The Nanoscopic Organization of Synapse Structures: A Common Basis for Cell Communication
Abstract
1. Introduction
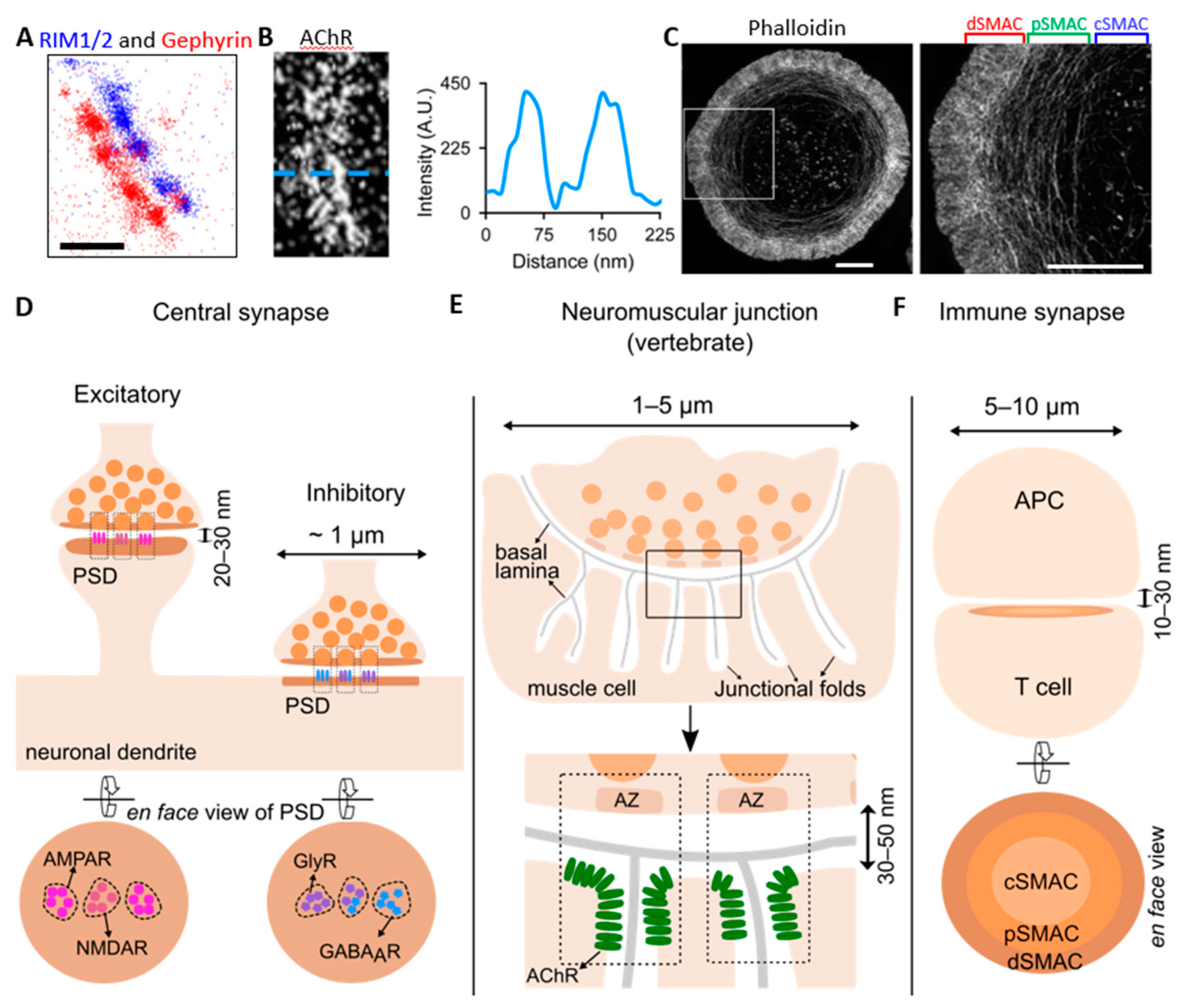
2. The Neuronal Synapse
2.1. Central Synapses
2.1.1. Laminar Organization at Post-Synaptic Density
2.1.2. Sub-Synaptic Domains (SSDs) in the Post-Synaptic Density
2.1.3. Sub-Synaptic Domains (SSDs) at the Pre-Synaptic Compartment
2.1.4. Trans-Synaptic Nanocolumns
2.2. Neuromuscular Junctions (NMJs)
3. Immune Synapses
4. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Tansey, E.M. Not committing barbarisms: Sherrington and the synapse, 1897. Brain Res. Bull. 1997, 44, 211–212. [Google Scholar] [CrossRef]
- Palay, S.L. Synapses in the central nervous system. J. Biophys. Bochem. Cytol 1956, 2, 193–207. [Google Scholar] [CrossRef]
- Gray, E.G. Electron microscopy of synaptic contants on dendrite spines of the cerebral cortex. Nature 1959, 183, 1592–1593. [Google Scholar] [CrossRef]
- Tsuji, S. René Couteaux (1909–1999) and the morphological identification of synapses. Biol. Cell 2006, 98, 503–509. [Google Scholar] [CrossRef]
- Norcross, M.A. A synaptic basis for T-lymphocyte activation. Ann. Immunol. 1984, 135D, 113–134. [Google Scholar] [CrossRef]
- Paul, W.E.; Seder, R.A. Lymphocyte responses and cytokines. Cell 1994, 76, 241–251. [Google Scholar] [CrossRef]
- Davis, D.M.; Chiu, I.; Fassett, M.; Cohen, G.B.; Mandelboim, O.; Strominger, J.L. The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. USA 1999, 96, 15062–15067. [Google Scholar] [CrossRef]
- Dustin, M.L. Signaling at neuro/immune synapses. J. Clin. Investig. 2012, 122, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L.; Colman, D.R. Neural and immunological synaptic relations. Science 2002, 298, 785–789. [Google Scholar] [CrossRef]
- Nishimune, H.; Shigemoto, K. Practical anatomy of the neuromuscular junction in health and disease. Neurol. Clin. 2018, 36, 231–240. [Google Scholar] [CrossRef]
- Biberfeld, P.; Johansson, A. Contact areas of cytotoxic lymphocytes and target cells. Exp. Cell Res. 1975, 94, 79–87. [Google Scholar] [CrossRef]
- McCann, F.E.; Vanherberghen, B.; Eleme, K.; Carlin, L.M.; Newsam, R.J.; Goulding, D.; Davis, D.M. The Size of the Synaptic Cleft and Distinct Distributions of Filamentous Actin, Ezrin, CD43, and CD45 at Activating and Inhibitory Human NK Cell Immune Synapses. J. Immunol. 2003, 170, 2862–2870. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.M.; Weinberg, R.J. Ultrastructure of Synapses in the Mammalian Brain. Cold Spring Harb Perspect Biol 2012, 4, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Specht, C.G. Subsynaptic domains in super-resolution microscopy: The treachery of images. Front. Mol. Neurosci. 2019, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Slater, C.R. The structure of human neuromuscular junctions: Some unanswered molecular questions. Int. J. Mol. Sci. 2017, 18, 2183. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A.; Harrison, C.; Eaton, S.L.; Llavero Hurtado, M.; Graham, L.C.; Alkhammash, L.; Oladiran, O.A.; Gale, A.; Lamont, D.J.; Simpson, H.; et al. Cellular and Molecular Anatomy of the Human Neuromuscular Junction. Cell Rep. 2017, 21, 2348–2356. [Google Scholar] [CrossRef]
- Curado, S.; Kumari, S.; Dustin, M.L. Cell biology meets physiology: Functional organization of vertebrate plasma membranes—The immunological synapse. In Current Topics in Membranes; Elsevier Inc.: New York, NY, USA, 2013; Volume 72, pp. 313–346. ISBN 9780124170278. [Google Scholar]
- Yang, X.; Corronc, H.L.; Legendre, P.; Triller, A.; Specht, C.G. Differential homeostatic regulation of glycinergic and GABAergic nanocolumns at mixed inhibitory synapses. bioRxiv 2020. [Google Scholar] [CrossRef]
- York, A.L.; Zheng, J.Q. Super-resolution microscopy reveals a nanoscale organization of acetylcholine receptors for trans-synaptic alignment at neuromuscular synapses. eNeuro 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Murugesan, S.; Hong, J.; Yi, J.; Li, D.; Beach, J.R.; Shao, L.; Meinhardt, J.; Madison, G.; Wu, X.; Betzig, E.; et al. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. J. Cell Biol. 2016, 215, 383–399. [Google Scholar] [CrossRef]
- Dustin, M.L. The immunological synapse. In Handbook of Cell Signaling, 2nd ed.; Elsevier Inc.: New York, NY, USA, 2010; Volume 1, pp. 71–75. ISBN 9780123741455. [Google Scholar]
- Griffiths, G.M.; Tsun, A.; Stinchcombe, J.C. The immunological synapse: A focal point for endocytosis and exocytosis. J. Cell Biol. 2010, 189, 399–406. [Google Scholar] [CrossRef]
- Emes, R.D.; Grant, S.G.N. Evolution of Synapse Complexity and Diversity. Annu. Rev. Neurosci. 2012, 35, 111–131. [Google Scholar] [CrossRef] [PubMed]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Sigal, Y.M.; Zhou, R.; Zhuang, X. Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361, 880–887. [Google Scholar] [CrossRef]
- Igarashi, M.; Nozumi, X.M.; Wu, L.; Zanacchi, F.C.; Istva, X.; Xu, P.; Zhang, M.; Xue, F.; Boyden, E. New observations in neuroscience using superresolution microscopy. J. Neurosci. 2018, 38, 9459–9467. [Google Scholar] [CrossRef]
- Gustafsson, M.G.L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef] [PubMed]
- Hell, S.W.; Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett 1994, 19, 780. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Stochastic optical reconstruction miscroscopy (STORM) provides sub-diffraction-limit image resolution. Nat. Methods 2006, 3, 793–795. [Google Scholar] [CrossRef]
- Heilemann, M.; van de Linde, S.; Schuttpelz, M.; Kasper, R.; Seefeldt, B.; Mukherjee, A.; Tinnefeld, P.; Sauer, M. Subdiffraction resolution imaging with conventional fluorescent probes. Angew. Chem Int. Ed. 2008, 47, 6172–6176. [Google Scholar] [CrossRef]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1646. [Google Scholar] [CrossRef]
- Hess, S.T.; Girirajan, T.P.K.K.; Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef]
- Sharonov, A.; Hochstrasser, R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA 2006, 103, 18911–18916. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Proppert, S.; Sauer, M. Eight years of single-molecule localization microscopy. Histochem. Cell Biol. 2014, 141, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Heuser, J.E.; Reese, T.S. Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J. Cell Biol. 1973, 57, 315–344. [Google Scholar] [CrossRef] [PubMed]
- Govind, C.K.; Meiss, D.E. Quantitative comparison of low- and high-output neuromuscular synapses from a motoneuron of the lobster (Homarus americanus). Cell Tissue Res. 1979, 198, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.L.; Winslow, J.L.; Govind, C.K.; Atwood, H.L. Synaptic structural complexity as a factor enhancing probability of calcium-mediated transmitter release. J. Neurophysiol. 1996, 75, 2451–2466. [Google Scholar] [CrossRef]
- Valtschanoff, J.G.; Weinberg, R.J. Laminar organization of the NMDA receptor complex within the postsynaptic density. J. Neurosci. 2001, 21, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.D.; Chen, X.; Vinade, L.; Dosemeci, A.; Lisman, J.E.; Reese, T.S. Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J. Neurosci. 2003, 23, 11270–11278. [Google Scholar] [CrossRef]
- Rostaing, P.; Real, E.; Siksou, L.; Lechaire, J.P.; Boudier, T.; Boeckers, T.M.; Gertler, F.; Gundelfinger, E.D.; Triller, A.; Marty, S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur. J. Neurosci. 2006, 24, 3463–3474. [Google Scholar] [CrossRef]
- Kuriu, T.; Inoue, A.; Bito, H.; Sobue, K.; Okabe, S. Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J. Neurosci. 2006, 26, 7693–7706. [Google Scholar] [CrossRef]
- Dani, A.; Huang, B.; Bergan, J.; Dulac, C.; Zhuang, X. Super-resolution Imaging of Chemical Synapses in the Brain. Neuron 2010, 68, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Winters, C.; Azzam, R.; Li, X.; Galbraith, J.A.; Leapman, R.D.; Reese, T.S. Organization of the core structure of the postsynaptic density. Proc. Natl. Acad. Sci. USA 2008, 105, 4453–4458. [Google Scholar] [CrossRef] [PubMed]
- DeGiorgis, J.A.; Galbraith, J.A.; Dosemeci, A.; Chen, X.; Reese, T.S. Distribution of the scaffolding proteins PSD-95, PSD-93, and SAP97 in isolated PSDs. Brain Cell Biol. 2006, 35, 239–250. [Google Scholar] [CrossRef]
- Macgillavry, H.D.; Song, Y.; Raghavachari, S.; Blanpied, T.A. Nanoscale Scaffolding Domains within the Postsynaptic Density Concentrate Synaptic AMPA Receptors. Neuron 2013, 78, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Specht, C.G.; Izeddin, I.; Rodriguez, P.C.; El Beheiry, M.; Rostaing, P.; Darzacq, X.; Dahan, M.; Triller, A.; Supe, N. Quantitative nanoscopy of inhibitory synapses: Counting gephyrin molecules and receptor binding sites. Neuron 2013, 79, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Pennacchietti, F.; Vascon, S.; Nieus, T.; Rosillo, C.; Das, S.; Tyagarajan, S.K.; Diaspro, A.; Del Bue, A.; Petrini, E.M.; Barberis, A.; et al. Nanoscale Molecular Reorganization of the Inhibitory Postsynaptic Density Is a Determinant of GABAergic Synaptic Potentiation. J. Neurosci. 2017, 37, 1747–1756. [Google Scholar] [CrossRef]
- Dzyubenko, E.; Rozenberg, A.; Hermann, D.M.; Faissner, A. Colocalization of synapse marker proteins evaluated by STED-microscopy reveals patterns of neuronal synapse distribution in vitro. J. Neurosci. Methods 2016, 273, 149–159. [Google Scholar] [CrossRef]
- Hruska, M.; Henderson, N.; Le Marchand, S.J.; Jafri, H.; Dalva, M.B. Synaptic nanomodules underlie the organization and plasticity of spine synapses. Nat. Neurosci. 2018, 21, 671–682. [Google Scholar] [CrossRef]
- Broadhead, M.J.; Horrocks, M.H.; Zhu, F.; Muresan, L.; Benavides-Piccione, R.; DeFelipe, J.; Fricker, D.; Kopanitsa, M.V.; Duncan, R.R.; Klenerman, D.; et al. PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Masch, J.-M.; Steffens, H.; Fischer, J.; Engelhardt, J.; Hubrich, J.; Keller-Findeisen, J.; D’Este, E.; Urban, N.T.; Grant, S.G.N.; Sahl, S.J.; et al. Robust nanoscopy of a synaptic protein in living mice by organic-fluorophore labeling. Proc. Natl. Acad. Sci. USA 2018, 115, 201807104. [Google Scholar] [CrossRef] [PubMed]
- Wegner, W.; Mott, A.C.; Grant, S.G.N.; Steffens, H.; Willig, K.I. In vivo STED microscopy visualizes PSD95 sub-structures and morphological changes over several hours in the mouse visual cortex. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Crosby, K.C.; Gookin, S.E.; Garcia, J.D.; Hahm, K.M.; Dell’Acqua, M.L.; Smith, K.R. Nanoscale subsynaptic domains underlie the organization of the inhibitory synapse. Cell Rep. 2019, 26, 3284–3297. [Google Scholar] [CrossRef]
- Nair, D.; Hosy, E.; Petersen, J.D.; Constals, A.; Giannone, G.; Choquet, D.; Sibarita, J.J.-B. Super-Resolution Imaging Reveals That AMPA Receptors Inside Synapses Are Dynamically Organized in Nanodomains Regulated by PSD95. J. Neurosci. 2013, 33, 13204–13224. [Google Scholar] [CrossRef]
- Triller, A.; Cluzeaud, F.; Pfeiffer, F.; Betz, H.; Korn, H. Distribution of glycine receptors at central synapses: An immunoelectron microscopy study. J. Cell Biol. 1985, 101, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, J.; Goncalves, J.; Bartol, T.M.; Camus, C.; Camus, C.; Levet, F.; Levet, F.; Levet, F.; Menegolla, A.P.; Menegolla, A.P.; et al. Nanoscale co-organization and coactivation of AMPAR, NMDAR, and mGluR at excitatory synapses. Proc. Natl. Acad. Sci. USA 2020, 117, 14503–14511. [Google Scholar] [CrossRef] [PubMed]
- Kharazia, V.N.; Weinberg, R.J. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci. Lett Lett 1997, 238, 41–44. [Google Scholar] [CrossRef]
- Racca, C.; Stephenson, F.A.; Streit, P.; Roberts, J.D.B.; Somogyi, P. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J. Neurosci. 2000, 20, 2512–2522. [Google Scholar] [CrossRef]
- Kennedy, M.B. Signal-processing machines at the postsynaptic density. Science 2000, 290, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Luján, R.; Nusser, Z.; Roberts, J.D.B.; Shigemoto, R.; Somogyi, P. Perisynaptic location of metabotropic glutamate receptors mGluR1 and mGluR5 on dendrites and dendritic spines in the rat hippocampus. Eur. J. Neurosci. 1996, 8, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Cognet, L.; Groc, L.; Lounis, B.; Choquet, D. Multiple routes for glutamate receptor trafficking: Surface diffusion and membrane traffic cooperate to bring receptors to synapses. Sci. STKE 2006, 2006, 1–5. [Google Scholar] [CrossRef]
- Triller, A.; Choquet, D. New Concepts in Synaptic Biology Derived from Single-Molecule Imaging. Neuron 2008, 59, 359–374. [Google Scholar] [CrossRef]
- Siddig, S.; Aufmkolk, S.; Doose, S.; Jobin, M.L.; Werner, C.; Sauer, M.; Calebiro, D. Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci. Adv. 2020, 6, eaay7193. [Google Scholar] [CrossRef] [PubMed]
- Triller, A.; Cluzeaud, F.; Korn, H. Gamma-Aminobutyric Acid-containing Terminals Can Be Apposed to Glycine Receptors at Central Synapses. J. Cell Biol. 1987, 104, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.J.; Watt, C.; Spike, R.C.; Sieghart, W. Colocalization of GABA, glycine, and their receptors at synapses in the rat spinal cord. J. Neurosci. 1996, 16, 974–982. [Google Scholar] [CrossRef]
- Jonas, P.; Bischofberger, J.; Sandkühler, J.; Jonas, P.; Josef Bischofberger, J.S. Corelease of two fast neurotransmitters at a central synapse. Science 1998, 281, 419–424. [Google Scholar] [CrossRef]
- Aubrey, K.R.; Supplisson, S. Heterogeneous Signaling at GABA and glycine co-releasing terminals. Front. Synaptic Neurosci. 2018, 10, 1–13. [Google Scholar] [CrossRef]
- Südhof, T.C. The Presynaptic Active Zone. Neuron 2012, 75, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Rizo, J. Mechanism of neurotransmitter release coming into focus. Protein Sci. 2018, 27, 1364–1391. [Google Scholar] [CrossRef]
- Nosov, G.; Kahms, M.; Klingauf, J. The Decade of Super-Resolution Microscopy of the Presynapse. Front Synaptic Neurosci 2020, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bademosi, A.T.; Lauwers, E.; Padmanabhan, P.; Odierna, L.; Chai, Y.J.; Papadopulos, A.; Goodhill, G.J.; Verstreken, P.; Van Swinderen, B.; Meunier, F.A. In vivo single molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat. Commun. 2017, 8, 13660. [Google Scholar] [CrossRef] [PubMed]
- Bademosi, A.T.; Steeves, J.; Karunanithi, S.; Zalucki, O.H.; Gormal, R.S.; Liu, S.; Lauwers, E.; Verstreken, P.; Anggono, V.; Meunier, F.A.; et al. Trapping of Syntaxin1a in Presynaptic Nanoclusters by a Clinically Relevant General Anesthetic. Cell Rep. 2018, 22, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Hosy, E.; Voigt, A.; Heine, M.; Schneider, R.; Hosy, E.; Kohl, J.; Klueva, J.; Choquet, D.; Thomas, U.; et al. Mobility of Calcium Channels in the Presynaptic Membrane. Neuron 2015, 86, 672–679. [Google Scholar] [CrossRef]
- Heck, J.; Parutto, P.; Ciuraszkiewicz, A.; Bikbaev, A.; Freund, R.; Mitlöhner, J.; Andres-Alonso, M.; Fejtova, A.; Holcman, D.; Heine, M. Transient Confinement of CaV2.1 Ca2+-Channel Splice Variants Shapes Synaptic Short-Term Plasticity. Neuron 2019, 103, 66–79.e12. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Kaufmann, W.A.; Malagon, G.; Gomez, L.; Tabuchi, K.; Watanabe, M.; Shigemoto, R.; Marty, A. Numbers of presynaptic Ca2+ channel clusters match those of functionally defined vesicular docking sites in single central synapses. Proc. Natl. Acad. Sci. USA 2017, 114, E5246–E5255. [Google Scholar] [CrossRef] [PubMed]
- Tang, A.H.; Chen, H.; Li, T.P.; Metzbower, S.R.; MacGillavry, H.D.; Blanpied, T.A. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 2016, 536, 210–214. [Google Scholar] [CrossRef]
- Glebov, O.O.; Jackson, R.E.; Winterflood, C.M.; Owen, D.M.; Barker, E.A.; Doherty, P.; Ewers, H.; Burrone, J.; Winter, C.M.; Owen, D.M.; et al. Nanoscale Structural Plasticity of the Active Zone Matrix Modulates Presynaptic Function. Cell Rep. 2017, 18, 2715–2728. [Google Scholar] [CrossRef]
- Willig, K.I.; Rizzoli, S.O.; Westphal, V.; Jahn, R.; Hell, S.W. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature 2006, 440, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Biederer, T.; Kaeser, P.S.; Blanpied, T.A. Trans-cellular nano-alignment of synaptic function. Neuron 2017, 96, 680–696. [Google Scholar] [CrossRef]
- Chen, H.; Tang, A.H.; Blanpied, T.A. Subsynaptic spatial organization as a regulator of synaptic strength and plasticity. Curr. Opin. Neurobiol. 2018, 51, 147–153. [Google Scholar] [CrossRef]
- Liu, K.K.L.; Hagan, M.F.; Lisman, J.E. Gradation (approx. 10 size states) of synaptic strength by quantal addition of structural modules. Philos Trans. R Soc. L. B Biol. Sci. 2017, 372, 20160328. [Google Scholar] [CrossRef]
- Scheefhals, N.; MacGillavry, H.D. Functional organization of postsynaptic glutamate receptors. Mol. Cell Neurosci. 2018, 91, 82–94. [Google Scholar] [CrossRef]
- Choquet, D.; Triller, A. The Dynamic Synapse. Neuron 2013, 80, 691–703. [Google Scholar] [CrossRef]
- Zuber, B.; Nikonenko, I.; Klauser, P.; Muller, D.; Dubochet, J. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. USA 2005, 102, 19192–19197. [Google Scholar] [CrossRef]
- High, B.; Cole, A.A.; Chen, X.; Reese, T.S. Electron microscopic tomography reveals discrete transcleft elements at excitatory and inhibitory synapses. Front. Synaptic Neurosci. 2015, 7, 1–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tao, C.-L.; Liu, Y.-T.; Sun, R.; Zhang, B.; Qi, L.; Shivakoti, S.; Tian, C.-L.; Zhang, P.; Lau, P.-M.; Zhou, Z.H.; et al. Differentiation and characterization of excitatory and inhibitory synapses by cryo-electron tomography and correlative microscopy. J. Neurosci. 2018, 38, 1548-17. [Google Scholar] [CrossRef]
- Rudenko, G. Neurexins—Versatile molecular platforms in the synaptic cleft. Curr. Opin. Struct. Biol. 2019, 54, 112–121. [Google Scholar] [CrossRef]
- Südhof, T.C. Neuroligins and Neurexins Link Synaptic Function to Cognitive Disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.T.; Compans, B.; Letellier, M.; Bartol, T.M.; Grillo-Bosch, D.; Sejnowski, T.J.; Sainlos, M.; Choquet, D.; Thoumine, O.; Hosy, E.; et al. Pre-post synaptic alignment through neuroligin-1 tunes synaptic transmission efficiency. Elife 2018, 7, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Chamma, I.; Letellier, M.; Butler, C.; Tessier, B.; Lim, K.-H.; Gauthereau, I.; Choquet, D.; Sibarita, J.-B.; Park, S.; Sainlos, M.; et al. Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Chamma, I.; Levet, F.; Sibarita, J.-B.; Sainlos, M.; Thoumine, O. Nanoscale organization of synaptic adhesion proteins revealed by single-molecule localization microscopy. Neurophotonics 2016, 3, 041810. [Google Scholar] [CrossRef] [PubMed]
- Trotter, J.H.; Hao, J.; Maxeiner, S.; Tsetsenis, T.; Liu, Z.; Zhuang, X.; Südhof, T.C. Synaptic neurexin-1 assembles into dynamically regulated active zone nanoclusters. J. Cell Biol. 2019, 218, 2677–2698. [Google Scholar] [CrossRef]
- Missler, M.; Südhof, T.C.; Biederer, T. Synaptic cell adhesion. Cold Spring Harb. Perspect. Biol. 2012, 4, a005694. [Google Scholar] [CrossRef] [PubMed]
- Perez de Arce, K.; Schrod, N.; Metzbower, S.W.R.; Allgeyer, E.; Kong, G.K.W.; Tang, A.-H.; Krupp, A.J.; Stein, V.; Liu, X.; Bewersdorf, J.; et al. Topographic mapping of the synaptic cleft into adhesive nanodomains. Neuron 2015, 88, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic Mapping of Mitochondria in Living Cells via Spatially- Restricted Enzymatic Tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Hung, V.; Zou, P.; Rhee, H.; Udeshi, N.D.; Cracan, V.; Svinkina, T.; Carr, S.A.; Mootha, V.K.; Ting, A.Y. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 2014, 55, 332–341. [Google Scholar] [CrossRef]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Cho, K.F.; Branon, T.C.; Rajeev, S.; Svinkina, T.; Udeshi, N.D.; Thoudam, T.; Kwak, C.; Rhee, H.-W.W.; Lee, I.-K.K.; Carr, S.A.; et al. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl. Acad. Sci. USA 2020, 117, 12143–12154. [Google Scholar] [CrossRef]
- Sanes, J.R. The synaptic cleft of the neuromuscular junction. Semin. Dev. Biol. 1995, 6, 163–173. [Google Scholar] [CrossRef]
- Patton, B.L. Basal lamina and the organization of neuromuscular synapses. J. Neurocytol. 2003, 32, 883–903. [Google Scholar] [CrossRef]
- Badawi, Y.; Nishimune, H. Super-resolution microscopy for analyzing neuromuscular junctions and synapses. Neurosci. Lett. 2020, 715, 134644. [Google Scholar] [CrossRef]
- Ehmann, N.; Sauer, M.; Kittel, R.J. Super-resolution microscopy of the synaptic active zone. Front. Cell Neurosc. 2015, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kittel, R.J.; Wichmann, C.; Rasse, T.M.; Fouquet, W.; Schmidt, M.; Schmid, A.; Wagh, D.A.; Pawlu, C.; Kellner, R.R.; Willig, K.I.; et al. Bruchpilot Promotes Active Zone Assembly, Ca2þ Channel Clustering, and Vesicle Release. Science 2006, 312, 1051–1054. [Google Scholar] [CrossRef]
- Ehmann, N.; Van De Linde, S.; Alon, A.; Ljaschenko, D.; Keung, X.Z.; Holm, T.; Rings, A.; DiAntonio, A.; Hallermann, S.; Ashery, U.; et al. Quantitative super-resolution imaging of Bruchpilot distinguishes active zone states. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Böhme, M.A.; Beis, C.; Reddy-Alla, S.; Reynolds, E.; Mampell, M.M.; Grasskamp, A.T.; Lützkendorf, J.; Bergeron, D.D.; Driller, J.H.; Babikir, H.; et al. Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca2+ channel-vesicle coupling. Nat. Neurosci. 2016, 19, 1311–1320. [Google Scholar] [CrossRef]
- Reddy-Alla, S.; Böhme, M.A.; Reynolds, E.; Beis, C.; Grasskamp, A.T.; Mampell, M.M.; Maglione, M.; Jusyte, M.; Rey, U.; Babikir, H.; et al. Stable Positioning of Unc13 Restricts Synaptic Vesicle Fusion to Defined Release Sites to Promote Synchronous Neurotransmission. Neuron 2017, 95, 1350–1364.e12. [Google Scholar] [CrossRef]
- Ghelani, T.; Sigrist, S.J. Coupling the Structural and Functional Assembly of Synaptic Release Sites. Front. Neuroanat. 2018, 12, 1–20. [Google Scholar] [CrossRef]
- Böhme, M.A.; McCarthy, A.W.; Grasskamp, A.T.; Beuschel, C.B.; Goel, P.; Jusyte, M.; Laber, D.; Huang, S.; Rey, U.; Petzoldt, A.G.; et al. Rapid active zone remodeling consolidates presynaptic potentiation. Nat. Commun. 2019, 10, 1–59. [Google Scholar] [CrossRef]
- Nishimune, H.; Badawi, Y.; Mori, S.; Shigemoto, K. Dual-color STED microscopy reveals a sandwich structure of Bassoon and Piccolo in active zones of adult and aged mice. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Badawi, Y.; Nishimune, H. Presynaptic active zones of mammalian neuromuscular junctions: Nanoarchitecture and selective impairments in aging. Neurosci. Res. 2018, 127, 78–88. [Google Scholar] [CrossRef]
- Patton, B.L.; Cunningham, J.M.; Thyboll, J.; Kortesmaa, J.; Westerblad, H.; Edström, L.; Tryggvason, K.; Sanes, J.R. Properly formed but improperly localized synaptic specializations in the absence of laminin α4. Nat. Neurosci. 2001, 4, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Sanes, J.R. The basement membrane/basal lamina of skeletal muscle. J. Biol. Chem. 2003, 278, 12601–12604. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L.; Olszowy, M.W.; Holdorf, A.D.; Li, J.; Bromley, S.; Desai, N.; Widder, P.; Rosenberger, F.; Van Der Merwe, P.A.; Allen, P.M.; et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell 1998, 94, 667–677. [Google Scholar] [CrossRef]
- Monks, C.R.F.; Freiberg, B.A.; Kupfer, H.; Sciaky, N.; Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 1998, 395, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Grakoui, A.; Bromley, S.K.; Sumen, C.; Davis, M.M.; Shaw, A.S.; Allen, P.M.; Dustin, M.L. The immunological synapse: A molecular machine controlling T cell activation. Science 1999, 285, 221–227. [Google Scholar] [CrossRef]
- Cullinan, P.; Sperling, A.I.; Burkhardt, J.K. The distal pole complex: A novel membrane domain distal to the immunological synapse. Immunol. Rev. Rev. 2002, 189, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L. Immunological Synapses. Encycl. Immunobiol. 2016, 3, 16–24. [Google Scholar]
- Dustin, M.L.; Depoil, D. New insights into the T cell synapse from single molecule techniques. Nat. Rev. Immunol. 2011, 11, 672–684. [Google Scholar] [CrossRef]
- Rossy, J.; Pageon, S.V.; Davis, D.M.; Gaus, K. Super-resolution microscopy of the immunological synapse. Curr. Opin. Immunol. 2013, 25, 307–312. [Google Scholar] [CrossRef]
- Xie, J.; Tato, C.M.; Davis, M.M. How the immune system talks to itself: The varied role of synapses. Immunol. Rev. 2013, 251, 65–79. [Google Scholar] [CrossRef]
- Goyette, J.; Nieves, D.J.; Ma, Y.; Gaus, K. How does T cell receptor clustering impact on signal transduction? J. Cell Sci. 2019, 132, jcs226423. [Google Scholar] [CrossRef]
- Lillemeier, B.F.; Mörtelmaier, M.A.; Forstner, M.B.; Huppa, J.B.; Groves, J.T.; Davis, M.M. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 2010, 11, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.; Barr, V.; Manley, S.; Patterson, G.; Balagopalan, L.; Akpan, I.; Regan, C.K.; Merrill, R.K.; Sommers, C.L.; Lippincott-Schwartz, J.; et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity 2011, 35, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Pageon, S.V.; Tabarin, T.; Yamamoto, Y.; Ma, Y.; Nicovich, P.R.; Bridgeman, J.S.; Cohnen, A.; Benzing, C.; Gao, Y.; Crowther, M.D.; et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. USA 2016, 113, E6905. [Google Scholar] [CrossRef]
- James, J.R.; White, S.S.; Clarke, R.W.; Johansen, A.M.; Dunne, P.D.; Sleep, D.L.; Fitzgerald, W.J.; Davis, S.J.; Klenerman, D. Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl. Acad. Sci. USA 2007, 104, 17662–17667. [Google Scholar] [CrossRef]
- Brameshuber, M.; Kellner, F.; Rossboth, B.K.; Ta, H.; Alge, K.; Sevcsik, E.; Göhring, J.; Axmann, M.; Baumgart, F.; Gascoigne, N.R.J.; et al. Monomeric TCRs drive T cell antigen recognition article. Nat. Immunol. 2018, 19, 487–496. [Google Scholar] [CrossRef]
- Williamson, D.J.; Owen, D.M.; Rossy, J.; Magenau, A.; Wehrmann, M.; Gooding, J.J.; Gaus, K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 2011, 12, 655–662. [Google Scholar] [CrossRef]
- Balagopalan, L.; Barr, V.A.; Kortum, R.L.; Park, A.K.; Samelson, L.E. Cutting Edge: Cell Surface Linker for Activation of T Cells Is Recruited to Microclusters and Is Active in Signaling. J. Immunol. 2013, 190, 3849–3853. [Google Scholar] [CrossRef]
- Feher, K.; Halstead, J.M.; Goyette, J.; Gaus, K. Can single molecule localization microscopy detect nanoclusters in T cells? Curr. Opin. Chem. Biol. 2019, 51, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.M.; Ponjavic, A.; Fritzsche, M.; Fernandes, R.A.; De La Serna, J.B.; Wilcock, M.J.; Schneider, F.; Urbančič, I.; McColl, J.; Anzilotti, C.; et al. Capturing resting T cells: The perils of PLL correspondence. Nat. Immunol. 2018, 19, 203–205. [Google Scholar] [CrossRef]
- Rak, G.D.; Mace, E.M.; Banerjee, P.P.; Svitkina, T.; Orange, J.S. Natural Killer cell lytic granule secretion occurs through a pervasive actin network at the immune synapse. PLoS Biol. 2011, 9, e1001151. [Google Scholar] [CrossRef]
- Brown, A.C.N.; Oddos, S.; Dobbie, I.M.; Alakoskela, J.M.; Parton, R.M.; Eissmann, P.; Neil, M.A.A.; Dunsby, C.; French, P.M.W.; Davis, I.; et al. Remodelling of cortical actin where lytic granules dock at Natural Killer cell immune synapses revealed by super-resolution microscopy. PLoS Biol. 2011, 9, e1001152. [Google Scholar] [CrossRef]
- Ashdown, G.W.; Burn, G.L.; Williamson, D.J.; Pandžić, E.; Peters, R.; Holden, M.; Ewers, H.; Shao, L.; Wiseman, P.W.; Owen, D.M. Live-Cell Super-resolution Reveals F-Actin and Plasma Membrane Dynamics at the T Cell Synapse. Biophys. J. 2017, 112, 1703–1713. [Google Scholar] [CrossRef] [PubMed]
- Fritzsche, M.; Fernandes, R.A.; Chang, V.T.; Colin-York, H.; Clausen, M.P.; Felce, J.H.; Galiani, S.; Erlenkämper, C.; Santos, A.M.; Heddleston, J.M.; et al. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 2017, 3, e1603032. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, D.; Burkhardt, J.K. Multiple actin networks coordinate mechanotransduction at the immunological synapse. J. Cell Biol. 2020, 219, 1–12. [Google Scholar] [CrossRef]
- Hammer, J.A.; Wang, J.C.; Saeed, M.; Pedrosa, A.T. Origin, Organization, Dynamics, and Function of Actin and Actomyosin Networks at the T Cell Immunological Synapse. Ann. Rev. Immunol. 2019, 37, 201–224. [Google Scholar] [CrossRef] [PubMed]
- Stefen, H.; Chaichim, C.; Power, J.; Fath, T. Regulation of the Postsynaptic Compartment of Excitatory Synapses by the Actin Cytoskeleton in Health and Its Disruption in Disease. Neural. Plast. 2016, 2016, 2371970. [Google Scholar] [CrossRef]
- Charrier, C.; Ehrensperger, M.M.-V.M.; Dahan, M.; Levi, S.; Triller, A. Cytoskeleton Regulation of Glycine Receptor Number at Synapses and Diffusion in the Plasma Membrane. J. Neurosci. 2006, 26, 8502–8511. [Google Scholar] [CrossRef]
- Jung, Y.; Riven, I.; Feigelson, S.W.; Kartvelishvily, E.; Tohya, K.; Miyasaka, M. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. USA 2016, 113, E5916–E5924. [Google Scholar] [CrossRef]
- Fernandes, R.A.; Ganzinger, K.A.; Tzou, J.C.; Jönsson, P.; Lee, S.F.; Palayret, M.; Santos, A.M.; Carr, A.R.; Ponjavic, A.; Chang, V.T.; et al. A cell topography-based mechanism for ligand discrimination by the T cell receptor. Proc. Natl. Acad. Sci. USA 2019, 116, 14002–14010. [Google Scholar] [CrossRef]
- Fernández-Orth, J.; Rolfes, L.; Gola, L.; Bittner, S.; Andronic, J.; Sukhorukov, V.L.; Sisario, D.; Landgraf, P.; Dieterich, D.C.; Cerina, M.; et al. A role for TASK2 channels in the human immunological synapse. Eur. J. Immunol. 2020, 51, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Pettmann, J.; Santos, A.M.; Dushek, O.; Davis, S.J. Membrane ultrastructure and T cell activation. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Orbach, R.; Su, X. Surfing on Membrane Waves: Microvilli, Curved Membranes, and Immune Signaling. Front. Immunol. 2020, 11, 2187. [Google Scholar] [CrossRef] [PubMed]
- Bartle, E.I.; Rao, T.C.; Urner, T.M.; Mattheyses, A.L. Bridging the gap: Super-resolution microscopy of epithelial cell junctions. Tissue Barriers 2018, 6, 1–13. [Google Scholar] [CrossRef]
- Gonschior, H.; Haucke, V.; Lehmann, M. Super-resolution imaging of tight and adherens junctions: Challenges and open questions. Int. J. Mol. Sci. 2020, 21, 744. [Google Scholar] [CrossRef]
- Otani, T.; Furuse, M. Tight Junction Structure and Function Revisited. Trends Cell Biol. 2020, 30, 805–817. [Google Scholar] [CrossRef]
- Mangeol, P.; Massey-Harroche, D.; Richard, F.; Lenne, P.-F.; Le Bivic, A. Super-resolution imaging uncovers the nanoscopic segregation of polarity proteins in epithelia. bioRxiv 2020. [Google Scholar] [CrossRef]
- McCutcheon, S.; Stout, R.F.; Spray, D.C. The dynamic Nexus: Gap junctions control protein localization and mobility in distinct and surprising ways. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Hoffman, D.P.; Shtengel, G.; Xu, C.S.; Campbell, K.R.; Freeman, M.; Wang, L.; Milkie, D.E.; Pasolli, H.A.; Iyer, N.; Bogovic, J.A.; et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 2020, 367, eaaz5357. [Google Scholar] [CrossRef]
- Sannerud, R.; Esselens, C.; Ejsmont, P.; Mattera, R.; Rochin, L.; Tharkeshwar, A.K.; De Baets, G.; De Wever, V.; Habets, R.; Baert, V.; et al. Restricted Location of PSEN2/g -Secretase Determines Substrate Specificity and Generates an Intracellular Ab Pool. Cell 2016, 166, 193–208. [Google Scholar] [CrossRef]
- Wolff, G.; Hagen, C.; Grünewald, K.; Kaufmann, R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol. Cell 2016, 108, 245–258. [Google Scholar] [CrossRef]
- Lu, C.-H.; Tang, W.-C.; Liu, Y.-T.; Chang, S.-W.; Wu, F.C.M.; Chen, C.-Y.; Tsai, Y.-C.; Yang, S.-M.; Kuo, C.-W.; Okada, Y.; et al. Lightsheet localization microscopy enables fast, large-scale, and three-dimensional super-resolution imaging. Commun. Biol. 2019, 2, 1–10. [Google Scholar] [CrossRef]
- Weiss, L.E.; Shalev Ezra, Y.; Goldberg, S.; Ferdman, B.; Adir, O.; Schroeder, A.; Alalouf, O.; Shechtman, Y. Three-dimensional localization microscopy in live flowing cells. Nat. Nanotechnol. 2020, 15, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Gwosch, K.C.; Pape, J.K.; Balzarotti, F.; Hoess, P.; Ellenberg, J.; Ries, J.; Hell, S.W. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 2020, 17, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.S.; Chen, Y.J.; Chang, C.L.; Lee, W.R.; Liou, J. Cortical actin contributes to spatial organization of ER-PM junctions. Mol. Biol. Cell 2017, 28, 3171–3180. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; Vaccaro, M.I.; Codogno, P.; Morel, E. ER–plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI 3P synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef]
- Shim, S.H.; Xia, C.; Zhong, G.; Babcock, H.P.; Vaughan, J.C.; Huang, B.; Wang, X.; Xu, C.; Bi, G.Q.; Zhuang, X. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA 2012, 109, 13978–13983. [Google Scholar] [CrossRef]
- Modi, S.; López-Doménech, G.; Halff, E.F.; Covill-Cooke, C.; Ivankovic, D.; Melandri, D.; Arancibia-Cárcamo, I.L.; Burden, J.J.; Lowe, A.R.; Kittler, J.T. Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 2019, 10, 4399. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Annaert, W. The Nanoscopic Organization of Synapse Structures: A Common Basis for Cell Communication. Membranes 2021, 11, 248. https://doi.org/10.3390/membranes11040248
Yang X, Annaert W. The Nanoscopic Organization of Synapse Structures: A Common Basis for Cell Communication. Membranes. 2021; 11(4):248. https://doi.org/10.3390/membranes11040248
Chicago/Turabian StyleYang, Xiaojuan, and Wim Annaert. 2021. "The Nanoscopic Organization of Synapse Structures: A Common Basis for Cell Communication" Membranes 11, no. 4: 248. https://doi.org/10.3390/membranes11040248
APA StyleYang, X., & Annaert, W. (2021). The Nanoscopic Organization of Synapse Structures: A Common Basis for Cell Communication. Membranes, 11(4), 248. https://doi.org/10.3390/membranes11040248





