Simulation Study of Chain-like Body Translocation through Conical Pores in Thick Membranes
Abstract
:1. Introduction
2. Model Description and Computational Framework
The Sequential Algorithm and Monte Carlo Experiments
3. Results and Discussion
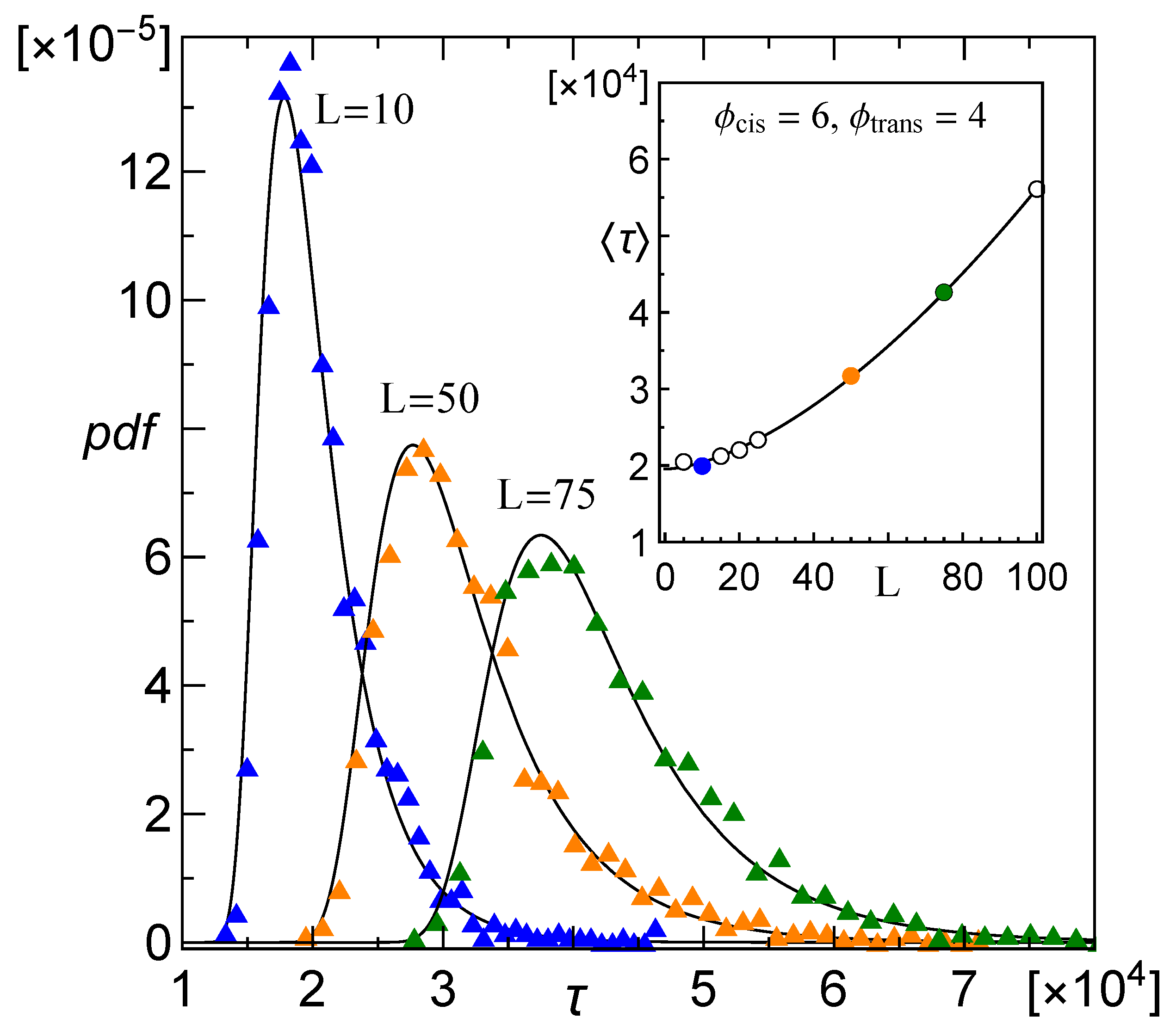
3.1. Probability Distribution of Translocation Time
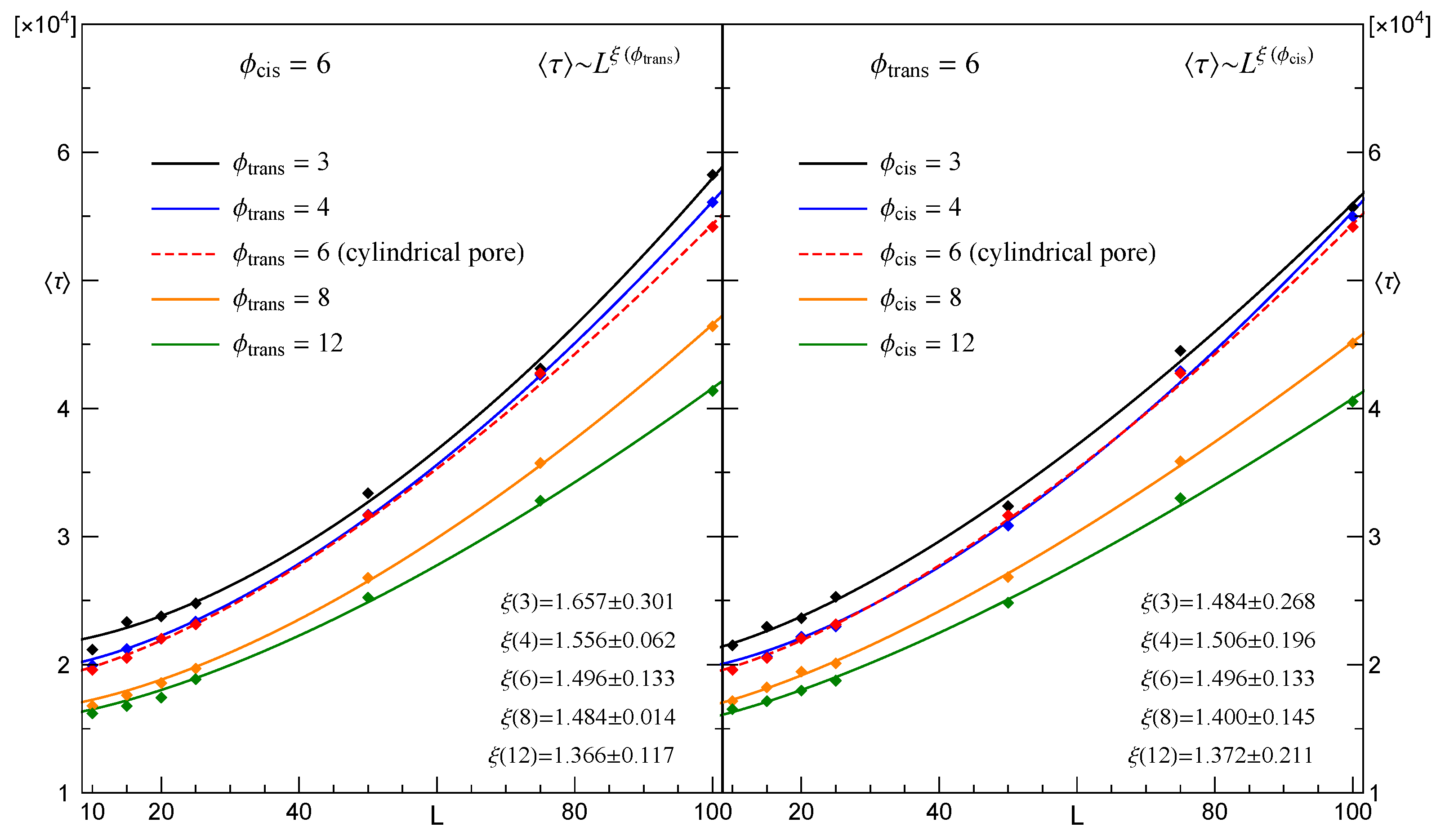
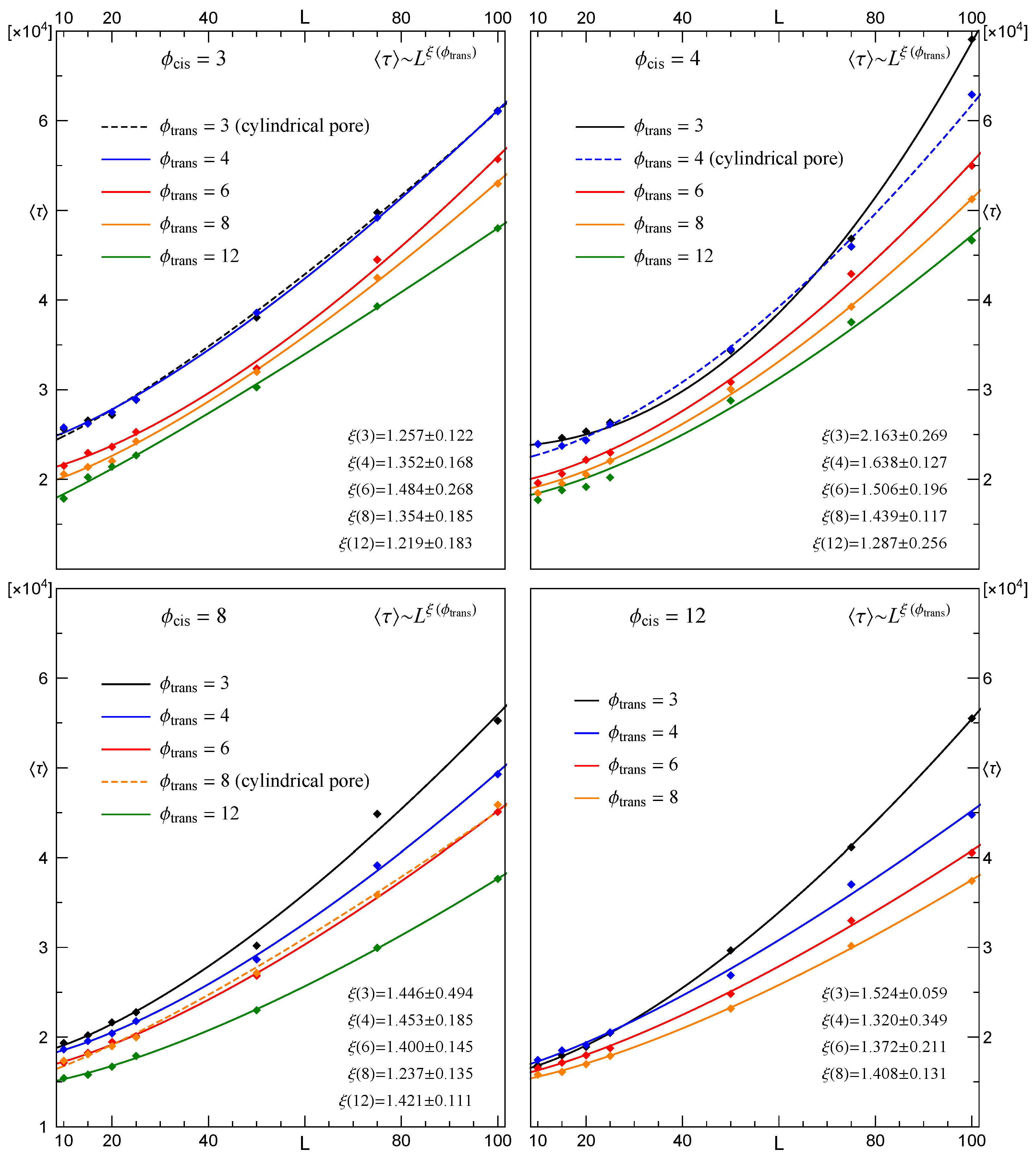
3.2. Dependence of the Mean Translocation Time on the Membrane Thickness
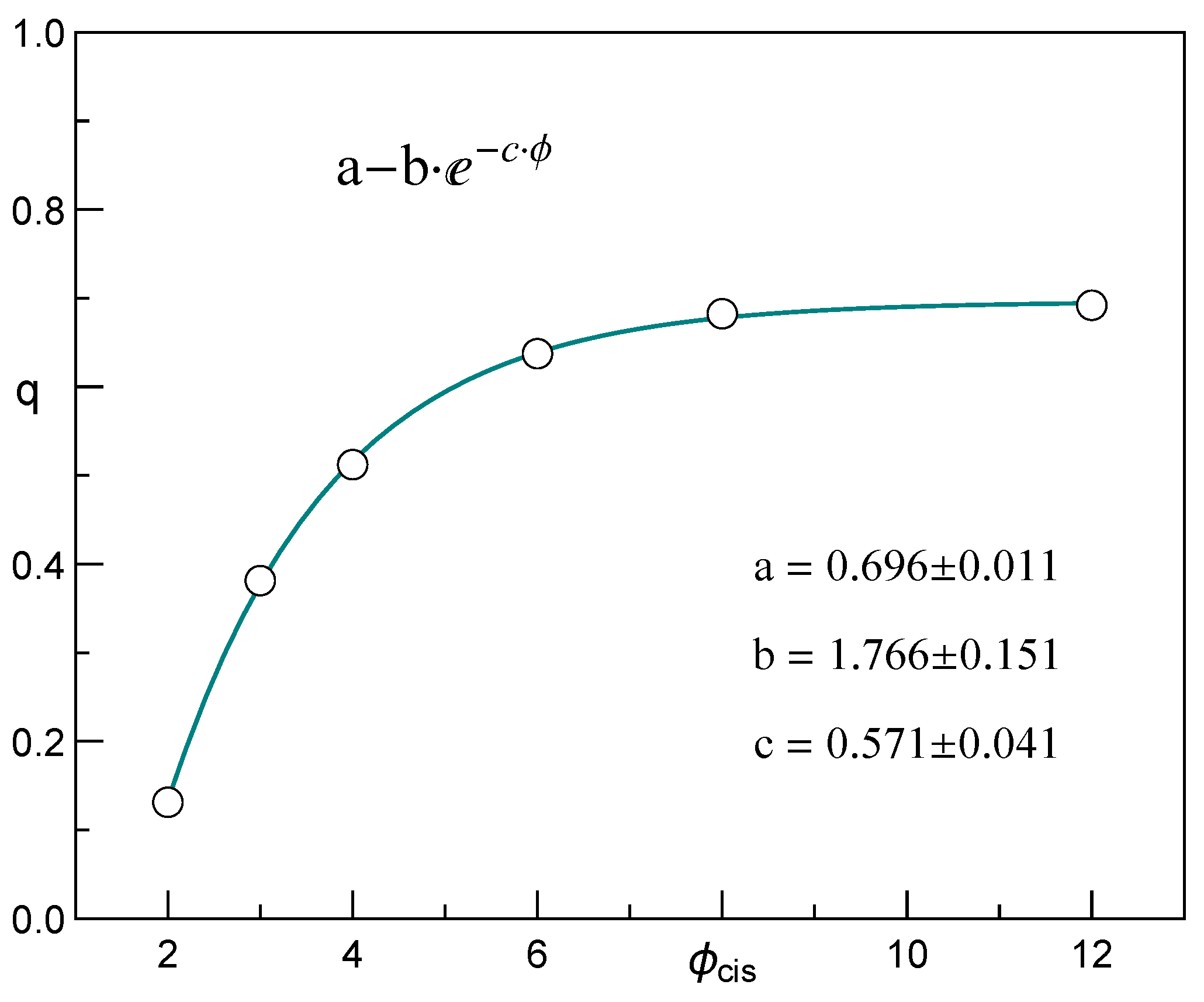
3.3. The Regression Models for the Translocation–Time-Distribution Parameters in Relation to Pore Diameter
4. Final Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Watson, H. Biological membranes. Essays Biochem. 2015, 15, 43–69. [Google Scholar] [CrossRef] [PubMed]
- Groves, J.T. Membrane Mechanics in Living Cells. Dev. Cell 2019, 48, 15–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, C.H. Biomimetic membranes for sensor and separation applications. Anal. Bioanal. Chem. 2009, 395, 697–718. [Google Scholar] [CrossRef] [PubMed]
- Makabe, R.; Ueyama, T.; Sakai, H.; Tanioka, A. Commercial Pressure Retarded Osmosis Systems for Seawater Desalination Plants. Membranes 2021, 11, 69. [Google Scholar] [CrossRef]
- Thabo, B.; Okoli, B.J.; Modise, S.J.; Nelana, S. Rejection Capacity of Nanofiltration Membranes for Nickel, Copper, Silver and Palladium at Various Oxidation States. Membranes 2021, 11, 653. [Google Scholar] [CrossRef]
- Marino, T.; Figoli, A. Arsenic Removal by Liquid Membranes. Membranes 2015, 5, 150–167. [Google Scholar] [CrossRef] [Green Version]
- Goyal, S.; Kim, Y.; Li, Y.; Iqbal, Y.L. Active and biomimetic nanofilters for selective protein separation. Biomed. Microdevices 2010, 12, 17–24. [Google Scholar] [CrossRef]
- Nunes, S.A.; Magalhaes, H.L.F.; de Farias Neto, S.R.; Lima, A.G.B.; Nascimento, L.P.C.; Farias, F.P.M.; Lima, E.S. Impact of Permeable Membrane on the Hydrocyclone Separation Performance for Oily Water Treatment. Membranes 2020, 10, 350. [Google Scholar] [CrossRef]
- Ileri, N.; Létant, S.; Britten, J.; Nguyen, H.; Larson, C.; Zaidi, S.; Palazoglu, A.; Faller, R.; Tringe, J.W.; Stroeve, P. Efficient Nanoporous Silicon Membranes for Integrated Microfluidic Separation and Sensing Systems. MRS Online Proc. Libr. 2011, 1191, OO09-02. [Google Scholar] [CrossRef]
- Gu, K.; Pang, S.; Zhou, Z.; Gao, C. Sodium Chloroacetate Modified Polyethyleneimine/Trimesic Acid Nanofiltration Membrane to Improve Antifouling Performance. Membranes 2020, 11, 705. [Google Scholar] [CrossRef]
- Kumar, M.; Stone, H.A. Membrane science emerging as a convergent scientific field with molecular origins and understanding, and global impact. Proc. Natl. Acad. Sci. USA 2021, 118, e2106494118. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yu, S.; Harrell, C.; Martin, C.R. Conical Nanopore Membranes. Preparation and Transport Properties. Anal. Chem. 2004, 76, 2025–2030. [Google Scholar] [CrossRef] [PubMed]
- Palyulin, V.V.; Ala-Nissila, T.; Metzler, R. Polymer translocation: The first two decades and the recent diversification. Soft Matter 2014, 10, 9016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.; Gen, Y.; Xie, H.; Jiang, Z.; Yang, Z. Translocation of a Polymer through a Crowded Channel under Electrical Force. BioMed Res. Int. 2017, 2017, 5267185. [Google Scholar] [CrossRef] [PubMed]
- Hamidabad, M.N.; Abdolvahab, R.H. translocation through a narrow pore under a pulling force. Sci. Rep. 2019, 9, 17885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Jou, I.; Ermann, N.; Muthukumar, M.; Keyser, U.F.; Bell, N.A.W. Dynamics of driven polymer transport through a nanopore. Nat. Phys. 2021, 17, 1043–1049. [Google Scholar] [CrossRef]
- Luan, B.; Martyna, G.; Stolovitzky, G. Characterizing and Controlling the Motion of ssDNA in a Solid-State Nanopore. Biophys. J. 2011, 101, 2214–2222. [Google Scholar] [CrossRef] [Green Version]
- Reynaud, L.; Bouchet-Spinelli, A.; Raillon, Z.; Buhot, A. Sensing with Nanopores and Aptamers: A Way Forward. Sensors 2020, 20, 4495. [Google Scholar] [CrossRef]
- Crnković, A.; Srnko, M.; Anderluh, G. Biological Nanopores: Engineering on Demand. Life 2021, 11, 27. [Google Scholar] [CrossRef]
- Gu, L.-Q.; Shim, J.W. Single Molecule Sensing by Nanopores and Nanopore Devices. Analyst 2010, 135, 441–451. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Baker, L.A.; Hillebrenner, H.; Martin, C.R. Biosensing with conically shaped nanopores and nanotubes. Phys. Chem. Chem. Phys. 2006, 8, 4976–4988. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, D.; Cui, W.; Zhang, H.; Pang, W.; Duan, X. Fabrications, Applications and Challenges of Solid-state Nanopores: A Mini Review. Nanomater. Nanotechnol. 2016, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, K.; Chen, S.B. Polyelectrolyte Translocation through a Corrugated Nanopore. Macromol. Theory Simul. 2020, 29, 2000042. [Google Scholar] [CrossRef]
- Mohan, A.; Kolomeisky, A.B.; Pasquali, M. Polymer translocation through pores with complex geometries. J. Chem. Phys. 2010, 133, 024902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdolvahab, R.H.; Hamidabad, M.N. Pore shapes effects on polymer translocation. Eur. Phys. J. E 2020, 43, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, Y.; Menard, L.D.; Panyukov, S.; Rubinstein, M.; Ramsey, J.M. Enhanced nanochannel translocation and localization of genomic DNA molecules using three-dimensional nanofunnels. Nat. Commun. 2017, 8, 807. [Google Scholar] [CrossRef] [Green Version]
- Menais, T.; Mossa, S.; Buhot, A. Polymer translocation through nano-pores in vibrating thin membranes. Sci. Rep. 2016, 6, 38558. [Google Scholar] [CrossRef] [Green Version]
- Ding, K.-J.; Cai, D.-Q.; Zhan, F.-R.; Wu, L.-J.; Wu, Y.-J.; Yu, Z.-L. Single long-polymer translocation through a long pore. Chin. Phys. 2006, 15, 0940-7. [Google Scholar] [CrossRef]
- Cohen, B.E. Membrane Thickness as a Key Factor Contributing to the Activation of Osmosensors and Essential Ras Signaling Pathways. Front. Cell Dev. Biol. 2018, 6, 76. [Google Scholar] [CrossRef] [Green Version]
- Li, L.B.; Vorobyov, I.; Allen, T.W. The role of membrane thickness in charged protein–lipid interactions. Biochim. Biophys. Acta 2011, 1818, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Lomora, M.; Dinu, I.A.; Itel, F.; Rigo, S.; Spulber, M.; Palivan, C.G. Does Membrane Thickness Affect the Transport of Selective Ions Mediated by Ionophores in Synthetic Membranes? Macrom. Rapid Commun. 2015, 36, 1929–1934. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Lloyd, K.; Sakashita, N.; Minato, S.; Ishida, K.; Mitsui, T. Clog and Release, and Reverse Motions of DNA in a Nanopore. Polymers 2019, 11, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Sensale, S.; Pan, Z.; Senapati, S.; Chang, H.-C. Slowing down DNA translocation through solid-state nanopores by edge-field leakage. Nat. Commun. 2021, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Nikoofard, N.; Fazli, H. A flexible polymer confined inside a cone-shaped nano-channel. Soft Matter 2015, 11, 4879–4887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.-Z.; Li, H.; Xu, X.; Luo, M.-B. Simulation study on the translocation of polyelectrolyte through conical nanopores. J. Phys. Condens. Matter 2018, 30, 495101. [Google Scholar] [CrossRef]
- Sun, L.-Z.; Cao, W.-P.; Wang, C.-H.; Xu, X. The translocation dynamics of the polymer through a conical pore: Non-stuck, weak-stuck, and strong-stuck modes. J. Chem. Phys. 2021, 154, 054903. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhuri, A.; Kapri, R. Driven translocation of a flexible polymer through an interacting conical pore. arXiv 2012, arXiv:2102.08067. [Google Scholar]
- Baschnagel, J.; Wittmer, J.P.; Meyer, H. Monte Carlo Simulation of Polymers: Coarse-Grained Models. In Computational Soft Matter: From Theory to Algorithms—Lecture Notes; Atting, A., Binder, K., Grubmuller, H., Kremer, K., Eds.; NIC Series: Julich, Germany, 2004; Volume 23, pp. 83–140. ISBN 3-00-12641-4. Available online: https://juser.fz-juelich.de/record/35298/files/nic-series-23.pdf (accessed on 15 December 2021).
- Binder, K. Monte Carlo and Molecular Dynamics Simulations in Polymer Science; Oxford University Press, Inc.: New York, NY, USA, 2015. [Google Scholar]
- Grzybowski, A.Z.; Domański, Z. A sequential algorithm with a built in tension-propagation mechanism for modeling the chain-like bodies dynamics. arXiv 2013, arXiv:1312.4206. [Google Scholar]
- Grzybowski, A.; Domański, Z.; Bartłomiejczyk, K. Algorithmization and simulation of the chain-like structures’ dynamics-interrelations between movement characteristics. Acta Electrotech. Inform. 2013, 13, 40–44. [Google Scholar] [CrossRef]
- Sarabadani, J.; Ala-Nissila, T. Theory of pore-driven and end-pulled polymer translocation dynamics through a nanopore: An overview. J. Phys. Condens. Matter 2018, 30, 274002. [Google Scholar] [CrossRef] [Green Version]
- Buyukdagli, S.; Sarabadani, J.; Ala-Nissila, T. Theoretical Modeling of Polymer Translocation: From the Electrohydrodynamics of Short Polymers to the Fluctuating Long Polymers. Polymers 2019, 11, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanunu, M.; Morrison, W.; Rabin, Y.; Grosberg, A.Y.; Meller, A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotechnol. 2010, 5, 160–165. [Google Scholar] [CrossRef] [Green Version]
- Rowghanian, P.; Grosberg, A.Y. Electrophoretic capture of a DNA chain into a nanopore. Phys. Rev. E 2013, 87, 042722. [Google Scholar] [CrossRef] [Green Version]
- Arnold, T.; Emerson, J. Nonparametric goodness-of-fit tests for discrete null distributions. R J. 2011, 3, 34–39. [Google Scholar] [CrossRef] [Green Version]
- Moyal, J.E. Theory of ionization fluctuations. Philos. Mag. 1955, 46, 263–280. [Google Scholar] [CrossRef]
- Domański, Z.; Grzybowski, A.Z. Probabilistic Model for Passage Time of Chain-Like Structure through Thick Membrane. Acta Phys. Pol. A 2021, 139, 513. [Google Scholar] [CrossRef]
- Lehtonen, J. The Lambert W function in ecological and evolutionary models. Methods Ecol. Evol. 2016, 7, 1110–1118. [Google Scholar] [CrossRef]
- Golicnik, M. On the Lambert W function and its utility in biochemical kinetics. Biochem. Eng. J. 2012, 63, 116–123. [Google Scholar] [CrossRef]

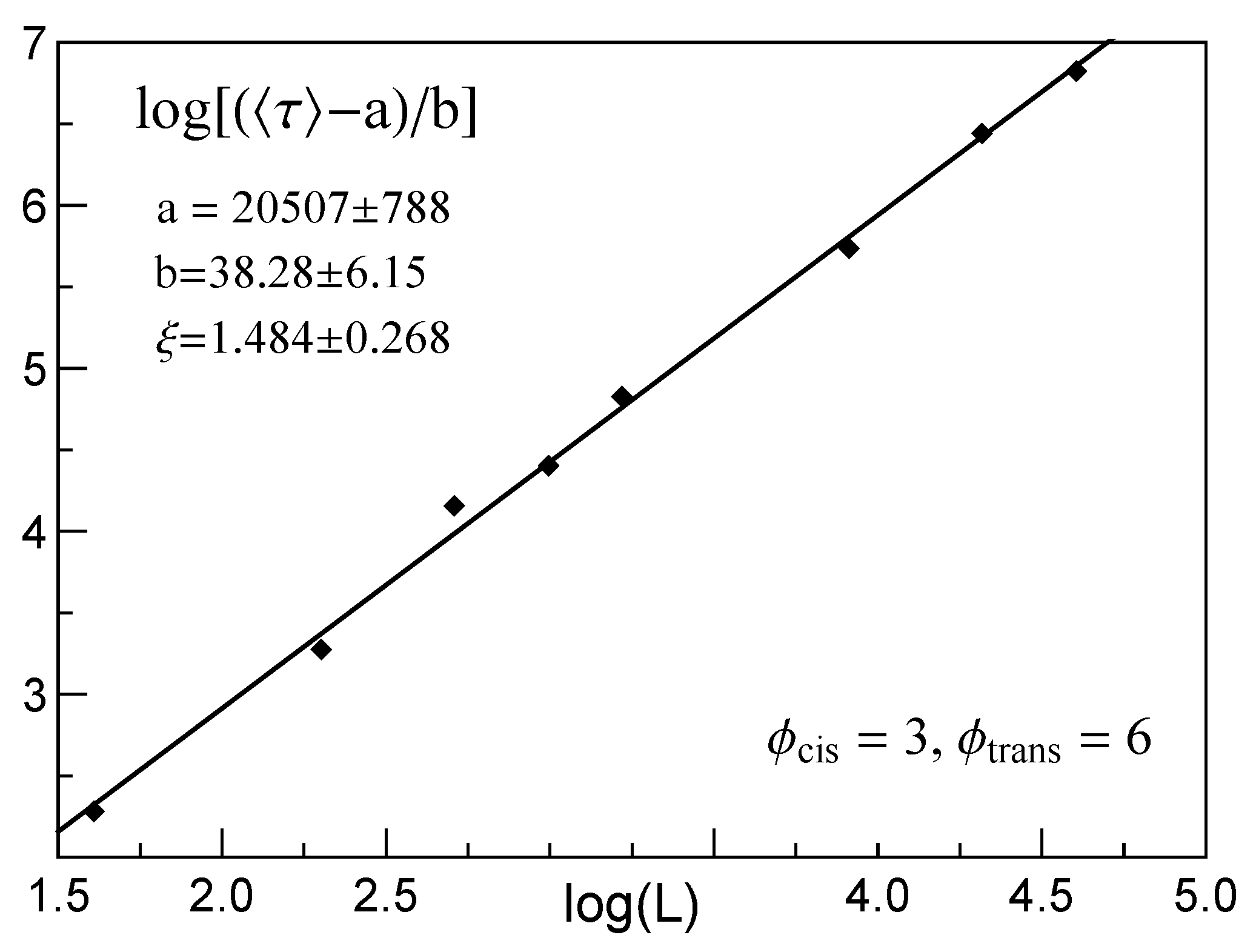


| Pore Length | |||||||
|---|---|---|---|---|---|---|---|
| 27637.8 | −522.2 | −687.7 | −614.9 | 0.837 | 1196.3 | 0.064 | |
| p-values: | 0.0000 | 0.0058 | 0.0002 | 0.3951 | |||
| 30,857.6 | −791.3 | −589.9 | 204.0 | 0.875 | 1143.9 | 0.051 | |
| p-values: | 0.0000 | 0.0001 | 0.0005 | 0.7656 | |||
| 39,421.5 | −1271.9 | −567.7 | 1566.3 | 0.888 | 1417.9 | 0.048 | |
| p values: | 0.0000 | 0.0000 | 0.0043 | 0.0787 | |||
| 52,573.9 | −1843.9 | −787.6 | 3123.7 | 0.943 | 1392.3 | 0.035 | |
| p values: | 0.0000 | 0.0000 | 0.0002 | 0.0015 | |||
| 70,436.7 | −2700.8 | −1266.1 | 4632.9 | 0.922 | 2502.7 | 0.049 | |
| p values: | 0.0000 | 0.0000 | 0.0006 | 0.0062 |
| Pore Length | |||||||
|---|---|---|---|---|---|---|---|
| 8.9963 | −0.08894 | −0.09771 | −0.06807 | 0.874 | 0.158 | 0.020 | |
| p values: | 0.0000 | 0.0008 | 0.0001 | 0.4751 | |||
| 9.1615 | −0.1368 | −0.0535 | 0.1113 | 0.932 | 0.115 | 0.014 | |
| p values: | 0.0000 | 0.0000 | 0.0014 | 0.1213 | |||
| 9.1120 | −0.1946 | 0.0060 | 0.4452 | 0.857 | 0.178 | 0.021 | |
| p values: | 0.0000 | 0.0000 | 0.7816 | 0.0006 | |||
| 9.6473 | −0.2338 | −0.0083 | 0.5692 | 0.860 | 0.220 | 0.025 | |
| p values: | 0.0000 | 0.0000 | 0.7575 | 0.0004 | |||
| 8.9997 | −0.3043 | 0.0870 | 1.1313 | 0.766 | 0.342 | 0.037 | |
| p values: | 0.0000 | 0.0000 | 0.0051 | 0.0000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domański, Z.; Grzybowski, A.Z. Simulation Study of Chain-like Body Translocation through Conical Pores in Thick Membranes. Membranes 2022, 12, 138. https://doi.org/10.3390/membranes12020138
Domański Z, Grzybowski AZ. Simulation Study of Chain-like Body Translocation through Conical Pores in Thick Membranes. Membranes. 2022; 12(2):138. https://doi.org/10.3390/membranes12020138
Chicago/Turabian StyleDomański, Zbigniew, and Andrzej Z. Grzybowski. 2022. "Simulation Study of Chain-like Body Translocation through Conical Pores in Thick Membranes" Membranes 12, no. 2: 138. https://doi.org/10.3390/membranes12020138
APA StyleDomański, Z., & Grzybowski, A. Z. (2022). Simulation Study of Chain-like Body Translocation through Conical Pores in Thick Membranes. Membranes, 12(2), 138. https://doi.org/10.3390/membranes12020138






