The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phylogenetic Analysis, Protein Sequence Alignment, Topology Prediction, and Protein Charge Prediction
2.2. Subcellular Localization Study
2.3. Yeast Isoflavone Uptake Assay
3. Results and Discussion
3.1. Phylogenetic and Protein Sequence Analyses of MATE Proteins
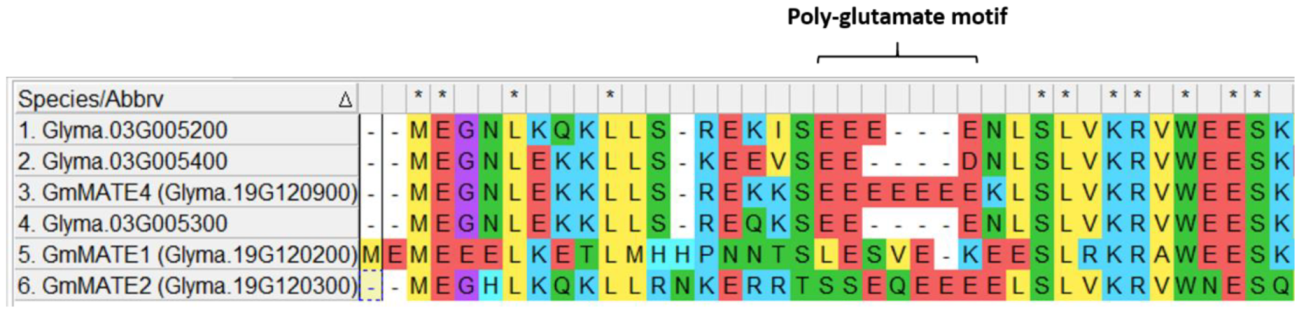
3.2. Identification of the Poly-Glutamate Motif in GmMATE4 by Sequence Alignment
3.3. The Effect of the Poly-Glutamate Motif on the Protein Net Charge
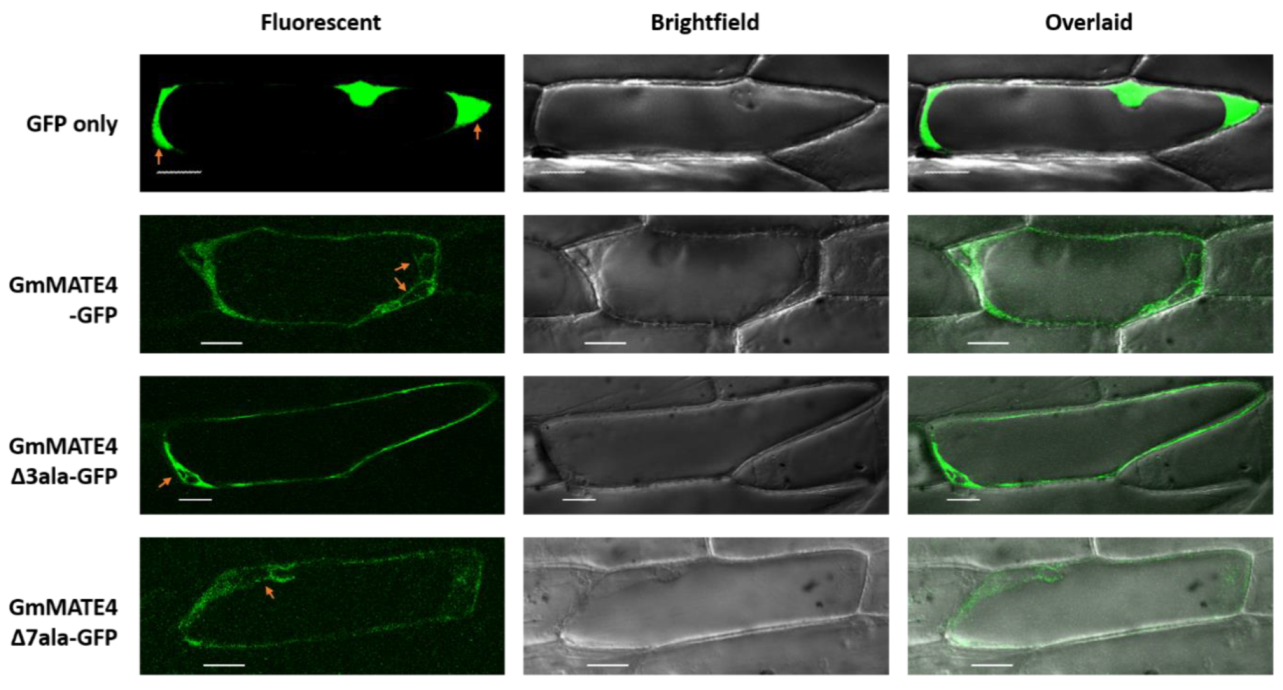
3.4. Subcellular Localization Study
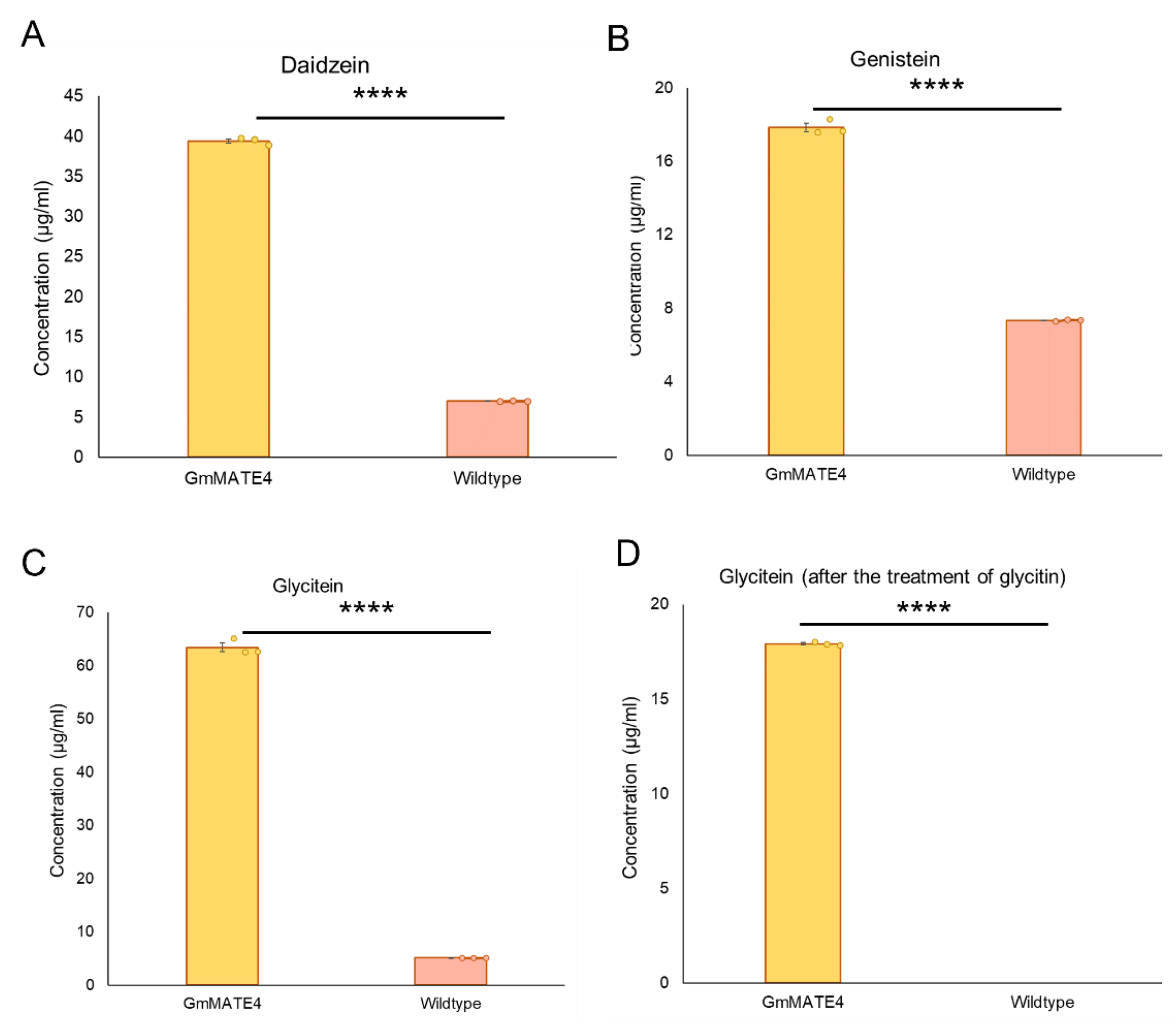
3.5. The Transporter Activity of GmMATE4
3.6. The Functional Significance of the Poly-Glutamate Motif on the Transporter Activity of GmMATE4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takanashi, K.; Shitan, N.; Yazaki, K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 2014, 31, 417–430. [Google Scholar] [CrossRef] [Green Version]
- Miyauchi, H.; Moriyama, S.; Kusakizako, T.; Kumazaki, K.; Nakane, T.; Yamashita, K.; Hirata, K.; Dohmae, N.; Nishizawa, T.; Ito, K.; et al. Structural basis for xenobiotic extrusion by eukaryotic MATE transporter. Nat. Commun. 2017, 8, 1633. [Google Scholar] [CrossRef]
- Lu, M.; Radchenko, M.; Symersky, J.; Nie, R.; Guo, Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 2013, 20, 1310–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; De Angeli, A.; Fernie, A.R.; Martinoia, E. Intra- and extra-cellular excretion of carboxylates. Trends Plant Sci. 2009, 15, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Shoji, T.; Inai, K.; Yazaki, Y.; Sato, Y.; Takase, H.; Shitan, N.; Yazaki, K.; Goto, Y.; Toyooka, K.; Matsuoka, K.; et al. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 2008, 149, 708–718. [Google Scholar] [CrossRef] [Green Version]
- Kusakizako, T.; Miyauchi, H.; Ishitani, R.; Nureki, O. Structural biology of the multidrug and toxic compound extrusion superfamily transporters. Biochim. Biophys. Acta-Biomembr. 2020, 1862, 183154. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.-S.; Ku, Y.-S.; Yung, W.-S.; Cheng, S.-S.; Man, C.-K.; Yang, L.; Song, S.; Chung, G.; Lam, H.-M. MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 2021, 22, 12017. [Google Scholar] [CrossRef] [PubMed]
- Debeaujon, I.; Peeters, A.J.M.; Léon-Kloosterziel, K.M.; Koornneef, M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 2001, 13, 853–872. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Dixon, R.A. MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 2009, 21, 2323–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, Y.; Wang, W.; Gai, J.; Li, Y. Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genom. 2016, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- dos Santos, A.L.; Chaves-Silva, S.; Yang, L.; Maia, L.G.S.; Chalfun-Júnior, A.; Sinharoy, S.; Zhao, J.; Benedito, V.A. Global analysis of the MATE gene family of metabolite transporters in tomato. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Singh, M.; Tripathi, R.D.; Trivedi, P.K. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 2014, 4, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; He, Z.; Pandey, G.K.; Tsuchiya, T.; Luan, S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 2002, 277, 5360–5368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagessar, K.L.; McHaourab, H.S.; Claxton, D.P. The N-terminal domain of an archaeal multidrug and toxin extrusion (MATE) transporter mediates proton coupling required for prokaryotic drug resistance. J. Biol. Chem. 2019, 294, 12807–12814. [Google Scholar] [CrossRef]
- Mikros, E.; Diallinas, G. Tales of tails in transporters. Open Biol. 2019, 9, 190083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diallinas, G. Dissection of transporter function: From genetics to structure. Trends Genet. 2016, 32, 576–590. [Google Scholar] [CrossRef] [PubMed]
- Li, M.W.; Muñoz, N.B.; Wong, C.F.; Wong, F.L.; Wong, K.-S.; Wong, J.W.-H.; Qi, X.; Li, K.-P.; Ng, M.-S.; Lam, H.-M. QTLs regulating the contents of antioxidants, phenolics, and flavonoids in soybean seeds share a common genomic region. Front. Plant Sci. 2016, 7, 854. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Stoeckert, C.J.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [Green Version]
- Prot-pi. Available online: https://www.protpi.ch/Calculator/ProteinTool (accessed on 22 December 2021).
- Halligan, B.D.; Ruotti, V.; Jin, W.; Laffoon, S.; Twigger, S.N.; Dratz, E.A. ProMoST (Protein Modification Screening Tool): A web-based tool for mapping protein modifications on two-dimensional gels. Nucleic Acids Res. 2004, 32, W638–W644. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; TriggsRaine, B.; Robbins, A.; Graham, K.C.; Woods, R.A. Identification of proteins that interact with a protein of interest: Applications of the yeast two-hybrid system. Mol. Cell. Biochem. 1997, 172, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Slatnar, A.; Jakopic, J.; Stampar, F.; Veberic, R.; Jamnik, P. The effect of bioactive compounds on in vitro and in vivo antioxidant activity of different berry juices. PLoS ONE 2012, 7, e47880. [Google Scholar] [CrossRef] [Green Version]
- Canelas, A.B.; ten Pierick, A.; Ras, C.; Seifar, R.M.; van Dam, J.C.; van Gulik, W.M.; Heijnen, J.J. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar] [CrossRef] [PubMed]
- Toro-Funes, N.; Odriozola-Serrano, I.; Bosch-Fusté, J.; Latorre-Moratalla, M.L.; Veciana-Nogués, M.T.; Izquierdo-Pulido, M.; Vidal-Carou, M.C. Fast simultaneous determination of free and conjugated isoflavones in soy milk by UHPLC–UV. Food Chem. 2012, 135, 2832–2838. [Google Scholar] [CrossRef]
- Griesmann, M.; Chang, Y.; Liu, X.; Song, Y.; Haberer, G.; Crook, M.B.; Billault-Penneteau, B.; Lauressergues, D.; Keller, J.; Imanishi, L.; et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 2018, 361, eaat1743. [Google Scholar] [CrossRef] [Green Version]
- Taiz, L. The plant vacuole. J. Exp. Biol. 1992, 172, 113–122. [Google Scholar] [CrossRef]
- Liao, Y.-Y.; Li, J.-L.; Pan, R.-L.; Chiou, T.-J. Structure–function analysis reveals amino acid residues of Arabidopsis phosphate transporter AtPHT1;1 crucial for its activity. Front. Plant Sci. 2019, 10, 1158. [Google Scholar] [CrossRef] [Green Version]
- Miller, C. ClC chloride channels viewed through a transporter lens. Nature 2006, 440, 484–489. [Google Scholar] [CrossRef]
- Miller, C. ClC channels: Reading eukaryotic function through prokaryotic spectacles. J. Gen. Physiol. 2003, 122, 129–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.; Lee, B.-C.; Lim, H.-H. Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+ antiporter. Proc. Natl. Acad. Sci. USA 2019, 116, 17345–17354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, L.; Campbell, E.B.; MacKinnon, R. Molecular mechanism of proton transport in CLC Cl-/H+ exchange transporters. Proc. Natl. Acad. Sci. USA 2012, 109, 11699–11704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmour, R.; Messner, P.; Guffanti, A.A.; Kent, R.; Scheberl, A.; Kendrick, N.; Krulwich, T.A. Two-dimensional gel electrophoresis analyses of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization of an S-layer protein with a role in alkaliphily. J. Bacteriol. 2000, 182, 5969–5981. [Google Scholar] [CrossRef] [Green Version]
- Matsuno, T.; Goto, T.; Ogami, S.; Morimoto, H.; Yamazaki, K.; Inoue, N.; Matsuyama, H.; Yoshimune, K.; Yumoto, I. Formation of proton motive force under low-aeration alkaline conditions in Alkaliphilic bacteria. Front. Microbiol. 2018, 9, 2331. [Google Scholar] [CrossRef] [Green Version]


| Net Charge of the Vacuolar N-Terminal Region | % of Acidic Amino Acids (No. of Acidic Amino Acids/Total Amino Acids) | ||
|---|---|---|---|
| pH 5 | pH 5.5 | ||
| Glyma.03G005200 | +0.773 | −0.37 | 24.2% (8/33) |
| Glyma.03G005400 | −1.881 | −3.245 | 29.4% (10/34) |
| Glyma.03G005300 | +0.794 | −0.363 | 23.5% (8/34) |
| GmMATE1 (Glyma.19G120200) | +0.3 | −1.534 | 27.5% (11/40) |
| GmMATE2 (Glyma.19G120300) | +2.708 | +1.426 | 21.6% (8/37) |
| GmMATE4 (Glyma.19G120900) | −0.348 | −2.043 | 33.3% (12/36) |
| GmMATE4Δ3ala | +1.992 | +0.712 | 25% (9/36) |
| GmMATE4Δ7ala | +5.113 | +4.385 | 13.9% (5/36) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, Y.-S.; Cheng, S.-S.; Cheung, M.-Y.; Niu, Y.; Liu, A.; Chung, G.; Lam, H.-M. The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity. Membranes 2022, 12, 206. https://doi.org/10.3390/membranes12020206
Ku Y-S, Cheng S-S, Cheung M-Y, Niu Y, Liu A, Chung G, Lam H-M. The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity. Membranes. 2022; 12(2):206. https://doi.org/10.3390/membranes12020206
Chicago/Turabian StyleKu, Yee-Shan, Sau-Shan Cheng, Ming-Yan Cheung, Yongchao Niu, Ailin Liu, Gyuhwa Chung, and Hon-Ming Lam. 2022. "The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity" Membranes 12, no. 2: 206. https://doi.org/10.3390/membranes12020206
APA StyleKu, Y.-S., Cheng, S.-S., Cheung, M.-Y., Niu, Y., Liu, A., Chung, G., & Lam, H.-M. (2022). The Poly-Glutamate Motif of GmMATE4 Regulates Its Isoflavone Transport Activity. Membranes, 12(2), 206. https://doi.org/10.3390/membranes12020206









