Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials
Abstract
1. Introduction
2. Biomimetic Bacterial Membrane Systems
2.1. Lipid Monolayer to Mimetic Bacterial Membrane
2.2. Lipid Bilayer to Mimetic Bacterial Membrane
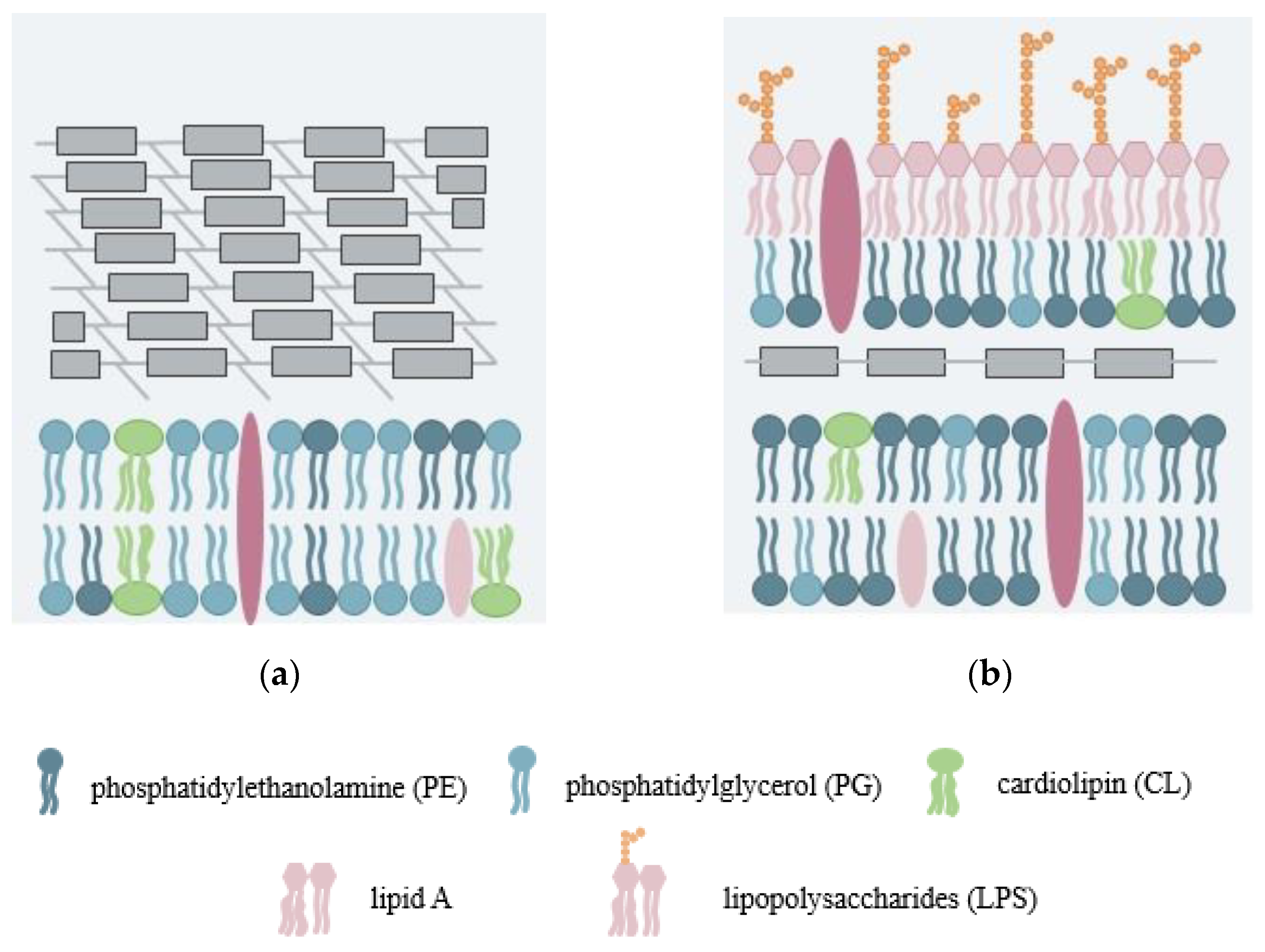
2.2.1. Symmetric Bilayer System
2.2.2. Asymmetric Lipid Bilayer
3. Formation of Various Bacterial Model Membranes
3.1. Langmuir–Blodgett Technique to Form Lipid Monolayer
3.2. Vesicle Fusion Method to Form Supported Lipid Bilayers
3.3. Langmuir–Blodgett and Langmuir–Schaefer Approach to Form Asymmetric Bilayers
4. Surface Characterization Techniques Used to Determine the Membrane Mimetic Systems
4.1. Quartz Crystal Microbalance with Dissipation
4.2. Surface Plasmon Resonance
4.3. Neutron Reflectometry
5. Applications: Gram-Positive Bacterial Membranes
5.1. Different Mimetic Gram-Positive Bacterial Membranes
5.2. Interaction of Antimicrobials with Gram-Positive Bacterial Membranes
6. Applications: Gram-Negative Bacterial Membranes
6.1. Different Mimetic Gram-Negative Bacterial Outer Membranes
6.2. Interaction of Antibiotics with Gram-Negative Bacterial Membranes
6.3. Bacterial Outer Membrane Protein Complex
6.4. Interaction of Nanoparticles with Gram-Negative Bacterial Membranes
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PE | phosphatidylethanolamines |
| PG | phosphatidylglycerols |
| CL | cardiolipins |
| QCM-D | Quartz Crystal Microbalance with Dissipation |
| SPR | Surface Plasmon Resonance |
| NR | Neutron reflectometry |
| L-PG | lysyl-phosphatidylglycerol |
| E. coli | Escherichia coli |
| ClcN | N-acetylglucosamines |
| LPS | polysaccharide |
| DMPC | 1,2-dimyristoyl-sn-glycero-3-phosphocholine |
| DMPG | 1,2-dimyristoyl-sn-glycero-3-phosphoryl-3’-rac-glycerol |
| PC | phosphatidylcholine |
| π | surface pressure |
| SLB | solid-supported lipid bilayer |
| DPPC | dipalmitoylphosphatidylcholine |
| LB | Langmuir–Blodgett |
| LS | Langmuir–Schaefer |
| POPG | 1-palmitoyl-2-oleoylphosphatidyl-glycerol |
| SLD | scattering length density |
| S. aureus | Staphylococcus aureus |
| POPE | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine |
| POPC | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| DPPG | 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol |
References
- Pajerski, W.; Ochonska, D.; Brzychczy-Wloch, M.; Indyka, P.; Jarosz, M.; Golda-Cepa, M.; Sojka, Z.; Kotarba, A. Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J. Nanopart. Res. 2019, 21, 186. [Google Scholar] [CrossRef]
- Cardon, S.; Sachon, E.; Carlier, L.; Drujon, T.; Walrant, A.; Alemán-Navarro, E.; Martínez-Osorio, V.; Guianvarc’h, D.; Sagan, S.; Fleury, Y.; et al. Peptidoglycan potentiates the membrane disrupting effect of the carboxyamidated form of DMS-DA6, a Gram-positive selective antimicrobial peptide isolated from Pachymedusa dacnicolor skin. PLoS ONE 2018, 13, e0205727. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Le Brun, A.P.; Clifton, L.A.; Halbert, C.E.; Lin, B.; Meron, M.; Holden, P.J.; Lakey, J.H.; Holt, S.A. Structural characterization of a model gram-negative bacterial surface using lipopolysaccharides from rough strains of Escherichia coli. Biomacromolecules 2013, 14, 2014–2022. [Google Scholar] [CrossRef]
- Paracini, N.; Schneck, E.; Imberty, A.; Micciulla, S. Lipopolysaccharides at Solid and Liquid Interfaces: Models for Biophysical Studies of the Gram-negative Bacterial Outer Membrane. Adv. Colloid Interface Sci. 2022, 301, 102603. [Google Scholar] [CrossRef] [PubMed]
- Dowhan, W. A retrospective: Use of Escherichia coli as a vehicle to study phospholipid synthesis and function. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 471–494. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Skoda, M.W.; Daulton, E.L.; Hughes, A.V.; Le Brun, A.P.; Lakey, J.H.; Holt, S.A. Asymmetric phospholipid: Lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J. R. Soc. Interface 2013, 10, 20130810. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Skoda, M.W.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of divalent cation removal on the structure of gram-negative bacterial outer membrane models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef]
- Carey, A.B.; Ashenden, A.; Köper, I. Model architectures for bacterial membranes. Biophys. Rev. 2022, 14, 111–143. [Google Scholar] [CrossRef]
- Barker, R.D.; McKinley, L.E.; Titmuss, S. Neutron Reflectivity as a Tool for Physics-Based Studies of Model Bacterial Membranes. In Biophysics of Infection; Leake, M.C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 261–282. [Google Scholar]
- Maget-Dana, R. The monolayer technique: A potent tool for studying the interfacial properties of antimicrobial and membrane-lytic peptides and their interactions with lipid membranes. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1462, 109–140. [Google Scholar] [CrossRef]
- Perczyk, P.; Broniatowski, M. Simultaneous action of microbial phospholipase C and lipase on model bacterial membranes—Modeling the processes crucial for bioaugmentation. Biochim. Biophys. Acta (BBA) Biomembr. 2021, 1863, 183620. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B.; Wydro, P.; Jańczyk, A. Probing the Modes of Antibacterial Activity of Chitosan. Effects of pH and Molecular Weight on Chitosan Interactions with Membrane Lipids in Langmuir Films. Biomacromolecules 2011, 12, 4144–4152. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Trefna, H.; Persson, M.; Kasemo, B.; Svedhem, S. Formation of supported lipid bilayers on silica: Relation to lipid phase transition temperature and liposome size. Soft Matter. 2013, 10, 187–195. [Google Scholar] [CrossRef]
- Swana, K.W.; Camesano, T.A.; Nagarajan, R. Formation of a Fully Anionic Supported Lipid Bilayer to Model Bacterial Inner Membrane for QCM-D Studies. Membranes 2022, 12, 558. [Google Scholar] [CrossRef]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef]
- Lind, T.K.; Skoda, M.W.A.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Phosphatidylethanolamine and Phosphatidylglycerol by Vesicle Fusion, a Simple but Relevant Model for Bacterial Membranes. ACS Omega 2019, 4, 10687–10694. [Google Scholar] [CrossRef]
- Hancock, R.E.; Nikaido, H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 1978, 136, 381–390. [Google Scholar] [CrossRef]
- Furusato, T.; Horie, F.; Matsubayashi, H.T.; Amikura, K.; Kuruma, Y.; Ueda, T. De Novo Synthesis of Basal Bacterial Cell Division Proteins FtsZ, FtsA, and ZipA Inside Giant Vesicles. ACS Synth. Biol. 2018, 7, 953–961. [Google Scholar] [CrossRef]
- Tuerkova, A.; Kabelka, I.; Králová, T.; Sukeník, L.; Pokorná, Š.; Hof, M.; Vácha, R. Effect of helical kink in antimicrobial peptides on membrane pore formation. Elife 2020, 9, e47946. [Google Scholar] [CrossRef]
- Montal, M.; Mueller, P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef] [PubMed]
- Paulowski, L.; Donoghue, A.; Nehls, C.; Groth, S.; Koistinen, M.; Hagge, S.O.; Böhling, A.; Winterhalter, M.; Gutsmann, T. The Beauty of Asymmetric Membranes: Reconstitution of the Outer Membrane of Gram-Negative Bacteria. Front. Cell Dev. Biol. 2020, 8, 586. [Google Scholar] [CrossRef]
- Gutsmann, T.; Heimburg, T.; Keyser, U.; Mahendran, K.R.; Winterhalter, M. Protein reconstitution into freestanding planar lipid membranes for electrophysiological characterization. Nat. Protoc. 2015, 10, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Hardy, G.J.; Nayak, R.; Zauscher, S. Model cell membranes: Techniques to form complex biomimetic supported lipid bilayers via vesicle fusion. Curr. Opin. Colloid Interface Sci. 2013, 18, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.W. DAPTOMYCIN, its membrane-active mechanism vs. that of other antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183395. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. [Google Scholar] [CrossRef]
- Martin, D.K. Nanobiotechnology of Biomimetic Membranes; Fundamental Biomedical Technologies; Springer: New York, NY, USA, 2007. [Google Scholar]
- Sarkis, J.; Vie, V. Biomimetic Models to Investigate Membrane Biophysics Affecting Lipid-Protein Interaction. Front. Bioeng. Biotechnol. 2020, 8, 270. [Google Scholar] [CrossRef]
- Rojewska, M.; Smułek, W.; Kaczorek, E.; Prochaska, K. Langmuir Monolayer Techniques for the Investigation of Model Bacterial Membranes and Antibiotic Biodegradation Mechanisms. Membranes 2021, 11, 707. [Google Scholar] [CrossRef]
- Broniatowski, M.; Flasiński, M.; Zięba, K.; Miśkowiec, P. Langmuir monolayer studies of the interaction of monoamphiphilic pentacyclic triterpenes with anionic mitochondrial and bacterial membrane phospholipids—Searching for the most active terpene. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 2460–2472. [Google Scholar] [CrossRef]
- Hoyo, J.; Torrent-Burgués, J.; Tzanov, T. Physical states and thermodynamic properties of model gram-negative bacterial inner membranes. Chem. Phys. Lipids 2019, 218, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Hoyo, J.; Torrent-Burgués, J.; Tzanov, T. Lipid-lipid interactions of Escherichia coli mimetic inner membrane at human physiological temperature. Gen. Physiol. Biophys. 2020, 39, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Jurak, M.; Szafran, K.; Cea, P.; Martín, S. Analysis of Molecular Interactions between Components in Phospholipid-Immunosuppressant-Antioxidant Mixed Langmuir Films. Langmuir 2021, 37, 5601–5616. [Google Scholar] [CrossRef] [PubMed]
- Jochelavicius, K.; Pereira, A.R.; Fiamingo, A.; Nobre, T.M.; Campana-Filho, S.P.; Oliveira, O.N., Jr. Chitosan effects on monolayers of zwitterionic, anionic and a natural lipid extract from E. coli at physiological pH. Colloids Surf. B Biointerfaces 2022, 209, 112146. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.A.; Deffune, E.; Oliveira, O.N., Jr.; Moraes, M.L. Penicillin-binding proteins (PBPs) determine antibiotic action in Langmuir monolayers as nanoarchitectonics mimetic membranes of methicillin-resistant Staphylococcus aureus. Colloids Surf. B Biointerfaces 2022, 214, 112447. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Y.; Hou, S.; Miao, Z.; Ma, Q. Interaction of amphotericin B and saturated or unsaturated phospholipid monolayers containing cholesterol or ergosterol at the air-water interface. Biophys. Chem. 2020, 258, 106317. [Google Scholar] [CrossRef]
- Wacklin, H. Neutron reflection from supported lipid membranes. Curr. Opin. Colloid Interface Sci. 2010, 15, 445–454. [Google Scholar] [CrossRef]
- Marquardt, D.; Geier, B.; Pabst, G. Asymmetric Lipid Membranes: Towards More Realistic Model Systems. Membranes 2015, 5, 180–196. [Google Scholar] [CrossRef]
- Luchini, A.; Cavasso, D.; Radulescu, A.; D’Errico, G.; Paduano, L.; Vitiello, G. Structural Organization of Cardiolipin-Containing Vesicles as Models of the Bacterial Cytoplasmic Membrane. Langmuir 2021, 37, 8508–8516. [Google Scholar] [CrossRef]
- Gong, H.; Hu, X.; Liao, M.; Fa, K.; Ciumac, D.; Clifton, L.A.; Sani, M.-A.; King, S.M.; Maestro, A.; Separovic, F.; et al. Structural Disruptions of the Outer Membranes of Gram-Negative Bacteria by Rationally Designed Amphiphilic Antimicrobial Peptides. ACS Appl. Mater. Interfaces 2021, 13, 16062–16074. [Google Scholar] [CrossRef]
- Boge, L.; Browning, K.L.; Nordström, R.; Campana, M.; Damgaard, L.S.E.; Seth Caous, J.; Hellsing, M.; Ringstad, L.; Andersson, M. Peptide-Loaded Cubosomes Functioning as an Antimicrobial Unit against Escherichia coli. ACS Appl. Mater. Interfaces 2019, 11, 21314–21322. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.; Mozsolits, H.; Aguilar, M.I. Surface plasmon resonance analysis of antimicrobial peptide–membrane interactions: Affinity & mechanism of action. Lett. Pept. Sci. 2003, 10, 475–485. [Google Scholar]
- Hancock, R.E. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997, 5, 37–42. [Google Scholar] [CrossRef]
- Le Brun, A.P.; Clifton, L.A.; Holt, S.A.; Holden, P.J.; Lakey, J.H. Deuterium Labeling Strategies for Creating Contrast in Structure-Function Studies of Model Bacterial Outer Membranes Using Neutron Reflectometry. Methods Enzymol. 2016, 566, 231–252. [Google Scholar] [PubMed]
- Lai, X.; Ding, Y.; Wu, C.M.; Chen, X.; Jiang, J.H.; Hsu, H.Y.; Wang, Y.; Le Brun, A.P.; Song, J.; Han, M.L.; et al. Phytantriol-Based Cubosome Formulation as an Antimicrobial against Lipopolysaccharide-Deficient Gram-Negative Bacteria. ACS Appl. Mater. Interfaces 2020, 12, 44485–44498. [Google Scholar] [CrossRef]
- Lai, X.; Han, M.L.; Ding, Y.; Chow, S.H.; Le Brun, A.P.; Wu, C.M.; Bergen, P.J.; Jiang, J.H.; Hsu, H.Y.; Muir, B.W.; et al. A polytherapy based approach to combat antimicrobial resistance using cubosomes. Nat. Commun. 2022, 13, 343. [Google Scholar] [CrossRef]
- Martin, D.K. Asymmetric Phospholipid LB Bilayers. In Nanobiotechnology of Biomimetic Membranes; Springer: New York, NY, USA, 2007; pp. 54–57. [Google Scholar]
- Freinkman, E.; Chng, S.-S.; Kahne, D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. USA 2011, 108, 2486–2491. [Google Scholar] [CrossRef]
- Arunmanee, W.; Pathania, M.; Solovyova, A.S.; Le Brun, A.P.; Ridley, H.; Baslé, A.; van den Berg, B.; Lakey, J.H. Gram-negative trimeric porins have specific LPS binding sites that are essential for porin biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E5034–E5043. [Google Scholar] [CrossRef]
- Nelson, N.; Opene, B.; Ernst, R.K.; Schwartz, D.K. Antimicrobial peptide activity is anticorrelated with lipid a leaflet affinity. PLoS ONE 2020, 15, e0242907. [Google Scholar] [CrossRef]
- Tamm, L.K.; McConnell, H.M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105–113. [Google Scholar] [CrossRef]
- Wagner, M.L.; Tamm, L.K. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: Silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys. J. 2000, 79, 1400–1414. [Google Scholar] [CrossRef]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Alghalayini, A.; Garcia, A.; Berry, T.; Cranfield, C.G. The Use of Tethered Bilayer Lipid Membranes to Identify the Mechanisms of Antimicrobial Peptide Interactions with Lipid Bilayers. Antibiotics 2019, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Andersson, J.; Knobloch, J.J.; Perkins, M.V.; Holt, S.A.; Köper, I. Synthesis and Characterization of Novel Anchorlipids for Tethered Bilayer Lipid Membranes. Langmuir 2017, 33, 4444–4451. [Google Scholar] [CrossRef] [PubMed]
- Raguse, B.; Braach-Maksvytis, V.; Cornell, B.A.; King, L.G.; Osman, P.D.J.; Pace, R.J.; Wieczorek, L. Tethered Lipid Bilayer Membranes: Formation and Ionic Reservoir Characterization. Langmuir 1998, 14, 648–659. [Google Scholar] [CrossRef]
- He, L.; Robertson, J.W.; Li, J.; Kärcher, I.; Schiller, S.M.; Knoll, W.; Naumann, R. Tethered bilayer lipid membranes based on monolayers of thiolipids mixed with a complementary dilution molecule. 1. Incorporation of channel peptides. Langmuir 2005, 21, 11666–11672. [Google Scholar] [CrossRef]
- Andersson, J.; Fuller, M.; Wood, K.; Holt, S.; Köper, I. A tethered bilayer lipid membrane that mimics microbial membranes. Phys. Chem. Chem. Physics. 2018, 20, 12958–12969. [Google Scholar] [CrossRef]
- Ciumac, D.; Gong, H.; Campbell, R.A.; Campana, M.; Xu, H.; Lu, J.R. Structural elucidation upon binding of antimicrobial peptides into binary mixed lipid monolayers mimicking bacterial membranes. J. Colloid Interface Sci. 2021, 598, 193–205. [Google Scholar] [CrossRef]
- Jeworrek, C.; Evers, F.; Howe, J.; Brandenburg, K.; Tolan, M.; Winter, R. Effects of specific versus nonspecific ionic interactions on the structure and lateral organization of lipopolysaccharides. Biophys. J. 2011, 100, 2169–2177. [Google Scholar] [CrossRef] [PubMed]
- Abraham, T.; Schooling, S.R.; Beveridge, T.J.; Katsaras, J. Monolayer Film Behavior of Lipopolysaccharide from Pseudomonas aeruginosa at the Air−Water Interface. Biomacromolecules 2008, 9, 2799–2804. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.G.; Schneck, E.; Quinn, B.E.; Konovalov, O.V.; Brandenburg, K.; Gutsmann, T.; Gill, T.; Hanna, C.B.; Pink, D.A.; Tanaka, M. Crucial roles of charged saccharide moieties in survival of gram negative bacteria against protamine revealed by combination of grazing incidence x-ray structural characterizations and Monte Carlo simulations. Phys. Rev. E 2010, 81, 041901. [Google Scholar] [CrossRef] [PubMed]
- Micciulla, S.; Gerelli, Y.; Schneck, E. Structure and Conformation of Wild-Type Bacterial Lipopolysaccharide Layers at Air-Water Interfaces. Biophys. J. 2019, 116, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Nowotarska, S.W.; Nowotarski, K.J.; Friedman, M.; Situ, C. Effect of structure on the interactions between five natural antimicrobial compounds and phospholipids of bacterial cell membrane on model monolayers. Molecules 2014, 19, 7497–7515. [Google Scholar] [CrossRef] [PubMed]
- Neville, F.; Hodges, C.S.; Liu, C.; Konovalov, O.; Gidalevitz, D. In situ characterization of lipid A interaction with antimicrobial peptides using surface X-ray scattering. Biochim. Biophys. Acta (BBA) Biomembr. 2006, 1758, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Thoma, J.; Abuillan, W.; Furikado, I.; Habe, T.; Yamamoto, A.; Gierlich, S.; Kaufmann, S.; Brandenburg, K.; Gutsmann, T.; Konovalov, O.; et al. Specific localisation of ions in bacterial membranes unravels physical mechanism of effective bacteria killing by sanitiser. Sci. Rep. 2020, 10, 12302. [Google Scholar] [CrossRef]
- Lind, T.K.; Wacklin, H.; Schiller, J.; Moulin, M.; Haertlein, M.; Pomorski, T.G.; Cárdenas, M. Formation and Characterization of Supported Lipid Bilayers Composed of Hydrogenated and Deuterated Escherichia coli Lipids. PLoS ONE 2015, 10, e0144671. [Google Scholar] [CrossRef]
- Lind, T.K.; Cárdenas, M.; Wacklin, H.P. Formation of Supported Lipid Bilayers by Vesicle Fusion: Effect of Deposition Temperature. Langmuir 2014, 30, 7259–7263. [Google Scholar] [CrossRef]
- Schönherr, H.; Johnson, J.M.; Lenz, P.; Frank, C.W.; Boxer, S.G. Vesicle adsorption and lipid bilayer formation on glass studied by atomic force microscopy. Langmuir ACS J. Surf. Colloids 2004, 20, 11600–11606. [Google Scholar] [CrossRef]
- Kalb, E.; Frey, S.; Tamm, L.K. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim. Biophys. Acta 1992, 1103, 307–316. [Google Scholar] [CrossRef]
- Figueira, T.; Freire, J.; Cunha-Santos, C.; Heras, M.; Goncalves, J.; Moscona, A.; Porotto, M.; Veiga, A.; Castanho, M. Quantitative analysis of molecular partition towards lipid membranes using surface plasmon resonance. Sci. Rep. 2017, 7, 1–10. [Google Scholar]
- Merz, C.; Knoll, W.; Textor, M.; Reimhult, E. Formation of supported bacterial lipid membrane mimics. Biointerphases 2008, 3, FA41–FA50. [Google Scholar] [CrossRef] [PubMed]
- Fragneto, G.; Charitat, T.; Graner, F.; Mecke, K.; Perino-Gallice, L.; Bellet-Amalric, E. A fluid floating bilayer. EPL (Europhys. Lett.) 2007, 53, 100. [Google Scholar] [CrossRef]
- Stidder, B.; Fragneto, G.; Roser, S.J. Structure and stability of DPPE planar bilayers. Soft Matter. 2007, 3, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Holt, S.A.; Hughes, A.V.; Daulton, E.L.; Arunmanee, W.; Heinrich, F.; Khalid, S.; Jefferies, D.; Charlton, T.R.; Webster, J.R.; et al. An accurate in vitro model of the E. coli envelope. Angew. Chem. Int. Ed. 2015, 54, 11952–11955. (In English) [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.V.; Patel, D.S.; Widmalm, G.; Klauda, J.B.; Clifton, L.A.; Im, W. Physical Properties of Bacterial Outer Membrane Models: Neutron Reflectometry & Molecular Simulation. Biophys. J. 2019, 116, 1095–1104. [Google Scholar]
- Sauerbrey, G. The Use of Quartz Oscillators for Weighing Thin Layers and for Microweighing. Z. Für Physik 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, G. Basic Principles of QCM-D. In QCM-D Studies on Polymer Behavior at Interfaces; Liu, G., Zhang, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–8. [Google Scholar]
- Saad, N.; Zaaba, S. Quartz Crystal Microbalance for Bacteria Application Review. In Proceedings of the 2014 2nd International Conference on Electronic Design (ICED), Penang, Malaysia, 19–21 August 2014. [Google Scholar]
- Hook, F.; Rodahl, M.; Keller, C.; Glasmastar, K.; Fredriksson, C.; Dahiqvist, P.; Kasemo, B. The dissipative QCM-D technique: Interfacial phenomena and sensor applications for proteins, biomembranes, living cells and polymers. In Proceedings of the 1999 Joint Meeting of the European Frequency and Time Forum and the IEEE International Frequency Control Symposium (Cat. No.99CH36313), Besancon, France, 13–16 April 1999. [Google Scholar]
- Alexander, T.E.; Lozeau, L.D.; Camesano, T.A. QCM-D characterization of time-dependence of bacterial adhesion. Cell Surface 2019, 5, 100024. [Google Scholar] [CrossRef]
- Edvardsson, M.; Svedhem, S.; Wang, G.; Richter, R.; Rodahl, M.; Kasemo, B. QCM-D and Reflectometry Instrument: Applications to Supported Lipid Structures and Their Biomolecular Interactions. Anal. Chem. 2008, 81, 349–361. [Google Scholar] [CrossRef]
- Guo, H.; Xing, Q.; Huang, R.; Lee, D.W.; Su, R.; Qi, W.; He, Z. Real-Time QCM-D Monitoring of Deposition of Gold Nanorods on a Supported Lipid Bilayer as a Model Cell Membrane. ACS Omega 2019, 4, 6059–6067. [Google Scholar] [CrossRef]
- Martin, D.K. Quartz Crystal Microbalance. In Nanobiotechnology of Biomimetic Membranes; Springer: New York, NY, USA, 2007; p. 96. [Google Scholar]
- Hsia, C.-Y.; Chen, L.; Singh, R.R.; DeLisa, M.P.; Daniel, S. A Molecularly Complete Planar Bacterial Outer Membrane Platform. Sci. Rep. 2016, 6, 32715. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Bakhtiar, R. Surface Plasmon Resonance Spectroscopy: A Versatile Technique in a Biochemist’s Toolbox. J. Chem. Educ. 2013, 90, 203–209. [Google Scholar] [CrossRef]
- Zhu, X.; Gao, T. Chapter 10—Spectrometry. In Nano-Inspired Biosensors for Protein Assay with Clinical Applications; Li, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 237–264. [Google Scholar]
- Galdiero, S.; Falanga, A.; Cantisani, M.; Vitiello, M.; Morelli, G.; Galdiero, M. Peptide-lipid interactions: Experiments and applications. Int. J. Mol. Sci. 2013, 14, 18758–18789. [Google Scholar] [CrossRef]
- Kinouchi, H.; Onishi, M.; Kamimori, H. Lipid membrane-binding properties of daptomycin using surface plasmon resonance. Anal. Sci. 2013, 29, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Neylon, C.; Lakey, J.H. Examining Protein–Lipid Complexes Using Neutron Scattering. In Lipid-Protein Interactions: Methods and Protocols; Kleinschmidt, J.H., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 119–150. [Google Scholar]
- Wood, M.H.; Clarke, S.M. Neutron Reflectometry for Studying Corrosion and Corrosion Inhibition. Metals 2017, 7, 304. [Google Scholar] [CrossRef]
- Torikai, N.; Yamada, N.L.; Noro, A.; Harada, M.; Kawaguchi, D.; Takano, A.; Matsushita, Y. Neutron Reflectometry on Interfacial Structures of the Thin Films of Polymer and Lipid. Polym. J. 2007, 39, 1238–1246. [Google Scholar] [CrossRef]
- Krueger, S.; Meuse, C.W.; Majkrzak, C.F.; Dura, J.A.; Berk, N.F.; Tarek, M.; Plant, A.L. Investigation of Hybrid Bilayer Membranes with Neutron Reflectometry: Probing the Interactions of Melittin. Langmuir 2001, 17, 511–521. [Google Scholar] [CrossRef]
- Majkrzak, C.F.; Berk, N.F. Advances in specular neutron reflectometry. Appl. Phys. A Mater. Sci. Process. 2002, 74, 67–69. [Google Scholar] [CrossRef]
- Cousin, F.; Fadda, G. An introduction to neutron reflectometry. EPJ Web Conf. 2020, 236, 04001. [Google Scholar] [CrossRef]
- Rajagopal, M.; Walker, S. Envelope Structures of Gram-Positive Bacteria. Curr. Top. Microbiol. Immunol. 2017, 404, 1–44. [Google Scholar]
- Rocha-Roa, C.; Orjuela, J.D.; Leidy, C.; Cossio, P.; Aponte-Santamaría, C. Cardiolipin prevents pore formation in phosphatidylglycerol bacterial membrane models. FEBS Lett. 2021, 595, 2701–2714. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.T.J.; Hale, J.D.; Elliott, M.; Hancock, R.E.W.; Straus, S.K. The importance of bacterial membrane composition in the structure and function of aurein 2.2 and selected variants. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-H.; Bhuiyan, M.S.; Shen, H.-H.; Cameron, D.R.; Rupasinghe, T.W.T.; Wu, C.-M.; Brun, A.P.L.; Kostoulias, X.; Domene, C.; Fulcher, A.J.; et al. Antibiotic resistance and host immune evasion in Staphylococcus aureus mediated by a metabolic adaptation. Proc. Natl. Acad. Sci. USA 2019, 116, 3722–3727. [Google Scholar] [CrossRef]
- Gray, D.A.; Wenzel, M. More Than a Pore: A Current Perspective on the In Vivo Mode of Action of the Lipopeptide Antibiotic Daptomycin. Antibiotics 2020, 9, 17. [Google Scholar] [CrossRef]
- Mescola, A.; Ragazzini, G.; Alessandrini, A. Daptomycin Strongly Affects the Phase Behavior of Model Lipid Bilayers. J. Phys. Chem. B 2020, 124, 8562–8571. [Google Scholar] [CrossRef]
- Juhaniewicz-Dębińska, J.; Dziubak, D.; Sęk, S. Physicochemical Characterization of Daptomycin Interaction with Negatively Charged Lipid Membranes. Langmuir 2020, 36, 5324–5335. [Google Scholar] [CrossRef] [PubMed]
- Khondker, A.; Bider, R.-C.; Passos-Gastaldo, I.; Wright, G.D.; Rheinstädter, M.C. Membrane interactions of non-membrane targeting antibiotics: The case of aminoglycosides, macrolides, and fluoroquinolones. Biochim. Biophys. Acta (BBA) Biomembr. 2021, 1863, 183448. [Google Scholar] [CrossRef]
- Juhaniewicz-Dębińska, J.; Lasek, R.; Tymecka, D.; Burdach, K.; Bartosik, D.; Sęk, S. Physicochemical and Biological Characterization of Novel Membrane-Active Cationic Lipopeptides with Antimicrobial Properties. Langmuir ACS J. Surf. Colloids 2020, 36, 12900–12910. [Google Scholar] [CrossRef]
- Wang, K.F.; Nagarajan, R.; Camesano, T.A. Antimicrobial peptide alamethicin insertion into lipid bilayer: A QCM-D exploration. Colloids Surf. B Biointerfaces 2014, 116, 472–481. [Google Scholar] [CrossRef]
- Cooper, M.A.; Williams, D.H.; Cho, Y.R. Surface plasmon resonance analysis of glycopeptide antibiotic activity at a model membrane surface. Chem. Commun. 1997, 17, 1625–1626. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Kabara, J.J.; Swieczkowski, D.M.; Conley, A.J.; Truant, J.P. Fatty acids and derivatives as antimicrobial agents. Antimicrob. Agents Chemother. 1972, 2, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kabara, J.J.; Vrable, R. Antimicrobial lipids: Natural and synthetic fatty acids and monoglycerides. Lipids 1977, 12, 753–759. [Google Scholar] [CrossRef]
- Batovska, D.; Todorova, I.; Tsvetkova, I.; Najdenski, H. Antibacterial Study of the Medium Chain Fatty Acids and Their 1-Monoglycerides: Individual Effects and Synergistic Relationships. Pol. J. Microbiol. Pol. Tow. Mikrobiol. Pol. Soc. Microbiol. 2009, 58, 43–47. [Google Scholar]
- Giger, K.; Lamberson, E.R.; Hovis, J.S. Formation of complex three-dimensional structures in supported lipid bilayers. Langmuir 2009, 25, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K.R.; Martin, L.L.; Ackland, M.L.; Torriero, A.A. Real-Time Quartz Crystal Microbalance Monitoring of Free Docosahexaenoic Acid Interactions with Supported Lipid Bilayers. Langmuir 2016, 32, 11717–11727. [Google Scholar] [CrossRef]
- Yoon, B.K.; Jackman, J.A.; Kim, M.C.; Cho, N.J. Spectrum of Membrane Morphological Responses to Antibacterial Fatty Acids and Related Surfactants. Langmuir 2015, 31, 10223–10232. [Google Scholar] [CrossRef]
- Bello, G.; Bodin, A.; Lawrence, M.J.; Barlow, D.; Mason, A.J.; Barker, R.D.; Harvey, R.D. The influence of rough lipopolysaccharide structure on molecular interactions with mammalian antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2016, 1858, 197–209. [Google Scholar] [CrossRef][Green Version]
- Perczyk, P.; Wójcik, A.; Hachlica, N.; Wydro, P.; Broniatowski, M. The composition of phospholipid model bacterial membranes determines their endurance to secretory phospholipase A2 attack—The role of cardiolipin. Biochim. Biophys. Acta (BBA)—Biomembr. 2020, 1862, 183239. [Google Scholar] [CrossRef]
- Sanders, M.R.; Clifton, L.A.; Frazier, R.A.; Green, R.J. Role of Lipid Composition on the Interaction between a Tryptophan-Rich Protein and Model Bacterial Membranes. Langmuir 2016, 32, 2050–2057. [Google Scholar] [CrossRef]
- Gong, H.; Sani, M.-A.; Hu, X.; Fa, K.; Hart, J.W.; Liao, M.; Hollowell, P.; Carter, J.; Clifton, L.A.; Campana, M.; et al. How do Self-Assembling Antimicrobial Lipopeptides Kill Bacteria? ACS Appl. Mater. Interfaces 2020, 12, 55675–55687. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Voo, Z.X.; Graham, B.; Spiccia, L.; Martin, L.L. Real-time examination of aminoglycoside activity towards bacterial mimetic membranes using Quartz Crystal Microbalance with Dissipation monitoring (QCM-D). Biochim. Biophys. Acta (BBA) 2015, 1848, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Sendecki, A.M.; Poyton, M.F.; Baxter, A.J.; Yang, T.; Cremer, P.S. Supported Lipid Bilayers with Phosphatidylethanolamine as the Major Component. Langmuir 2017, 33, 13423–13429. [Google Scholar] [CrossRef] [PubMed]
- Horne, J.E.; Brockwell, D.J.; Radford, S.E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 2020, 295, 10340–10367. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Raetz, C.R. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 1978, 42, 614–659. [Google Scholar] [CrossRef]
- Clifton, L.A.; Ciesielski, F.; Skoda, M.W.; Paracini, N.; Holt, S.A.; Lakey, J.H. The Effect of Lipopolysaccharide Core Oligosaccharide Size on the Electrostatic Binding of Antimicrobial Proteins to Models of the Gram Negative Bacterial Outer Membrane. Langmuir 2016, 32, 3485–3494. [Google Scholar] [CrossRef]
- Dupuy, F.G.; Pagano, I.; Andenoro, K.; Peralta, M.F.; Elhady, Y.; Heinrich, F.; Tristram-Nagle, S. Selective Interaction of Colistin with Lipid Model Membranes. Biophys. J. 2018, 114, 919–928. [Google Scholar] [CrossRef]
- Han, M.-L.; Shen, H.-H.; Hansford, K.A.; Schneider, E.K.; Sivanesan, S.; Roberts, K.D.; Thompson, P.E.; Le Brun, A.P.; Zhu, Y.; Sani, M.-A.; et al. Investigating the Interaction of Octapeptin A3 with Model Bacterial Membranes. ACS Infect. Dis. 2017, 3, 606–619. [Google Scholar] [CrossRef]
- Han, M.L.; Velkov, T.; Zhu, Y.; Roberts, K.D.; Le Brun, A.P.; Chow, S.H.; Gutu, A.D.; Moskowitz, S.M.; Shen, H.H.; Li, J. Polymyxin-Induced Lipid A Deacylation in Pseudomonas aeruginosa Perturbs Polymyxin Penetration and Confers High-Level Resistance. ACS Chem. Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef]
- Thormar, H.; Hilmarsson, H.; Bergsson, G. Stable concentrated emulsions of the 1-monoglyceride of capric acid (monocaprin) with microbicidal activities against the food-borne bacteria Campylobacter jejuni, Salmonella spp., and Escherichia coli. Appl. Environ. Microbiol. 2006, 72, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Yoon, B.K.; Jackman, J.A.; Kim, M.C.; Sut, T.N.; Cho, N.J. Correlating Membrane Morphological Responses with Micellar Aggregation Behavior of Capric Acid and Monocaprin. Langmuir 2017, 33, 2750–2759. [Google Scholar] [CrossRef] [PubMed]
- Hyldgaard, M.; Sutherland, D.S.; Sundh, M.; Mygind, T.; Meyer, R.L. Antimicrobial mechanism of monocaprylate. Appl. Environ. Microbiol. 2012, 78, 2957–2965. [Google Scholar] [CrossRef] [PubMed]
- Bishop, R.E. Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 1881–1896. [Google Scholar] [CrossRef]
- Webb, C.T.; Heinz, E.; Lithgow, T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 2012, 20, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.-H.; Leyton, D.L.; Shiota, T.; Belousoff, M.J.; Noinaj, N.; Lu, J.; Holt, S.A.; Tan, K.; Selkrig, J.; Webb, C.T.; et al. Reconstitution of a nanomachine driving the assembly of proteins into bacterial outer membranes. Nat. Commun. 2014, 5, 5078. [Google Scholar] [CrossRef]
- Gruss, F.; Zähringer, F.; Jakob, R.P.; Burmann, B.M.; Hiller, S.; Maier, T. The structural basis of autotransporter translocation by TamA. Nat. Struct. Mol. Biol. 2013, 20, 1318–1320. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Bamert, R.S.; Le Brun, A.P.; Duff, A.P.; Wu, C.-M.; Hsu, H.-Y.; Shiota, T.; Lithgow, T.; Shen, H.-H. Substrate-dependent arrangements of the subunits of the BAM complex determined by neutron reflectometry. Biochim. Biophys. Acta (BBA) Biomembr. 2021, 1863, 183587. [Google Scholar] [CrossRef]
- Grage, S.L.; Keleshian, A.M.; Turdzeladze, T.; Battle, A.R.; Tay, W.C.; May, R.P.; Holt, S.A.; Contera, S.A.; Haertlein, M.; Moulin, M.; et al. Bilayer-mediated clustering and functional interaction of MscL channels. Biophys. J. 2011, 100, 1252–1260. [Google Scholar] [CrossRef]
- Tan, C.; Hosseini, S.F.; Jafari, S.M. Cubosomes and Hexosomes as Novel Nanocarriers for Bioactive Compounds. J. Agric. Food Chem. 2022, 70, 1423–1437. [Google Scholar] [CrossRef]
- Yaghmur, A.; Mu, H. Recent advances in drug delivery applications of cubosomes, hexosomes, and solid lipid nanoparticles. Acta Pharm. Sin. B 2021, 11, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Li, M.-H. Recent Progress in Polymer Cubosomes and Hexosomes. Macromol. Rapid Commun. 2021, 42, 2100194. [Google Scholar] [CrossRef] [PubMed]






| Model Membrane Types | Bacterial Membrane System | Applications | Observations | Ref |
|---|---|---|---|---|
| monolayer | DPPG | Interaction with antimicrobial peptides |
| [120] |
| Ra-LPS | Effects of divalent cations Ca2+ | In the presence of Ca2+: Thickness of: Ra-LPS tail: 13.8 ± 0.1 Å Inner core oligosaccharide: 23.5 ± 0.5 Å Outer core oligosaccharide: 8.6 ± 0.5 Å In the presence of EDTA: Thickness of: Ra-LPS tails: 12.7 ± 0.2 Å Inner core oligosaccharide: 21.2 ± 0.5 Å Outer core oligosaccharide: 8.5 ± 0.2 Å There were 5.3 Ca2+ per Ra-LPS headgroup. The presence of EDTA rendered a less ordered Ra-LPS monolayer. | [9] | |
| Ra-LPS/DPPC | Interaction with antimicrobial peptides (LL37, LFb) |
| [117] | |
| Rc-LPS/DPPC | Interaction with antimicrobial peptides (LL37, LFb) |
After adding LL37 or LFb peptides:
| [117] | |
| Rc-LPS | Interaction with antimicrobial peptides (G3, C8G3) |
| [42] | |
| Symmetry bilayer | POPE/POPG | NA |
| [18] |
| Lipid extracts of E. coli | NA |
| [69] | |
| DMPC/DMPG | Interaction with LL-37-loaded cubosomes |
| [43] | |
| POPE/POPG/TOCL | Interaction with antimicrobial peptides (colistin) |
| [127] | |
| LPS/DLPG | Interaction with antimicrobial peptides (colistin) |
| [127] | |
| Asymmetry bilayer | Ra-LPS | NA |
| [77] |
| Effect of divalent cations | In the presence of Ca2+ solution:
| [9] | ||
| NA |
| [8] | ||
| Rc-LPS | NA | The asymmetry (Rc-LPS/ DPPC) of the outer leaflet ratio: 25:58 and the inner leaflet ratio: 28:57
| [8] | |
| Interaction with antimicrobial peptides (G3, C8G3) |
| [42] | ||
| Lipid A | NA |
| [8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Ren, R.; Lyu, L.; Song, J.; Wang, Y.; Lin, T.-W.; Brun, A.L.; Hsu, H.-Y.; Shen, H.-H. Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials. Membranes 2022, 12, 906. https://doi.org/10.3390/membranes12100906
Li S, Ren R, Lyu L, Song J, Wang Y, Lin T-W, Brun AL, Hsu H-Y, Shen H-H. Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials. Membranes. 2022; 12(10):906. https://doi.org/10.3390/membranes12100906
Chicago/Turabian StyleLi, Shiqi, Ruohua Ren, Letian Lyu, Jiangning Song, Yajun Wang, Tsung-Wu Lin, Anton Le Brun, Hsien-Yi Hsu, and Hsin-Hui Shen. 2022. "Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials" Membranes 12, no. 10: 906. https://doi.org/10.3390/membranes12100906
APA StyleLi, S., Ren, R., Lyu, L., Song, J., Wang, Y., Lin, T.-W., Brun, A. L., Hsu, H.-Y., & Shen, H.-H. (2022). Solid and Liquid Surface-Supported Bacterial Membrane Mimetics as a Platform for the Functional and Structural Studies of Antimicrobials. Membranes, 12(10), 906. https://doi.org/10.3390/membranes12100906









