AFM Study of Nanoscale Membrane Perturbation Induced by Antimicrobial Lipopeptide C14 KYR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sample Preparation for FTIR Analysis
2.3. Synchrotron Radiation FTIR Microspectroscopy (SR-FTIR)
2.4. SR-FTIR Data Processing and Data Analysis
2.5. Sample Preparation for Morphology Studies
2.6. AFM Imaging and Force Measurements
3. Results and Discussion
3.1. Biochemical Alteration of E. coli Membrane Detected by the SR-FTIR Microspectroscopy
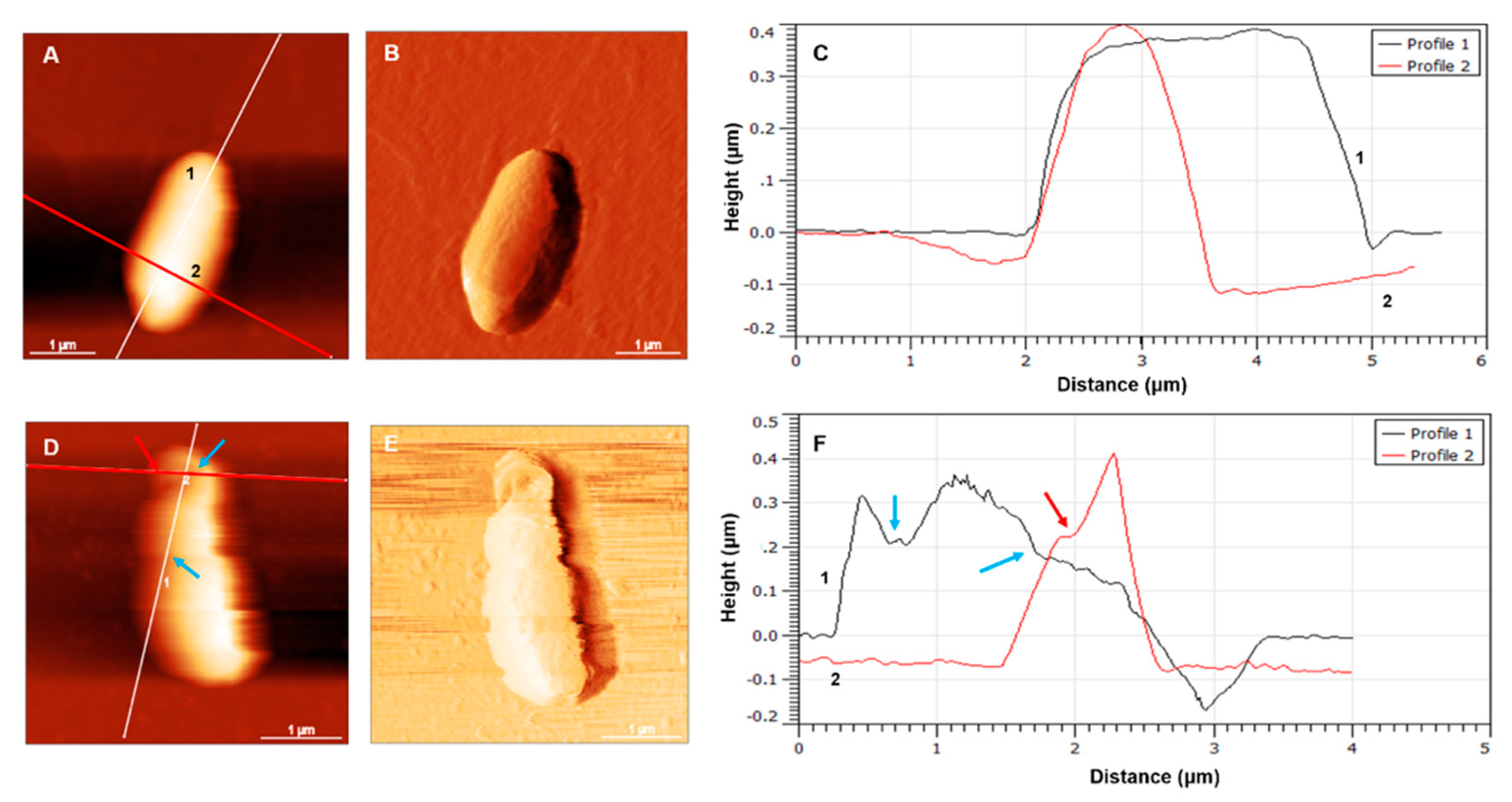
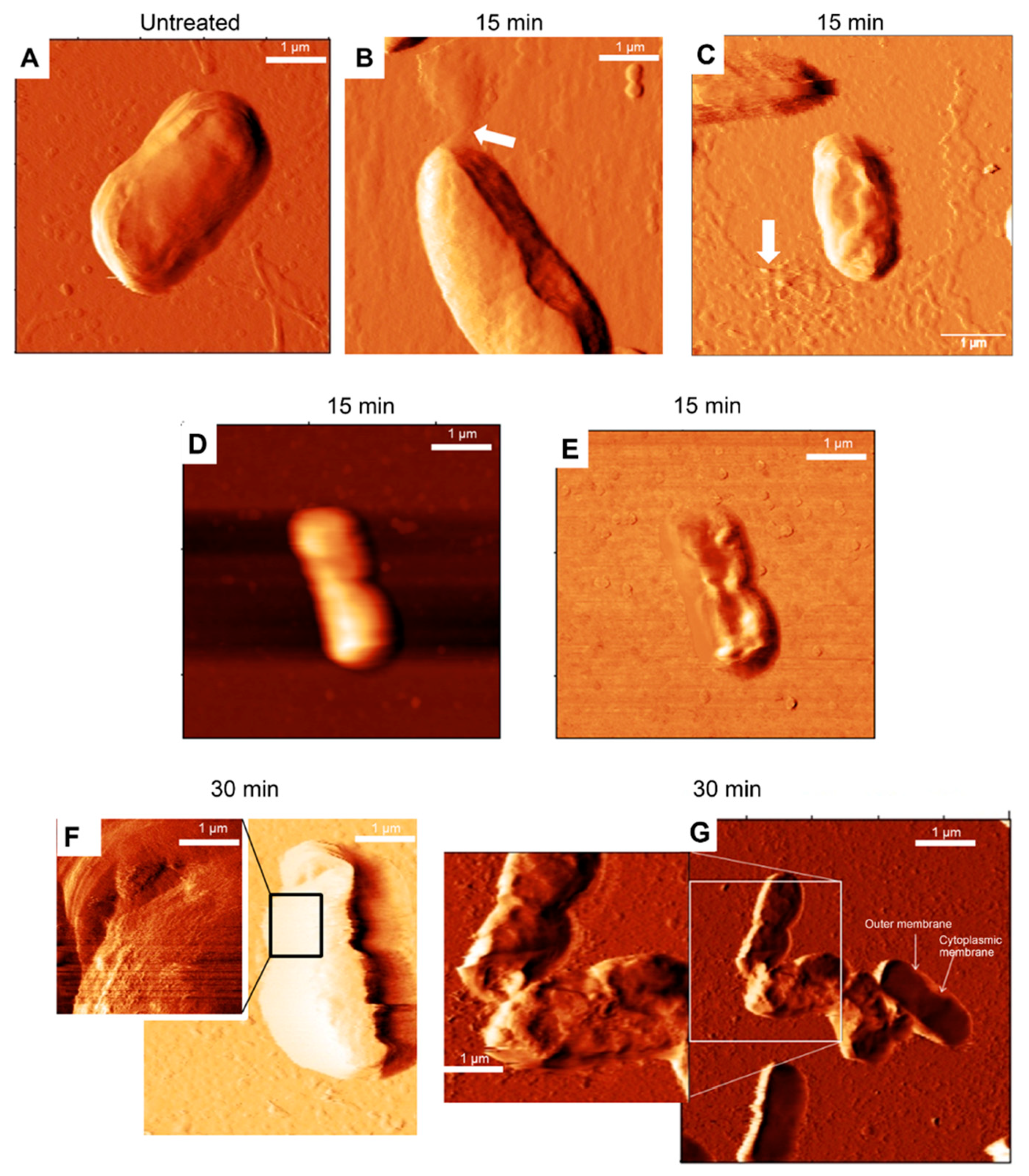
3.2. Morphology of E. coli HB101 after Treatment with KYR and C14-KYR Peptides
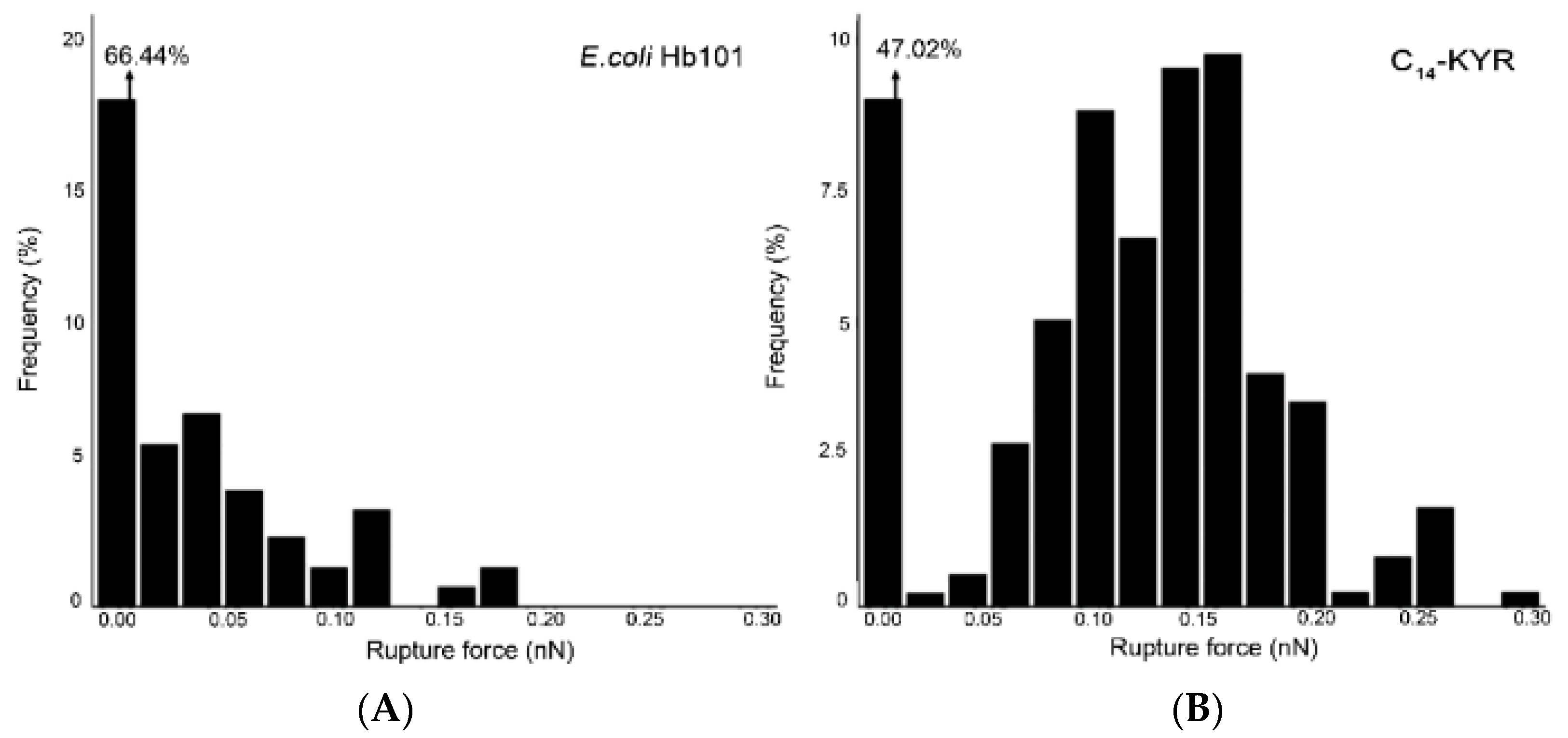
3.3. Rupture Force Measurements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mangoni, M.L.; Shai, Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: Similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011, 68, 2267–2280. [Google Scholar] [CrossRef]
- Sumi, C.D.; Yang, B.W.; Yeo, I.-C.; Hahm, Y.T. Antimicrobial peptides of the genus Bacillus: A new era for antibiotics. Can. J. Microbiol. 2015, 61, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Grau-Campistany, A.; Manresa, Á.; Pujol, M.; Rabanal, F.; Cajal, Y.; Grau, A. Tryptophan-containing lipopeptide antibiotics derived from polymyxin B with activity against Gram positive and Gram negative bacteria. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Greber, K.E.; Zielińska, J.; Nierzwicki, Ł.; Ciura, K.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Are the short cationic lipopeptides bacterial membrane disruptors? Structure-Activity Relationship and molecular dynamic evaluation. Biochim. Biophys. Acta (BBA) Biomembr. 2019, 1861, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Nasompag, S.; Dechsiri, P.; Hongsing, N.; Phonimdaeng, P.; Daduang, S.; Klaynongsruang, S.; Camesano, T.A.; Patramanon, R. Effect of acyl chain length on therapeutic activity and mode of action of the CX-KYR-NH2 antimicrobial lipopeptide. Biochim. Biophys. Acta (BBA) Biomembr. 2015, 1848, 2351–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straus, S.K.; Hancock, R.E. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Kamarajan, P.; Hayami, T.; Matte, B.; Liu, Y.; Danciu, T.; Ramamoorthy, A.; Worden, F.; Kapila, S.; Kapila, Y.; Nisin, Z.P. A Bacteriocin and Food Preservative, Inhibits Head and Neck Cancer Tumorigenesis and Prolongs Survival. PLoS ONE 2015, 10, 0131008. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yang, W.; Wang, Q.; Li, H.; Wang, K.; Yang, L.; Liu, W. Atomic force microscopy investigation of the characteristic effects of silver ions on Escherichia coli and Staphylococcus epidermidis. Talanta 2010, 81, 1508–1512. [Google Scholar] [CrossRef] [PubMed]
- Dorobantu, L.S.; Goss, G.G.; Burrell, R. Atomic force microscopy: A nanoscopic view of microbial cell surfaces. Micron 2012, 43, 1312–1322. [Google Scholar] [CrossRef]
- Alsteens, D.; Trabelsi, H.; Soumillion, P.; Dufrêne, Y. Multiparametric atomic force microscopy imaging of single bacteriophages extruding from living bacteria. Nat. Commun. 2013, 4, 2926. [Google Scholar] [CrossRef]
- Pittman, A.; Marsh, B.P.; King, G.M. Conformations and Dynamic Transitions of a Melittin Derivative that Forms Macromolecule-Sized Pores in Lipid Bilayers. Langmuir 2018, 34, 8393–8399. [Google Scholar] [CrossRef]
- Touhami, A.; Nysten, B.; Dufrêne, Y.F. Nanoscale Mapping of the Elasticity of Microbial Cells by Atomic Force Microscopy. Langmuir 2003, 19, 4539–4543. [Google Scholar] [CrossRef]
- Domingues, M.; Silva, P.M.; Franquelim, H.; Carvalho, F.A.; Castanho, M.; Santos, N.C. Antimicrobial protein rBPI21-induced surface changes on Gram-negative and Gram-positive bacteria. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 543–551. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Atomic Force Microscopy, a Powerful Tool in Microbiology. J. Bacteriol. 2002, 184, 5205–5213. [Google Scholar] [CrossRef] [Green Version]
- Andre, G.; Kulakauskas, S.; Chapot-Chartier, M.-P.; Navet, B.; Deghorain, M.; Bernard, E.; Hols, P.; Dufrêne, Y.F. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat. Commun. 2010, 1, 27–28. [Google Scholar] [CrossRef]
- Schaer-Zammaretti, P.; Ubbink, J. Imaging of lactic acid bacteria with AFM—elasticity and adhesion maps and their relationship to biological and structural data. Ultramicroscopy 2003, 97, 199–208. [Google Scholar] [CrossRef]
- Dinarelli, S.; Girasole, M.; Longo, G. FC_analysis: A tool for investigating atomic force microscopy maps of force curves. BMC Bioinform. 2018, 19, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaussart, A.; Péchoux, C.; Trieu-Cuot, P.; Hols, P.; Mistou, M.-Y.; Dufrêne, Y.F. Molecular mapping of the cell wall polysaccharides of the human pathogen Streptococcus agalactiae. Nanoscale 2014, 6, 14820–14827. [Google Scholar] [CrossRef] [PubMed]
- Dufrêne, Y.F. Towards nanomicrobiology using atomic force microscopy. Nat. Rev. Genet. 2008, 6, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Viljoen, A.; Foster, S.J.; Fantner, G.E.; Hobbs, J.K.; Dufrêne, Y.F. Scratching the Surface: Bacterial Cell Envelopes at the Nanoscale. mBio 2020, 11, e03020-19. [Google Scholar] [CrossRef] [Green Version]
- Fantner, G.E.; Barbero, R.J.; Gray, D.S.; Belcher, A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 2010, 5, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Formosa, C.; Grare, M.; Jauvert, E.; Coutable, A.; Regnouf-De-Vains, J.B.; Mourer, M.; Duval, R.; Dague, E. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain. Sci. Rep. 2012, 2, 575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srisayam, M.; Weerapreeyakul, N.; Barusrux, S.; Tanthanuch, W.; Thumanu, K. Application of FTIR microspectroscopy for characterization of biomolecular changes in human melanoma cells treated by sesamol and kojic acid. J. Dermatol. Sci. 2014, 73, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Junhom, C.; Weerapreeyakul, N.; Tanthanuch, W.; Thumanu, K. FTIR microspectroscopy defines early drug resistant human hepatocellular carcinoma (HepG2) cells. Exp. Cell Res. 2016, 340, 71–80. [Google Scholar] [CrossRef]
- Machana, S.; Weerapreeyakul, N.; Barusrux, S.; Thumanu, K.; Tanthanuch, W. FTIR microspectroscopy discriminates anticancer action on human leukemic cells by extracts of Pinus kesiya; Cratoxylum formosum ssp. pruniflorum and melphalan. Talanta 2012, 93, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Pocasap, P.; Weerapreeyakul, N.; Junhom, C.; Phiboonchaiyanan, P.P.; Srisayam, M.; Nonpunya, A.; Siriwarin, B.; Khamphio, M.; Nanok, C.; Thumanu, K.; et al. FTIR Microspectroscopy for the Assessment of Mycoplasmas in HepG2 Cell Culture. Appl. Sci. 2020, 10, 3766. [Google Scholar] [CrossRef]
- Ye, D.; Tanthanuch, W.; Thumanu, K.; Sangmalee, A.; Parnpai, R.; Heraud, P. Discrimination of functional hepatocytes derived from mesenchymal stem cells using FTIR microspectroscopy. Analyst 2012, 137, 4774–4784. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Camesano, T.A. ACS Symposium Series Ch. 10. In Microbial Surfaces; American Chemical Society: Washington, DC, USA, 2008; Volume 984, pp. 163–188. [Google Scholar]
- Ivanov, I.E.; Boyd, C.D.; Newell, P.D.; Schwartz, M.E.; Turnbull, L.; Johnson, M.S.; Whitchurch, C.B.; O’Toole, G.A.; Camesano, T.A. Atomic force and super-resolution microscopy support a role for LapA as a cell-surface biofilm adhesin of Pseudomonas fluorescens. Res. Microbiol. 2012, 163, 685–691. [Google Scholar] [CrossRef] [Green Version]
- Atabek, A.; Camesano, T.A. Atomic Force Microscopy Study of the Effect of Lipopolysaccharides and Extracellular Polymers on Adhesion of Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 8503–8509. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Ordóñez, A.; Mouwen, D.; López, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Microbiol. Methods 2011, 84, 369–378. [Google Scholar] [CrossRef]
- Ojeda, J.J.; Dittrich, M. Fourier Transform Infrared Spectroscopy for Molecular Analysis of Microbial Cells. In Methods in Molecular Biology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2012; Volume 881, pp. 187–211. [Google Scholar]
- Corte, L.; Tiecco, M.; Roscini, L.; De Vincenzi, S.; Colabella, C.; Germani, R.; Tascini, C.; Cardinali, G. FTIR Metabolomic Fingerprint Reveals Different Modes of Action Exerted by Structural Variants of N-Alkyltropinium Bromide Surfactants on Escherichia coli and Listeria innocua Cells. PLoS ONE 2015, 10, 0115275. [Google Scholar] [CrossRef] [Green Version]
- Faghihzadeh, F.; Anaya, N.M.; Schifman, L.; Oyanedel-Craver, V. Fourier transform infrared spectroscopy to assess molecular-level changes in microorganisms exposed to nanoparticles. Nanotechnol. Environ. Eng. 2016, 1, 1–16. [Google Scholar] [CrossRef]
- Rieppo, L.; Saarakkala, S.; Närhi, T.; Helminen, H.; Jurvelin, J.; Rieppo, J. Application of second derivative spectroscopy for increasing molecular specificity of fourier transform infrared spectroscopic imaging of articular cartilage. Osteoarthr. Cartil. 2012, 20, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ami, D.; Mereghetti, P.; Leri, M.; Giorgetti, S.; Natalello, A.; Doglia, S.M.; Stefani, M.; Bucciantini, M. A FTIR microspectroscopy study of the structural and biochemical perturbations induced by natively folded and aggregated transthyretin in HL-1 cardiomyocytes. Sci. Rep. 2018, 8, 12508. [Google Scholar] [CrossRef] [PubMed]
- Zarnowiec, P.; Lechowicz, Ł.; Czerwonka, G.; Kaca, W. Fourier Transform Infrared Spectroscopy (FTIR) as a Tool for the Identification and Differentiation of Pathogenic Bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef] [PubMed]
- Leitner, M.; Fantner, G.E.; Fantner, E.J.; Ivanova, K.; Ivanov, T.; Rangelow, I.; Ebner, A.; Rangl, M.; Tang, J.; Hinterdorfer, P. Increased imaging speed and force sensitivity for bio-applications with small cantilevers using a conventional AFM setup. Micron 2012, 43, 1399–1407. [Google Scholar] [CrossRef]
- Strauss, J.; Burnham, N.A.; Camesano, T.A. Atomic force microscopy study of the role of LPS O-antigen on adhesion of E. coli. J. Mol. Recognit. 2009, 22, 347–355. [Google Scholar] [CrossRef]
- Gafri, H.F.S.; Zuki, F.M.; Aroua, M.K.; Hashim, N.A. Mechanism of bacterial adhesion on ultrafiltration membrane modified by natural antimicrobial polymers (chitosan) and combination with activated carbon (PAC). Rev. Chem. Eng. 2019, 35, 421–443. [Google Scholar] [CrossRef]
- Liu, L.; Chen, S.; Zhang, X.; Xue, Z.; Cui, S.; Hua, X.; Yang, B.; Yan, H.; Liu, C.; Wang, J.; et al. Mechanical penetration of β-lactam–resistant Gram-negative bacteria by programmable nanowires. Sci. Adv. 2020, 6, eabb9593. [Google Scholar] [CrossRef]

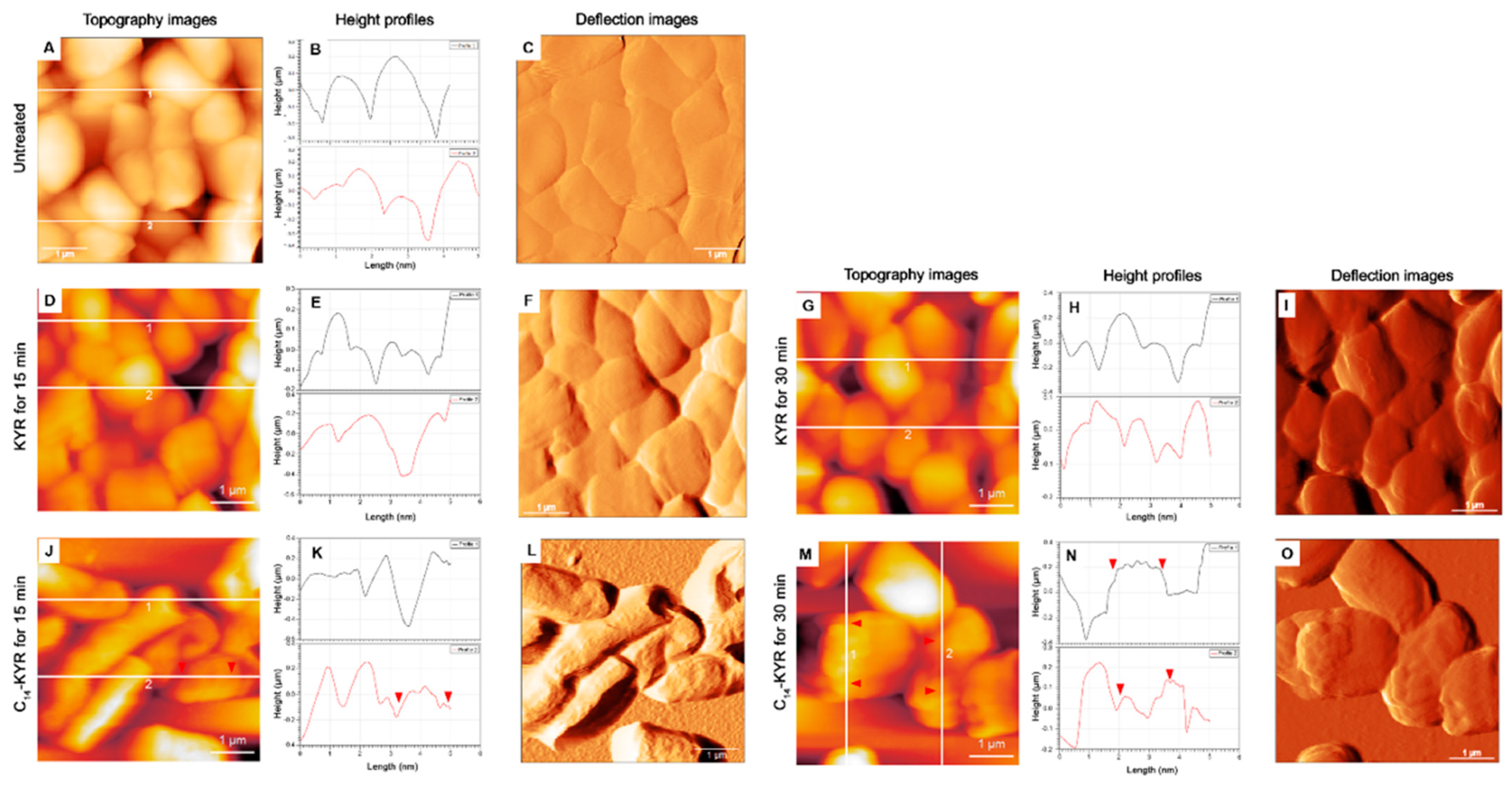
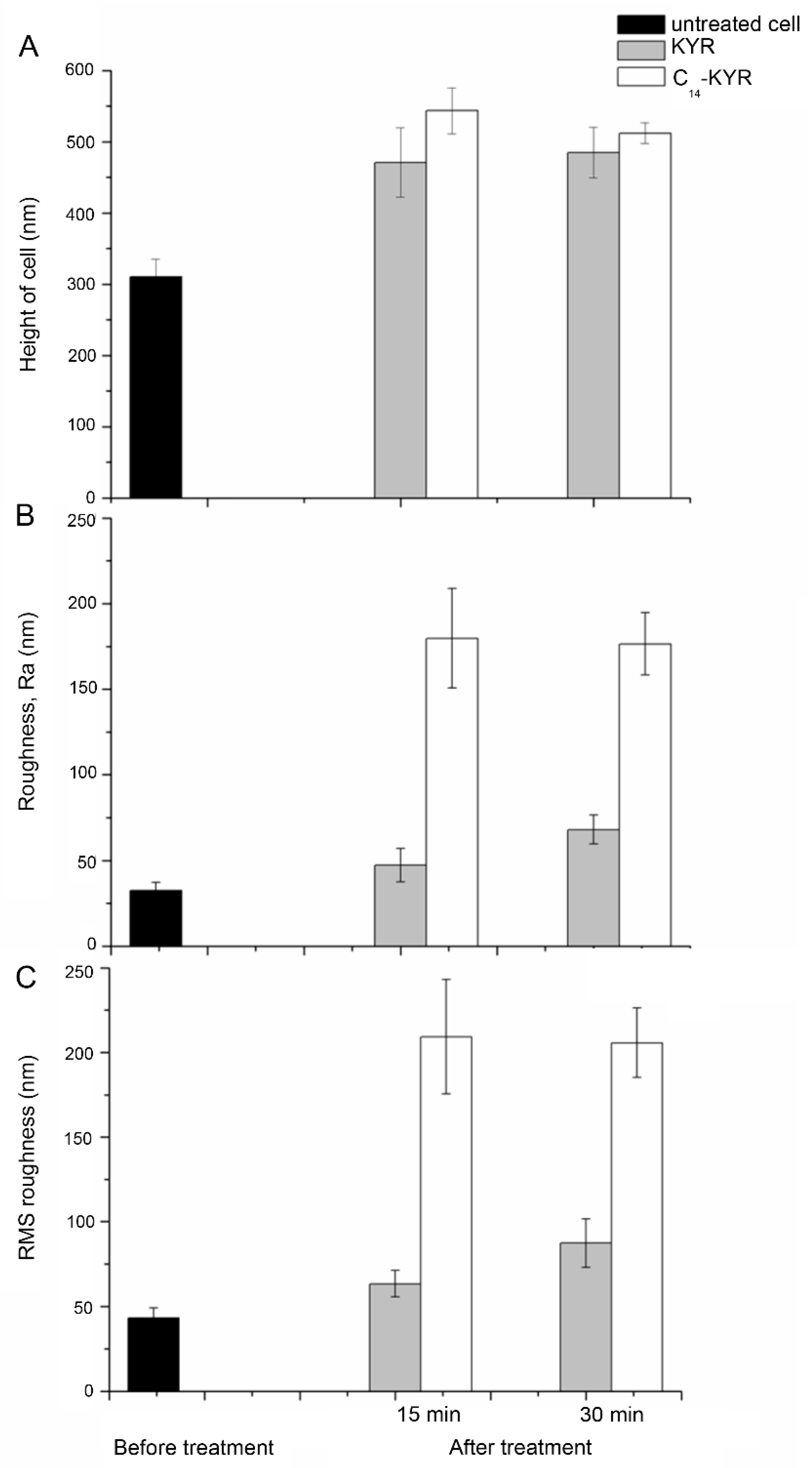
| Treatment Time | Sample | Size (µm) | Appearance Description of Cell Surface | Measurement (nm) | ||
|---|---|---|---|---|---|---|
| Mean of Height, h | The Average Roughness, Ra 1 | RMS Roughness 2 | ||||
| 0 min | Untreated (n = 251) | L: 3.4 ± 0.3 W: 1.43 ± 0.4 | Smooth | 310.4 ± 25.2 | 32.68 ± 4.7 | 43.03 ± 5.9 |
| 15 min | KYR (n = 176) | L: 2.8 ± 0.7 W: 1.28 ± 0.3 | Smooth | 471.0 ± 48.4 | 47.25 ± 9.7 | 63.3 ± 7.9 |
| C14-KYR (n = 213) | L: 2.4 ± 0.4 W: 1.34 ± 0.3 | Rough | 543.5 ± 32.2 | 179.67 ± 29.1 | 209.33 ± 33.8 | |
| 30 min | KYR (n = 225) | L: 3.1 ± 0.3 W: 1.07 ± 0.1 | slightly deformed surface | 485.1 ± 35.3 | 68.03 ± 8.4 | 87.3 ± 14.3 |
| C14-KYR (n = 228) | L: 2.8 ± 0.5 W: 1.18 ± 0.12 | Porous/Cell shrinkage | 512.3 ± 14.2 | 176.44 ± 18.2 | 205.7 ± 20.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nasompag, S.; Siritongsuk, P.; Thammawithan, S.; Srichaiyapol, O.; Prangkio, P.; Camesano, T.A.; Sinthuvanich, C.; Patramanon, R. AFM Study of Nanoscale Membrane Perturbation Induced by Antimicrobial Lipopeptide C14 KYR. Membranes 2021, 11, 495. https://doi.org/10.3390/membranes11070495
Nasompag S, Siritongsuk P, Thammawithan S, Srichaiyapol O, Prangkio P, Camesano TA, Sinthuvanich C, Patramanon R. AFM Study of Nanoscale Membrane Perturbation Induced by Antimicrobial Lipopeptide C14 KYR. Membranes. 2021; 11(7):495. https://doi.org/10.3390/membranes11070495
Chicago/Turabian StyleNasompag, Sawinee, Pawinee Siritongsuk, Saengrawee Thammawithan, Oranee Srichaiyapol, Panchika Prangkio, Terri A. Camesano, Chomdao Sinthuvanich, and Rina Patramanon. 2021. "AFM Study of Nanoscale Membrane Perturbation Induced by Antimicrobial Lipopeptide C14 KYR" Membranes 11, no. 7: 495. https://doi.org/10.3390/membranes11070495
APA StyleNasompag, S., Siritongsuk, P., Thammawithan, S., Srichaiyapol, O., Prangkio, P., Camesano, T. A., Sinthuvanich, C., & Patramanon, R. (2021). AFM Study of Nanoscale Membrane Perturbation Induced by Antimicrobial Lipopeptide C14 KYR. Membranes, 11(7), 495. https://doi.org/10.3390/membranes11070495






