. 2015, 489, 153–159. [Google Scholar] [CrossRef]
- Gregurec, D.; Olszyna, M.; Politakos, N.; Yate, L.; Dahne, L.; Moya, S.E. Stability of polyelectrolyte multilayers in oxidizing media: A critical issue for the development of multilayer based membranes for nanofiltration. Colloid Polym. Sci. 2015, 293, 381–388. [Google Scholar] [CrossRef]
- Stange, C.; Tiehm, A. Occurrence of antibiotic resistance genes and microbial source tracking markers in the water of a karst spring in Germany. Sci. Total Environ. 2020, 742, 140529. [Google Scholar] [CrossRef]
- Voigt, A.M.; Zacharias, N.; Timm, C.; Wasser, F.; Sib, E.; Skutlarek, D.; Parcina, M.; Schmithausen, R.M.; Schwartz, T.; Hembach, N.; et al. Association between antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in anthropogenic wastewater—An evaluation of clinical influences. Chemosphere 2020, 241, 125032. [Google Scholar] [CrossRef]
- Sidhu, J.P.S.; Gupta, V.V.S.R.; Stange, C.; Ho, J.; Harris, N.; Barry, K.; Gonzalez, D.; van Nostrand, J.D.; Zhou, J.; Page, D.; et al. Prevalence of antibiotic resistance and virulence genes in the biofilms from an aquifer recharged with stormwater. Water Res. 2020, 185, 116269. [Google Scholar] [CrossRef]
- Stange, C.; Yin, D.; Xu, T.; Guo, X.; Schäfer, C.; Tiehm, A. Distribution of clinically relevant antibiotic resistance genes in Lake Tai, China. Sci. Total Environ. 2019, 655, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.C.; Borowska, E.; Peschke, R.; Schwartz, T.; Horn, H. Decay of elevated antibiotic resistance genes in natural river sediments after sedimentation of wastewater particles. Sci. Total Environ. 2020, 705, 135861. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.C.; Borowska, E.; Schwartz, T.; Horn, H. Impact of the particulate matter from wastewater discharge on the abundance of antibiotic resistance genes and facultative pathogenic bacteria in downstream river sediments. Sci. Total Environ. 2019, 649, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, S.; Boopathy, R.; Nathaniel, R.; Corbin, A.; LaFleur, G. Presence of antibiotic resistant bacteria and antibiotic resistance genes in raw source water and treated drinking water. Int. Biodeterior. Biodegrad. 2015, 102, 370–374. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Yang, F.; Yang, J.; Yin, D. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 2014, 493, 626–631. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, L.; Zhang, T.; Jin, L.; Han, Q.; Zhang, D.; Lin, K.; Cui, C. Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. J. Hazard. Mater. 2018, 360, 364–372. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Antibiotic resistance and virulence genes in coliform water isolates. Int. J. Hyg. Environ. Health 2016, 219, 823–831. [Google Scholar] [CrossRef] [Green Version]
- Stange, C.; Sidhu, J.P.S.; Toze, S.; Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 2019, 222, 541–548. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, T.; Jiang, L.; Luo, Y.; Yao, S.; Zhang, D.; Lin, K.; Cui, C. Occurrence and reduction of antibiotic resistance genes in conventional and advanced drinking water treatment processes. Sci. Total Environ. 2019, 669, 777–784. [Google Scholar] [CrossRef]
- Torti, A.; Lever, M.A.; Jørgensen, B.B. Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar. Genom. 2015, 24, 185–196. [Google Scholar] [CrossRef] [PubMed]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Sheng, G.-P.; Lu, Y.-Z.; Zeng, R.J.; Yu, H.-Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. 2017, 111, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Slipko, K.; Reif, D.; Wögerbauer, M.; Hufnagl, P.; Krampe, J.; Kreuzinger, N. Removal of extracellular free DNA and antibiotic resistance genes from water and wastewater by membranes ranging from microfiltration to reverse osmosis. Water Res. 2019, 164, 114916. [Google Scholar] [CrossRef] [PubMed]
- Le, T.-H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.-H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef]
- Sun, L.; Gao, C.; He, N.; Yang, B.; Duan, X.; Chen, T. The removal of antibiotic resistance genes in secondary effluent by the combined process of PAC-UF. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2019, 54, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Lipp, P.; Sacher, F.; Baldauf, G. Removal of organic micro-pollutants during drinking water treatment by nanofiltration and reverse osmosis. Desalin. Water Treat. 2012, 13, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Ming Chew, C.; David Ng, K.M.; Richard Ooi, H.H.; Ismail, W.M.Z.W. Malaysia’s largest river bank filtration and ultrafiltration systems for municipal drinking water production. Water Pract. Technol. 2015, 10, 59–65. [Google Scholar] [CrossRef]
- Xia, S.; Nan, J.; Liu, R.; Li, G. Study of drinking water treatment by ultrafiltration of surface water and its application to China. Desalination 2004, 170, 41–47. [Google Scholar] [CrossRef]
- Hoslett, J.; Massara, T.M.; Malamis, S.; Ahmad, D.; van den Boogaert, I.; Katsou, E.; Ahmad, B.; Ghazal, H.; Simons, S.; Wrobel, L.; et al. Surface water filtration using granular media and membranes: A review. Sci. Total Environ. 2018, 639, 1268–1282. [Google Scholar] [CrossRef]
- Breazeal, M.V.R.; Novak, J.T.; Vikesland, P.J.; Pruden, A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2013, 47, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Maroni, P.; Ruiz-Cabello, F.J.M.; Tiraferri, A. Studying the role of surface chemistry on polyelectrolyte adsorption using gold-thiol self-assembled monolayer with optical reflectivity. Soft Matter 2014, 10, 9220–9225. [Google Scholar] [CrossRef] [PubMed]
- Deibele, L.; Dohrn, R. Miniplant-Technik. In der Prozessindustrie; Wiley-VCH: Weinheim, Germany, 2012; ISBN 3527660372. [Google Scholar]
- Belfort, G. Synthetic Membrane Process: Fundamentals and Water Applications; Elsevier Science: Oxford, UK, 1984; ISBN 0323145817. [Google Scholar]
- German Environmental Agency. Antibiotics and AntibioticResistances in the Environment; German Environmental Agency: Dessau-Roßlau, Germany, 2015. [Google Scholar]
- Larsson, D.G.J.; Andremont, A.; Bengtsson-Palme, J.; Brandt, K.K.; de Roda Husman, A.M.; Fagerstedt, P.; Fick, J.; Flach, C.-F.; Gaze, W.H.; Kuroda, M.; et al. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018, 117, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Westphal-Settele, K.; Konradi, S.; Balzer, F.; Schönfeld, J.; Schmithausen, R. Die Umwelt als Reservoir für Antibiotikaresistenzen: Ein wachsendes Problem für die öffentliche Gesundheit? Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2018, 61, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef]
- Hendrickson, E.R.; Payne, J.A.; Young, R.M.; Starr, M.G.; Perry, M.P.; Fahnestock, S.; Ellis, D.E.; Ebersole, R.C. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 2002, 68, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Clark, N.C.; Cooksey, R.C.; Hill, B.C.; Swenson, J.M.; Tenover, F.C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 1993, 37, 2311–2317. [Google Scholar] [CrossRef] [Green Version]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Danielsen, M.; Huys, G.; Swings, J. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 2003, 69, 1270–1275. [Google Scholar] [CrossRef] [Green Version]
- Barraud, O.; Baclet, M.C.; Denis, F.; Ploy, M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010, 65, 1642–1645. [Google Scholar] [CrossRef] [Green Version]
- Volkmann, H.; Schwartz, T.; Bischoff, P.; Kirchen, S.; Obst, U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 2004, 56, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Ager, K.; Latulippe, D.R.; Zydney, A.L. Plasmid DNA transmission through charged ultrafiltration membranes. J. Membr. Sci. 2009, 344, 123–128. [Google Scholar] [CrossRef]
- Latulippe, D.R.; Ager, K.; Zydney, A.L. Flux-dependent transmission of supercoiled plasmid DNA through ultrafiltration membranes. J. Membr. Sci. 2007, 294, 169–177. [Google Scholar] [CrossRef]
- Roy, Y.; Warsinger, D.M.; Lienhard, J.H. Effect of temperature on ion transport in nanofiltration membranes: Diffusion, convection and electromigration. Desalination 2017, 420, 241–257. [Google Scholar] [CrossRef]
- Du, J.R.; Zhang, X.; Feng, X.; Wu, Y.; Cheng, F.; Ali, M.E. Desalination of high salinity brackish water by an NF-RO hybrid system. Desalination 2020, 491, 114445. [Google Scholar] [CrossRef]
- Schaep, J.; Vandecasteele, C.; Mohammad, A.W.; Bowen, W.R. Analysis of the Salt Retention of Nanofiltration Membranes Using the Donnan–Steric Partitioning Pore Model. Sep. Sci. Technol. 1999, 34, 3009–3030. [Google Scholar] [CrossRef]
- Kumar, A.; Chordia, N. In silico PCR primer designing and validation. Methods Mol. Biol. (Clifton N. J.) 2015, 1275, 143–151. [Google Scholar] [CrossRef]
- Arkhangelsky, E.; Steubing, B.; Ben-Dov, E.; Kushmaro, A.; Gitis, V. Influence of pH and ionic strength on transmission of plasmid DNA through ultrafiltration membranes. Desalination 2008, 227, 111–119. [Google Scholar] [CrossRef]
- Reif, D.; Slipko, K.; Krampe, J.; Kreuzinger, N. Nachgeschaltete Membranfiltration zur Entfernung antibiotikaresistenter Bakterien und Gene aus Kläranlagenabläufen. Wiener Mitt. 2019, 249, 245–258. [Google Scholar]




| PEG Molecular Weight (Da) | Concentration (g·L−1) |
|---|---|
| 62 | 0.52 |
| 106 | 0.39 |
| 150 | 0.34 |
| 200 | 0.30 |
| 300 | 0.25 |
| 400 | 0.22 |
| 600 | 0.18 |
| 1000 | 0.30 |
| 1500 | 0.24 |
| 2000 | 0.22 |
| Gene | Fragment Size (nt) | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Reference |
|---|---|---|---|---|
| 16S rDNA | 1500 | agagtttgatcctggctcag | ggttaccttgttacgactt | [39] |
| vanA | 1030 | catgaatagaataaaagttgcaata | cccctttaacgctaatacgatcaa | [40] |
| sul2 | 722 | cggcatcgtcaacataacc | gtgtgcggatgaagtcag | [41] |
| ermB | 405 | catttaacgacgaaactggc | ggaacatctgtggtatggcg | [42] |
| intl1 | 196 | gccttgatgttacccgagag | gatcggtcgaatgcgtgt | [43] |
| mecA | 91 | cgcaacgttcaatttaattttgttaa | tggtctttctgcattcctgga | [44] |
| Gene | Amplicon (nt) | TA (°C) | tE (s) |
|---|---|---|---|
| 16S rDNA | 1500 | 58 | 50 |
| vanA | 1030 | 60 | 35 |
| sul2 | 722 | 65 | 25 |
| ermB | 405 | 63 | 20 |
| intl1 | 196 | 63 | 20 |
| mecA | 91 | 63 | 20 |
| Permeate Collection Time Interval (h) | Rejection Rate at 90% |
|---|---|
| 3.5 | 383 Da |
| 4.5 | 385 Da |
| 5.5 | 384 Da |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niestroj-Pahl, R.; Stelmaszyk, L.; ElSherbiny, I.M.A.; Abuelgasim, H.; Krug, M.; Staaks, C.; Birkholz, G.; Horn, H.; Li, T.; Dong, B.; et al. Performance of Layer-by-Layer-Modified Multibore® Ultrafiltration Capillary Membranes for Salt Retention and Removal of Antibiotic Resistance Genes. Membranes 2020, 10, 398. https://doi.org/10.3390/membranes10120398
Niestroj-Pahl R, Stelmaszyk L, ElSherbiny IMA, Abuelgasim H, Krug M, Staaks C, Birkholz G, Horn H, Li T, Dong B, et al. Performance of Layer-by-Layer-Modified Multibore® Ultrafiltration Capillary Membranes for Salt Retention and Removal of Antibiotic Resistance Genes. Membranes. 2020; 10(12):398. https://doi.org/10.3390/membranes10120398
Chicago/Turabian StyleNiestroj-Pahl, Robert, Lara Stelmaszyk, Ibrahim M. A. ElSherbiny, Hussein Abuelgasim, Michaela Krug, Christian Staaks, Greta Birkholz, Harald Horn, Tian Li, Bingzhi Dong, and et al. 2020. "Performance of Layer-by-Layer-Modified Multibore® Ultrafiltration Capillary Membranes for Salt Retention and Removal of Antibiotic Resistance Genes" Membranes 10, no. 12: 398. https://doi.org/10.3390/membranes10120398








