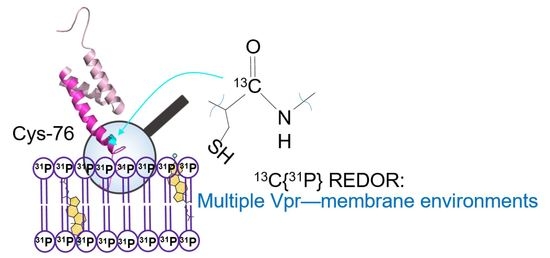
Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Vpr Protein Production and Purification
2.3. DPC Solubilized Vpr Sample for Sequence-Specific Resonance Assignments
2.4. Vpr Proteoliposomes Preparation
2.5. Calcein Release Assay
2.6. Spectroscopic Characterizations
3. Results and Discussions
3.1. Secondary Structure Analysis
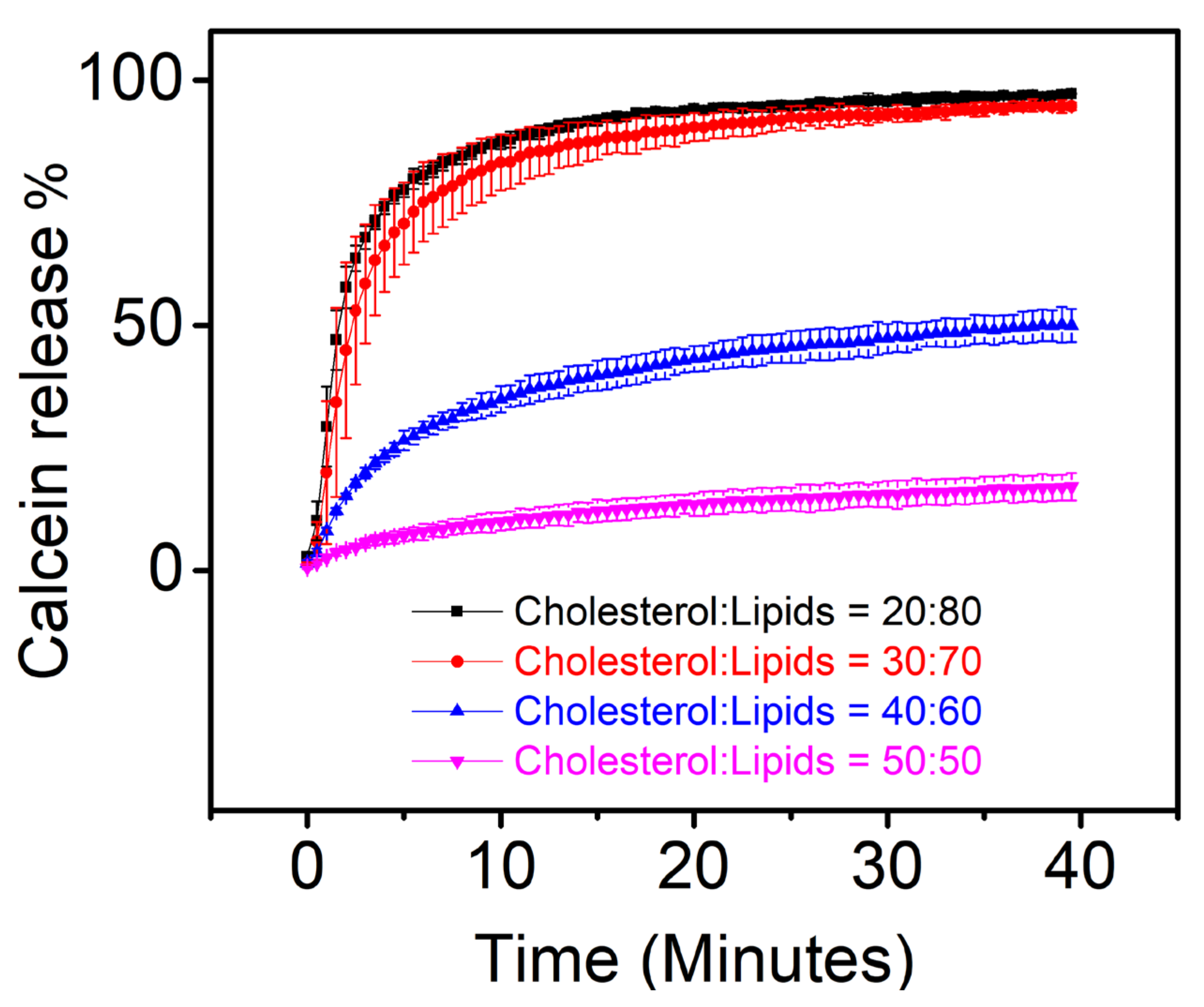
3.2. Calcein Release Assay
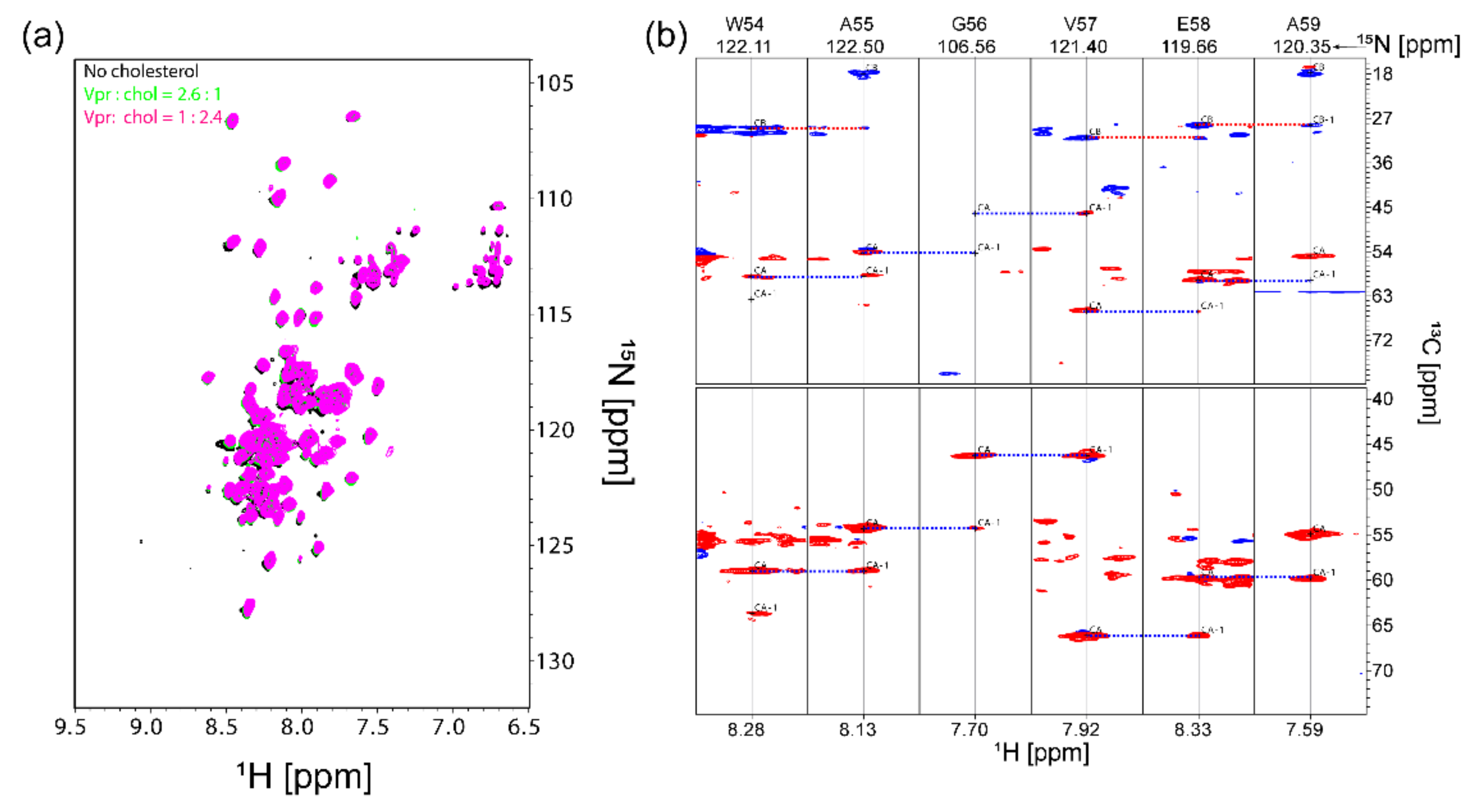
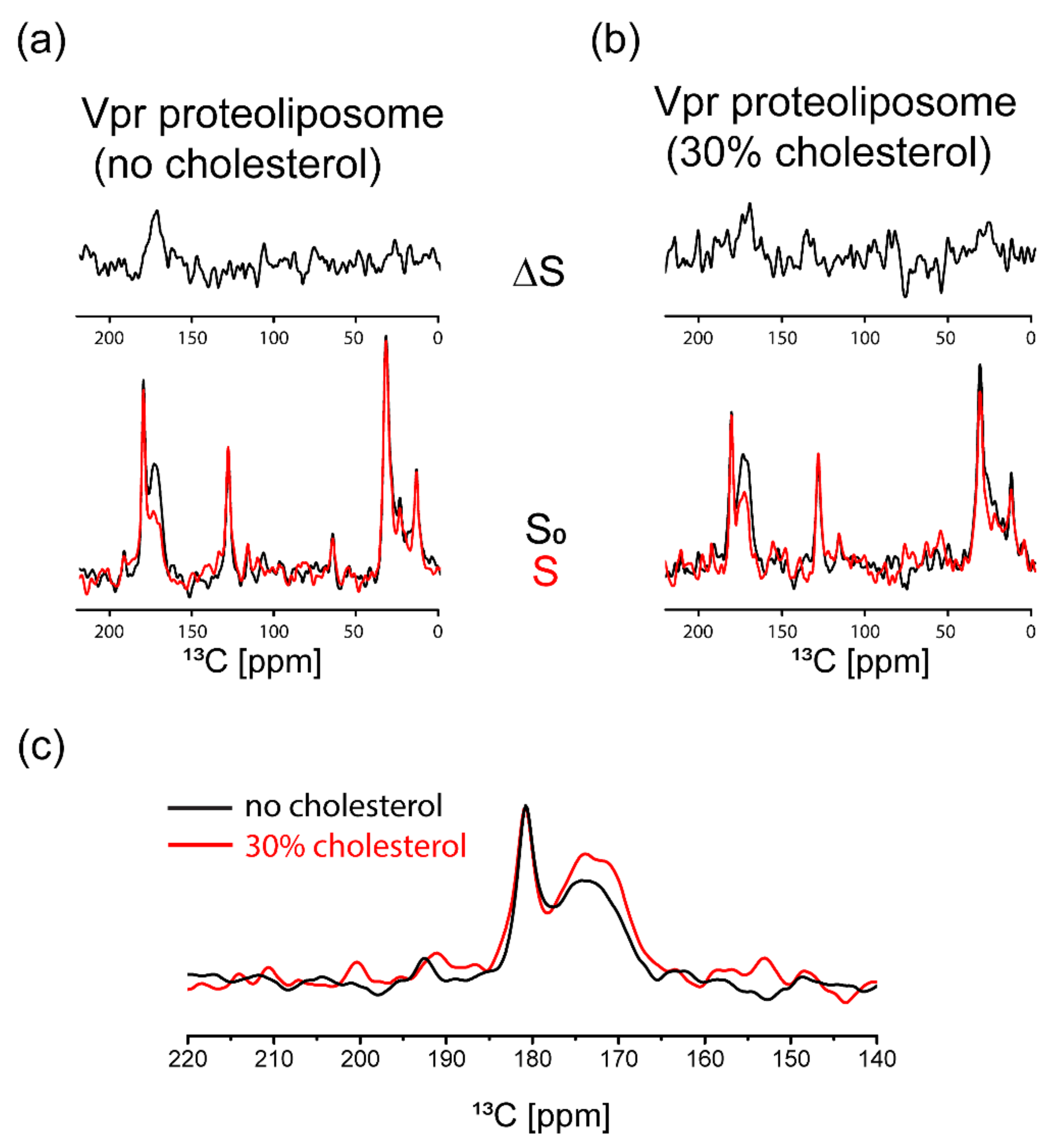
3.3. NMR Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Important Derivatives of Cholesterol Include Bile Salts and Steroid Hormones. In Biochemistry, 5th ed.; Freeman, W.H.: New York, NY, USA, 2002; Section 26.4. [Google Scholar]
- Takahashi, T.; Suzuki, T. Function of membrane rafts in viral lifecycles and host cellular response. Biochem. Res. Int. 2011, 2011, 245090. [Google Scholar] [CrossRef]
- Sun, X.; Whittaker, G.R. Role for influenza virus envelope cholesterol in virus entry and infection. J. Virol. 2003, 77, 12543–12551. [Google Scholar] [CrossRef]
- Tang, Q.; Liu, P.; Chen, M.; Qin, Y. Virion-Associated Cholesterol Regulates the Infection of Human Parainfluenza Virus Type 3. Viruses 2019, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Aloia, R.C.; Tian, H.; Jensen, F.C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 1993, 90, 5181–5185. [Google Scholar] [CrossRef] [PubMed]
- Brugger, B.; Glass, B.; Haberkant, P.; Leibrecht, I.; Wieland, F.T.; Krausslich, H.G. The HIV lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. USA 2006, 103, 2641–2646. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.M.; Crowe, S.M.; Mak, J. Lipid rafts and HIV-1: From viral entry to assembly of progeny virions. J. Clin. Virol. 2001, 22, 217–227. [Google Scholar] [CrossRef]
- Campbell, S.M.; Crowe, S.M.; Mak, J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS 2002, 16, 2253–2261. [Google Scholar] [CrossRef]
- Ono, A.; Waheed, A.A.; Freed, E.O. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology 2007, 360, 27–35. [Google Scholar] [CrossRef]
- Dick, R.A.; Goh, S.L.; Feigenson, G.W.; Vogt, V.M. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. USA 2012, 109, 18761–18766. [Google Scholar] [CrossRef]
- Yandrapalli, N.; Lubart, Q.; Tanwar, H.S.; Picart, C.; Mak, J.; Muriaux, D.; Favard, C. Self assembly of HIV-1 Gag protein on lipid membranes generates PI(4,5)P2/Cholesterol nanoclusters. Sci. Rep. 2016, 6, 39332. [Google Scholar] [CrossRef]
- Meher, G.; Sinha, S.; Pattnaik, G.P.; Ghosh Dastidar, S.; Chakraborty, H. Cholesterol Modulates Membrane Properties and the Interaction of gp41 Fusion Peptide To Promote Membrane Fusion. J. Phys. Chem. B 2019, 123, 7113–7122. [Google Scholar] [CrossRef]
- Kwon, B.; Mandal, T.; Elkins, M.R.; Oh, Y.; Cui, Q.; Hong, M. Cholesterol Interaction with the Trimeric HIV Fusion Protein gp41 in Lipid Bilayers Investigated by Solid-State NMR Spectroscopy and Molecular Dynamics Simulations. J. Mol. Biol. 2020, 432, 4705–4721. [Google Scholar] [CrossRef]
- Popov, S.; Rexach, M.; Zybarth, G.; Reiling, N.; Lee, M.A.; Ratner, L.; Lane, C.M.; Moore, M.S.; Blobel, G.; Bukrinsky, M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998, 17, 909–917. [Google Scholar] [CrossRef]
- Kamata, M.; Nitahara-Kasahara, Y.; Miyamoto, Y.; Yoneda, Y.; Aida, Y. Importin-alpha promotes passage through the nuclear pore complex of human immunodeficiency virus type 1 Vpr. J. Virol. 2005, 79, 3557–3564. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jenkins, Y.; McEntee, M.; Weis, K.; Greene, W.C. Characterization of HIV-1 vpr nuclear import: Analysis of signals and pathways. J. Cell Biol. 1998, 143, 875–885. [Google Scholar] [CrossRef]
- He, J.; Choe, S.; Walker, R.; Di Marzio, P.; Morgan, D.O.; Landau, N.R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 1995, 69, 6705–6711. [Google Scholar] [CrossRef] [PubMed]
- Sabbah, E.N.; Druillennec, S.; Morellet, N.; Bouaziz, S.; Kroemer, G.; Roques, B.P. Interaction between the HIV-1 protein Vpr and the adenine nucleotide translocator. Chem. Biol. Drug Des. 2006, 67, 145–154. [Google Scholar] [CrossRef]
- Qiao, H.; McMillan, J.R. Gelsolin segment 5 inhibits HIV-induced T-cell apoptosis via Vpr-binding to VDAC. FEBS Lett. 2007, 581, 535–540. [Google Scholar] [CrossRef]
- Yasuda, J.; Miyao, T.; Kamata, M.; Aida, Y.; Iwakura, Y. T cell apoptosis causes peripheral T cell depletion in mice transgenic for the HIV-1 vpr gene. Virology 2001, 285, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Kogan, M.; Rappaport, J. HIV-1 accessory protein Vpr: Relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 2011, 8, 25. [Google Scholar] [CrossRef]
- Guenzel, C.A.; Herate, C.; Benichou, S. HIV-1 Vpr-a still "enigmatic multitasker". Front. Microbiol. 2014, 5, 127. [Google Scholar] [CrossRef]
- Piller, S.C.; Ewart, G.D.; Premkumar, A.; Cox, G.B.; Gage, P.W. Vpr protein of human immunodeficiency virus type 1 forms cation-selective channels in planar lipid bilayers. Proc. Natl. Acad. Sci. USA 1996, 93, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.S.; Lackner, A.A.; Lang, S.M.; Simon, M.A.; Sehgal, P.K.; Daniel, M.D.; Desrosiers, R.C. Progression to Aids in the Absence of a Gene for Vpr or Vpx. J. Virol. 1995, 69, 2378–2383. [Google Scholar] [CrossRef]
- Rice, A.P.; Kimata, J.T. Subversion of Cell Cycle Regulatory Mechanisms by HIV. Cell Host Microbe 2015, 17, 736–740. [Google Scholar] [CrossRef]
- Gross, D.A.; Leborgne, C.; Chappert, P.; Masurier, C.; Leboeuf, M.; Monteilhet, V.; Boutin, S.; Lemonnier, F.A.; Davoust, J.; Kichler, A. Induction of tumor-specific CTL responses using the C-terminal fragment of Viral protein R as cell penetrating peptide. Sci. Rep. 2019, 9, 3937. [Google Scholar] [CrossRef]
- Henklein, P.; Bruns, K.; Sherman, M.P.; Tessmer, U.; Licha, K.; Kopp, J.; de Noronha, C.M.; Greene, W.C.; Wray, V.; Schubert, U. Functional and structural characterization of synthetic HIV-1 Vpr that transduces cells, localizes to the nucleus, and induces G2 cell cycle arrest. J. Biol. Chem. 2000, 275, 32016–32026. [Google Scholar] [CrossRef]
- Coeytaux, E.; Coulaud, D.; Le Cam, E.; Danos, O.; Kichler, A. The cationic amphipathic alpha-helix of HIV-1 viral protein R (Vpr) binds to nucleic acids, permeabilizes membranes, and efficiently transfects cells. J. Biol. Chem. 2003, 278, 18110–18116. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.B.; Liu, C.H.; Chen, Y.T.; Yu, T.Y. The Study of HIV-1 Vpr-Membrane and Vpr-hVDAC-1 Interactions by Graphene Field-Effect Transistor Biosensors. ACS Appl. Bio Mater. 2020, 3, 6351–6357. [Google Scholar] [CrossRef]
- Ono, A.; Freed, E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 2001, 98, 13925–13930. [Google Scholar] [CrossRef] [PubMed]
- Kalyana Sundaram, R.V.; Li, H.; Bailey, L.; Rashad, A.A.; Aneja, R.; Weiss, K.; Huynh, J.; Bastian, A.R.; Papazoglou, E.; Abrams, C.; et al. Impact of HIV-1 Membrane Cholesterol on Cell-Independent Lytic Inactivation and Cellular Infectivity. Biochemistry 2016, 55, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Muller, B.; Tessmer, U.; Schubert, U.; Krausslich, H.G. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than Gag and is phosphorylated in infected cells. J. Virol. 2000, 74, 9727–9731. [Google Scholar] [CrossRef]
- Fritz, J.V.; Dujardin, D.; Godet, J.; Didier, P.; De Mey, J.; Darlix, J.L.; Mely, Y.; de Rocquigny, H. HIV-1 Vpr oligomerization but not that of Gag directs the interaction between Vpr and Gag. J. Virol. 2010, 84, 1585–1596. [Google Scholar] [CrossRef]
- Solbak, S.M.; Reksten, T.R.; Hahn, F.; Wray, V.; Henklein, P.; Henklein, P.; Halskau, O.; Schubert, U.; Fossen, T. HIV-1 p6—A structured to flexible multifunctional membrane-interacting protein. Biochim. Biophys. Acta 2013, 1828, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Nicol, F.; Nir, S.; Szoka, F.C., Jr. Effect of cholesterol and charge on pore formation in bilayer vesicles by a pH-sensitive peptide. Biophys. J. 1996, 71, 3288–3301. [Google Scholar] [CrossRef]
- Pae, J.; Saalik, P.; Liivamagi, L.; Lubenets, D.; Arukuusk, P.; Langel, U.; Pooga, M. Translocation of cell-penetrating peptides across the plasma membrane is controlled by cholesterol and microenvironment created by membranous proteins. J. Control. Release 2014, 192, 103–113. [Google Scholar] [CrossRef]
- Keller, R. The Computer-Aided Resonance Assignment Tutorial; Cantina Verlag: Goldau, Switzerland, 2004; CARA can be download from cara.nmr.ch. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Morellet, N.; Bouaziz, S.; Petitjean, P.; Roques, B.P. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 2003, 327, 215–227. [Google Scholar] [CrossRef]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Takahashi, H.; Nakanishi, T.; Kami, K.; Arata, Y.; Shimada, I. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 2000, 7, 220–223. [Google Scholar] [PubMed]
- Jia, L.H.; Liang, S.; Sackett, K.; Xie, L.; Ghosh, U.; Weliky, D.P. REDOR solid-state NMR as a probe of the membrane locations of membrane-associated peptides and proteins. J. Magn. Reson. 2015, 253, 154–165. [Google Scholar] [CrossRef][Green Version]
- Ratnayake, P.U.; Sackett, K.; Nethercott, M.J.; Weliky, D.P. pH-dependent vesicle fusion induced by the ectodomain of the human immunodeficiency virus membrane fusion protein gp41: Two kinetically distinct processes and fully-membrane-associated gp41 with predominant beta sheet fusion peptide conformation. Biochim. Biophys. Acta 2015, 1848, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Schmick, S.D.; Weliky, D.P. Major antiparallel and minor parallel beta sheet populations detected in the membrane-associated human immunodeficiency virus fusion peptide. Biochemistry 2010, 49, 10623–10635. [Google Scholar] [CrossRef]
- Morcombe, C.R.; Zilm, K.W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003, 162, 479–486. [Google Scholar] [CrossRef]
- Marsh, D. Protein modulation of lipids, and vice-versa, in membranes. Biochim. Biophys. Acta 2008, 1778, 1545–1575. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Modulation of membrane function by cholesterol. Biochimie 1991, 73, 1303–1310. [Google Scholar] [CrossRef]
- Laganowsky, A.; Reading, E.; Allison, T.M.; Ulmschneider, M.B.; Degiacomi, M.T.; Baldwin, A.J.; Robinson, C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Robertson, J.L. The Role of the Membrane in Transporter Folding and Activity. J. Mol. Biol. 2021, 433, 167103. [Google Scholar] [CrossRef]
- Jodaitis, L.; van Oene, T.; Martens, C. Assessing the Role of Lipids in the Molecular Mechanism of Membrane Proteins. Int. J. Mol. Sci. 2021, 22, 7267. [Google Scholar] [CrossRef]
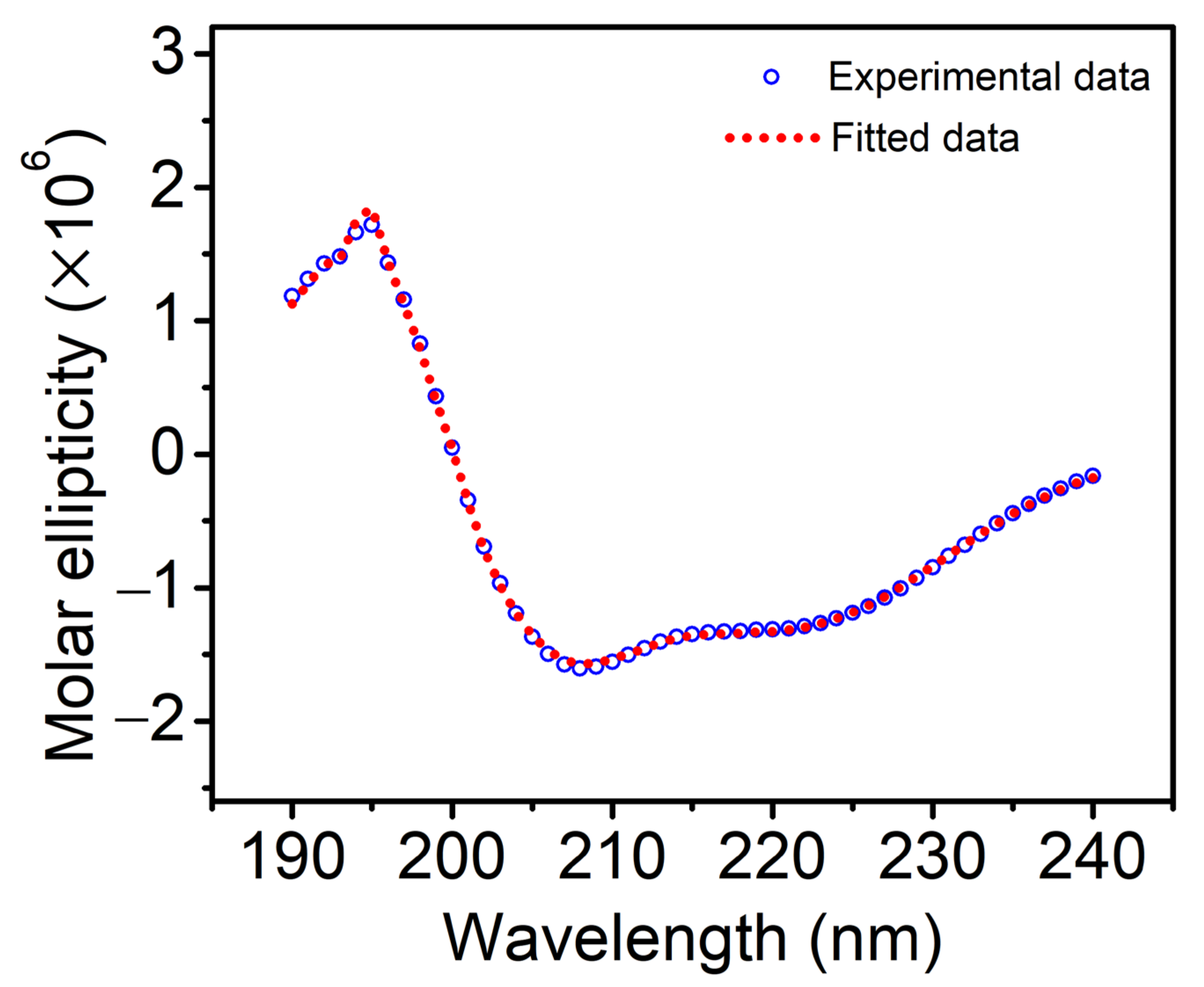
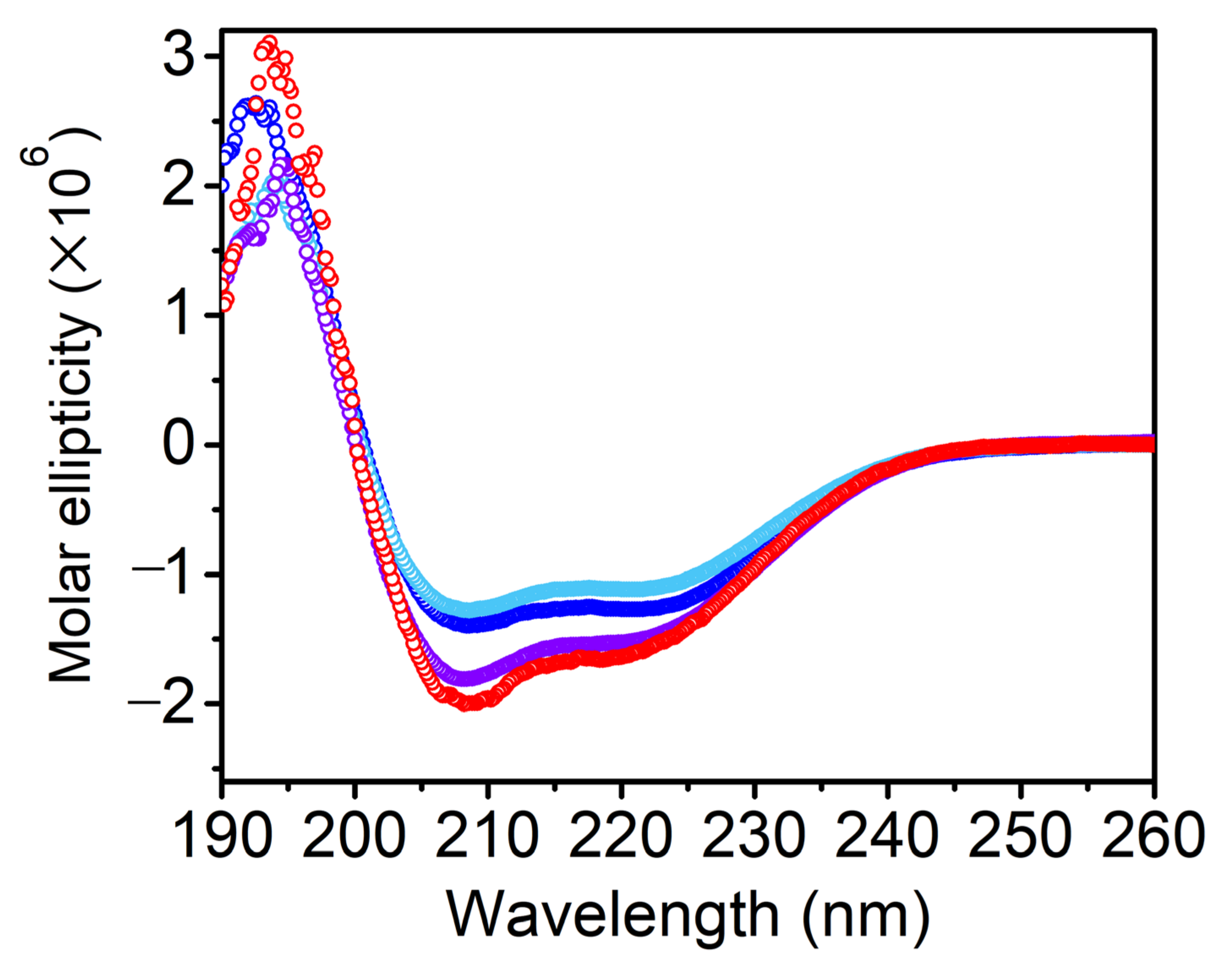
| Helix 1 | Helix 2 | Strand 1 | Strand 2 | Turns | Unordered |
|---|---|---|---|---|---|
| 0.31 | 0.20 | 0.03 | 0.04 | 0.11 | 0.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-H.; Huang, S.-J.; Yu, T.-Y. Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane. Membranes 2021, 11, 784. https://doi.org/10.3390/membranes11100784
Liu C-H, Huang S-J, Yu T-Y. Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane. Membranes. 2021; 11(10):784. https://doi.org/10.3390/membranes11100784
Chicago/Turabian StyleLiu, Chun-Hao, Shing-Jong Huang, and Tsyr-Yan Yu. 2021. "Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane" Membranes 11, no. 10: 784. https://doi.org/10.3390/membranes11100784
APA StyleLiu, C.-H., Huang, S.-J., & Yu, T.-Y. (2021). Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane. Membranes, 11(10), 784. https://doi.org/10.3390/membranes11100784