Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins
Abstract
:1. Introduction
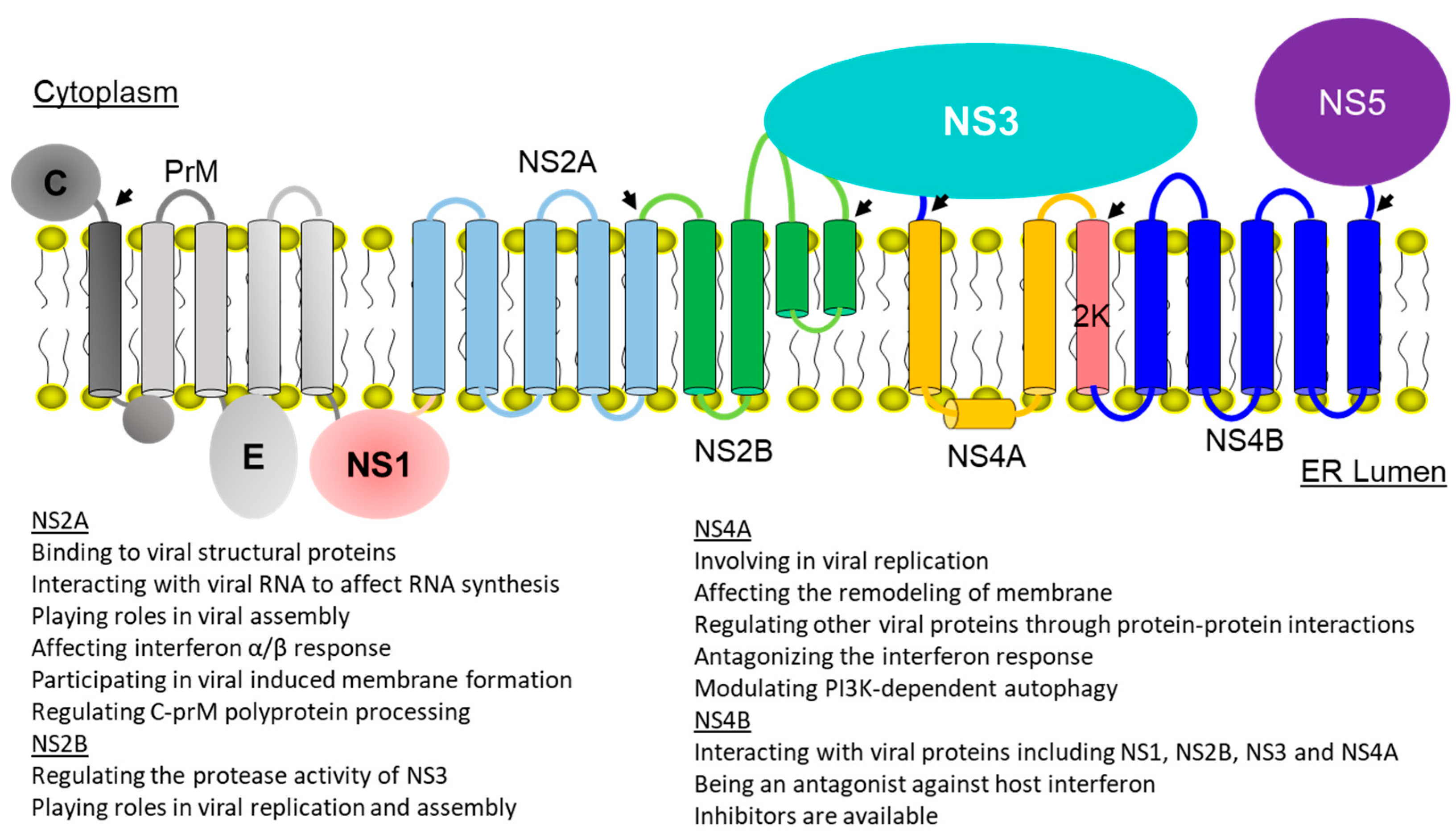
2. Membrane Topologies and Functions of Viral Membrane Proteins
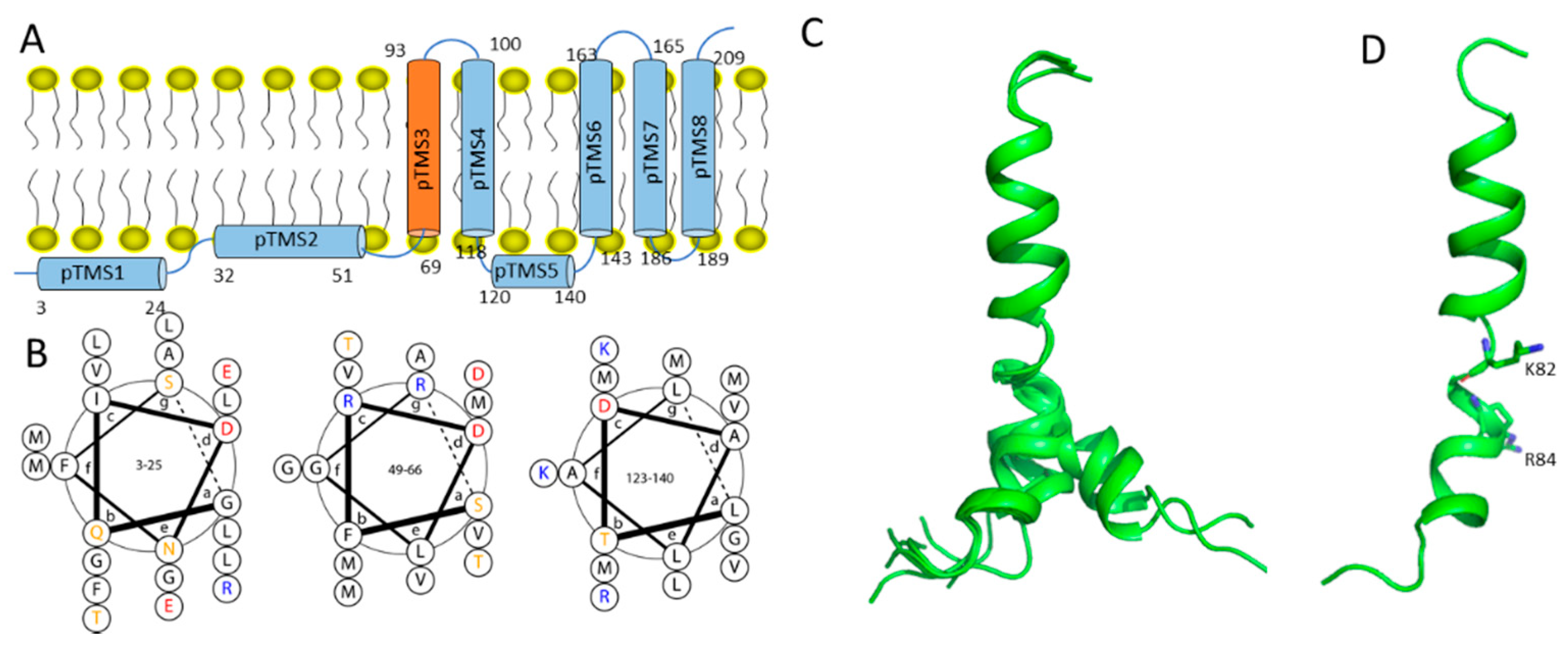
2.1. Dengue NS2A
2.2. Dengue NS2B

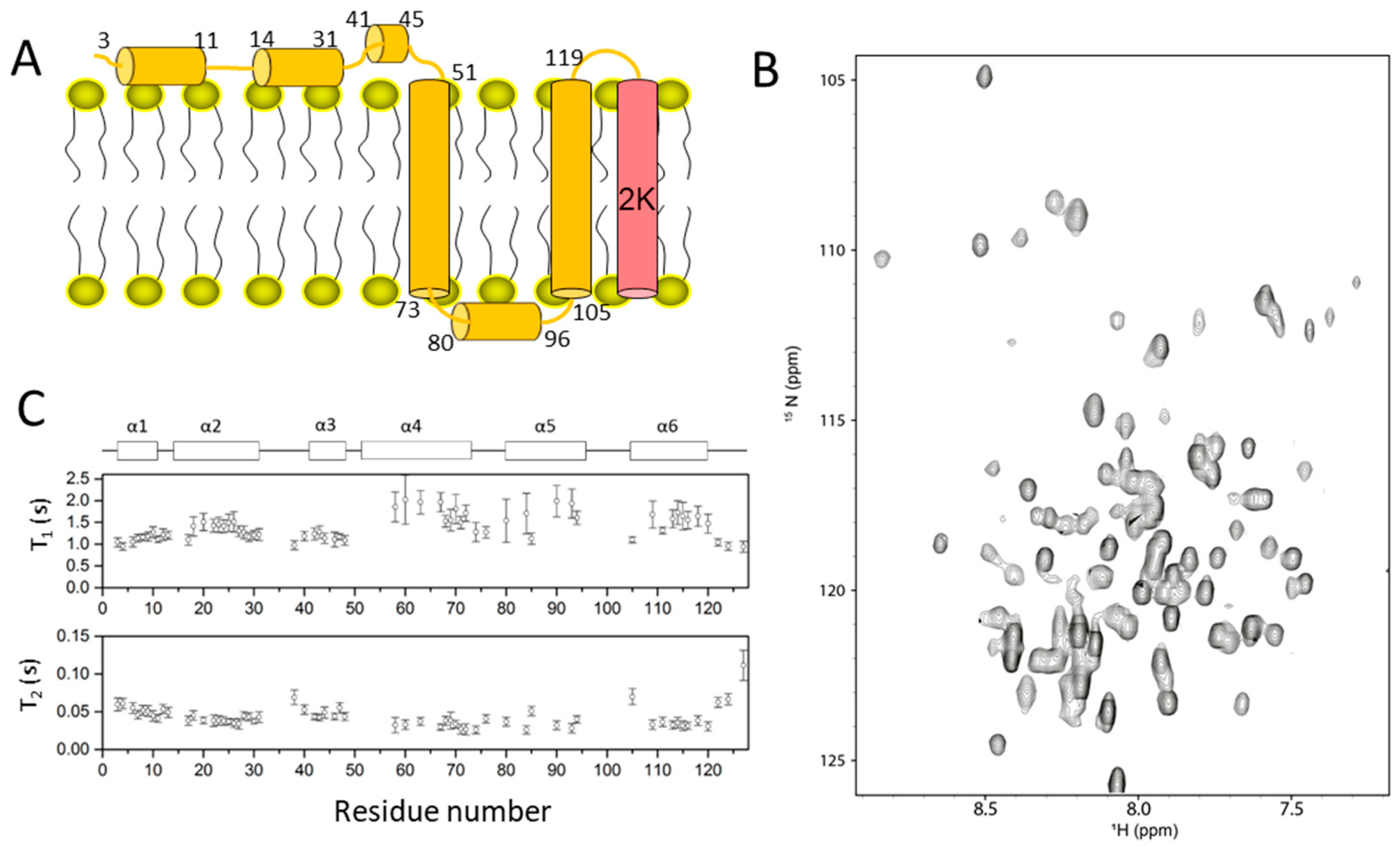
2.3. Dengue NS4A
2.4. Dengue Virus NS4B
3. Dengue Membrane Proteins as a Drug Target
3.1. NS2A Inhibitors
3.2. NS2B Inhibitors
3.3. NS4A Inhibitors
3.4. NS4B Inhibitors
4. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Wang, Q.Y.; Noble, C.G.; Chen, Y.L.; Dong, H.; Zou, B.; Yokokawa, F.; Nilar, S.; Smith, P.; Beer, D.; et al. Ten years of dengue drug discovery: Progress and prospects. Antivir. Res. 2013, 100, 500–519. [Google Scholar] [CrossRef] [PubMed]
- Villar, L.; Dayan, G.H.; Arredondo-García, J.L.; Rivera, D.M.; Cunha, R.; Deseda, C.; Reynales, H.; Costa, M.S.; Morales-Ramírez, J.O.; Carrasquilla, G.; et al. Efficacy of a Tetravalent Dengue Vaccine in Children in Latin America. N. Engl. J. Med. 2015, 372, 113–123. [Google Scholar] [PubMed]
- Nitsche, C.; Holloway, S.; Schirmeister, T.; Klein, C.D. Biochemistry and Medicinal Chemistry of the Dengue Virus Protease. Chem. Rev. 2014, 114, 11348–11381. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, S.M.; Pryor, M.J.; Wright, P.J.; Jans, D.A. Dengue virus RNA polymerase NS5: A potential therapeutic target? Curr. Drug Targets 2006, 7, 1623–1638. [Google Scholar] [CrossRef]
- Behnam, M.A.M.; Nitsche, C.; Boldescu, V.; Klein, C.D. The Medicinal Chemistry of Dengue. Virus J. Med. Chem. 2016, 59, 5622–5649. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Zou, J.; Wang, Q.Y.; Shi, P.Y. Targeting dengue virus NS4B protein for drug discovery. Antivir. Res. 2015, 118, 39–45. [Google Scholar] [CrossRef]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antivir. Res. 2015, 118, 148–158. [Google Scholar] [CrossRef]
- Lim, S.P.; Noble, C.G.; Shi, P.Y. The dengue virus NS5 protein as a target for drug discovery. Antivir. Res. 2015, 119, 57–67. [Google Scholar]
- Lim, S.P.; Shi, P.Y. West Nile virus drug discovery. Viruses 2013, 5, 2977–3006. [Google Scholar] [CrossRef] [Green Version]
- Brecher, M.; Zhang, J.; Li, H. The flavivirus protease as a target for drug discovery. Virol. Sin. 2013, 28, 326–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noble, C.G.; Shi, P.Y. Structural biology of dengue virus enzymes: Towards rational design of therapeutics. Antivir. Res. 2012, 96, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Sampath, A.; Padmanabhan, R. Molecular targets for flavivirus drug discovery. Antivir. Res. 2009, 81, 6–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, R.; Kuhn, R.J. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 2008, 11, 369–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malet, H.; Masse, N.; Selisko, B.; Romette, J.L.; Alvarez, K.; Guillemot, J.C.; Tolou, H.; Yap, T.L.; Vasudevan, S.; Lescar, J.; et al. The flavivirus polymerase as a target for drug discovery. Antivir. Res. 2008, 80, 23–35. [Google Scholar] [CrossRef]
- Melino, S.; Paci, M. Progress for dengue virus diseases. Towards the NS2B-NS3pro inhibition for a therapeutic-based approach. FEBS J. 2007, 274, 2986–3002. [Google Scholar] [CrossRef]
- Mass, N.; Selisko, B.; Malet, H.; Peyrane, F.; Debarnot, C.; Decroly, E.; Benarroch, D.; Egloff, M.P.; Guillernot, J.C.; Alvarez, K.; et al. Dengue virus: Viral targets and antiviral drugs. Virologie 2007, 11, 121–133. [Google Scholar]
- Aleshin, A.E.; Shiryaev, S.A.; Strongin, A.Y.; Liddington, R.C. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007, 16, 795–806. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Shi, P.-Y. Strategies for Zika drug discovery. Curr. Opin. Virol. 2019, 35, 19–26. [Google Scholar] [CrossRef]
- Pierson, T.C.; Kielian, M. Flaviviruses: Braking the entering. Curr. Opin. Virol. 2013, 3, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Barnard, T.R.; Abram, Q.H.; Lin, Q.F.; Wang, A.B.; Sagan, S.M. Molecular Determinants of Flavivirus Virion Assembly. Trends Biochem. Sci. 2021, 46, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Wen, D.; Yap, T.L.; Yan, C.K.; Lescar, J.; Vasudevan, S.G. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antivir. Res. 2008, 80, 360–369. [Google Scholar] [PubMed]
- Yon, C.; Teramoto, T.; Mueller, N.; Phelan, J.; Ganesh, V.K.; Murthy, K.H.M.; Padmanabhan, R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′ -RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005, 280, 27412–27419. [Google Scholar] [PubMed] [Green Version]
- Nasar, S.; Rashid, N.; Iftikhar, S. Dengue proteins with their role in pathogenesis, and strategies for developing an effective anti-dengue treatment: A review. J. Med. Virol. 2020, 92, 941–955. [Google Scholar] [CrossRef]
- Vossmann, S.; Wieseler, J.; Kerber, R.; Kummerer, B.M. A basic cluster in the N terminus of yellow fever virus NS2A contributes to infectious particle production. J. Virol. 2015, 89, 4951–4965. [Google Scholar] [CrossRef] [Green Version]
- Leung, J.Y.; Pijlman, G.P.; Kondratieva, N.; Hyde, J.; Mackenzie, J.M.; Khromykh, A.A. Role of nonstructural protein NS2A in flavivirus assembly. J. Virol. 2008, 82, 4731–4741. [Google Scholar] [CrossRef] [Green Version]
- Swarbrick, C.M.D.; Basavannacharya, C.; Chan, K.W.K.; Chan, S.-A.; Singh, D.; Wei, N.; Phoo, W.W.; Luo, D.; Lescar, J.; Vasudevan, S.G. NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res. 2017, 45, 12904–12920. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Xu, T.; Watson, R.P.; Scherer-Becker, D.; Sampath, A.; Jahnke, W.; Yeong, S.S.; Wang, C.H.; Lim, S.P.; Strongin, A.; et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008, 27, 3209–3219. [Google Scholar] [CrossRef] [Green Version]
- Lescar, J.; Luo, D.; Xu, T.; Sampath, A.; Lim, S.P.; Canard, B.; Vasudevan, S.G. Towards the design of antiviral inhibitors against flaviviruses: The case for the multifunctional NS3 protein from Dengue virus as a target. Antivir. Res. 2008, 80, 94–101. [Google Scholar] [CrossRef]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Luo, D.; Chene, P.; Vasudevan, S.G.; Lescar, J. Towards the design of flavivirus helicase/NTPase inhibitors: Crystallographic and mutagenesis studies of the dengue virus NS3 helicase catalytic domain. Novartis Found. Symp. 2006, 277, 87–97; discussion 97–101, 103–251. [Google Scholar]
- Umareddy, I.; Chao, A.; Sampath, A.; Gu, F.; Vasudevan, S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006, 87, 2605–2614. [Google Scholar] [CrossRef] [PubMed]
- Prikhod’ko, G.G.; Prikhod’ko, E.A.; Pletnev, A.G.; Cohen, J.I. Langat flavivirus protease NS3 binds caspase-8 and induces apoptosis. J. Virol. 2002, 76, 5807–5812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, G.; Chen, Y.L.; Dong, H.; Lim, C.C.; Yap, L.J.; Yau, Y.H.; Shochat, S.G.; Lescar, J.; Shi, P.Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 2011, 286, 14362–14372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Xu, T.; Hunke, C.; Gruber, G.; Vasudevan, S.G.; Lescar, J. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008, 82, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Shiryaev, S.A.; Ratnikov, B.I.; Aleshin, A.E.; Kozlov, I.A.; Nelson, N.A.; Lebl, M.; Smith, J.W.; Liddington, R.C.; Strongin, A.Y. Switching the substrate specificity of the two-component NS2B-NS3 flavivirus proteinase by structure-based mutagenesis. J. Virol. 2007, 81, 4501–4509. [Google Scholar] [CrossRef] [Green Version]
- Shiryaev, S.A.; Aleshin, A.E.; Ratnikov, B.I.; Smith, J.W.; Liddington, R.C.; Strongin, A.Y. Expression and purification of a two-component flaviviral proteinase resistant to autocleavage at the NS2B-NS3 junction region. Protein Expr. Purif. 2007, 52, 334–339. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Lim, S.P.; Patel, S.; Patel, V.; Beer, D.; Ma, N.L.; Vasudevan, S.; Keller, T. Targeting the protease activity of Dengue virus NS3. Acta Pharmacol. Sin. 2006, 27, 251. [Google Scholar]
- Kroschewski, H.; Lim, S.P.; Butcher, R.E.; Yap, T.L.; Lescar, J.; Wright, P.J.; Vasudevan, S.G.; Davidson, A.D. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J. Biol. Chem. 2008, 283, 19410–19421. [Google Scholar] [CrossRef] [Green Version]
- Poulsen, A.; Kang, C.; Keller, T.H. Drug design for flavivirus proteases: What are we missing? Curr. Pharm. Des. 2014, 20, 3422–3427. [Google Scholar] [CrossRef]
- Kang, C.; Gayen, S.; Wang, W.; Severin, R.; Chen, A.S.; Lim, H.A.; Chia, C.S.; Schuller, A.; Doan, D.N.; Poulsen, A.; et al. Exploring the binding of peptidic West Nile virus NS2B-NS3 protease inhibitors by NMR. Antivir. Res. 2013, 97, 137–144. [Google Scholar] [CrossRef]
- Lim, S.P. Dengue drug discovery: Progress, challenges and outlook. Antivir. Res. 2019, 163, 156–178. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, C. Strategies Towards Protease Inhibitors for Emerging Flaviviruses. In Dengue and Zika: Control and Antiviral Treatment Strategies; Hilgenfeld, R., Vasudevan, S.G., Eds.; Springer: Singapore, 2018; pp. 175–186. [Google Scholar]
- Timiri, A.K.; Sinha, B.N.; Jayaprakash, V. Progress and prospects on DENV protease inhibitors. Eur. J. Med. Chem. 2016, 117, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Moquin, S.A.; Simon, O.; Karuna, R.; Lakshminarayana, S.B.; Yokokawa, F.; Wang, F.; Saravanan, C.; Zhang, J.; Day, C.W.; Chan, K.; et al. NITD-688, a pan-serotype inhibitor of the dengue virus NS4B protein, shows favorable pharmacokinetics and efficacy in preclinical animal models. Sci. Transl. Med. 2021, 13, eabb2181. [Google Scholar] [CrossRef] [PubMed]
- Kaptein, S.J.; Goethals, O.; Kiemel, D.; Marchand, A.; Kesteleyn, B.; Bonfanti, J.-F.; Bardiot, D.; Stoops, B.; Jonckers, T.H.M.; Dallmeier, K.; et al. A pan-serotype dengue virus inhibitor targeting the NS3–NS4B interaction. Nature 2021, 598, 504–509. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Dong, H.; Zou, B.; Karuna, R.; Wan, K.F.; Zou, J.; Susila, A.; Yip, A.; Shan, C.; Yeo, K.L.; et al. Discovery of Dengue Virus NS4B Inhibitors. J. Virol. 2015, 89, 8233–8244. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary Structure and Membrane Topology of the Full-Length Dengue Virus NS4B in Micelles. Angew. Chem. Int. Ed. 2016, 55, 12068–12072. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Wong, Y.L.; Liew, L.S.; Kang, C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta 2015, 1848, 2244–2252. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kim, Y.M.; Zou, J.; Wang, Q.Y.; Gayen, S.; Wong, Y.L.; Lee, L.T.; Xie, X.; Huang, Q.; Lescar, J.; et al. Secondary structure and membrane topology of dengue virus NS4B N-terminal 125 amino acids. Biochim. Biophys. Acta 2015, 1848, 3150–3157. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lee, M.Y.; Loh, Y.R.; Kang, C. Secondary structure and membrane topology of dengue virus NS4A protein in micelles. Biochim. Biophys. Acta Biomembr. 2018, 1860, 442–450. [Google Scholar] [CrossRef]
- Reddy, S.B.G.; Chin, W.-X.; Shivananju, N.S. Dengue virus NS2 and NS4: Minor proteins, mammoth roles. Biochem. Pharmacol. 2018, 154, 54–63. [Google Scholar] [CrossRef]
- Huang, Q.; Li, Q.; Joy, J.; Chen, A.S.; Ruiz-Carrillo, D.; Hill, J.; Lescar, J.; Kang, C. Lyso-myristoyl phosphatidylcholine micelles sustain the activity of Dengue non-structural (NS) protein 3 protease domain fused with the full-length NS2B. Protein Expr. Purif. 2013, 92, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Chen, A.S.; Li, Q.; Kang, C. Expression, purification, and initial structural characterization of nonstructural protein 2B, an integral membrane protein of Dengue-2 virus, in detergent micelles. Protein Expr. Purif. 2011, 80, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Li, Q. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 2011, 15, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ng, H.Q.; Kang, C. Secondary structure and topology of the transmembrane domain of Syndecan-2 in detergent micelles. FEBS Lett. 2019, 593, 554–561. [Google Scholar] [CrossRef]
- Ng, E.Y.; Loh, Y.R.; Li, Y.; Li, Q.; Kang, C. Expression, purification of Zika virus membrane protein-NS2B in detergent micelles for NMR studies. Protein Expr. Purif. 2019, 154, 1–6. [Google Scholar] [CrossRef]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.Y. Membrane topology and function of dengue virus NS2A protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef]
- Taniguchi, T.; Takaoka, A. The interferon-α/β system in antiviral responses: A multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 2002, 14, 111–116. [Google Scholar] [CrossRef]
- Muñoz-Jordán, J.L.; Sánchez-Burgos, G.G.; Laurent-Rolle, M.; García-Sastre, A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 2003, 100, 14333–14338. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Zou, J.; Zhang, X.; Zhou, Y.; Routh, A.L.; Kang, C.; Popov, V.L.; Chen, X.; Wang, Q.Y.; Dong, H.; et al. Dengue NS2A Protein Orchestrates Virus Assembly. Cell Host Microbe 2019, 26, 606–622.e608. [Google Scholar] [CrossRef]
- Bañó-Polo, M.; Baeza-Delgado, C.; Orzáez, M.; Marti-Renom, M.A.; Abad, C.; Mingarro, I. Polar/Ionizable Residues in Transmembrane Segments: Effects on Helix-Helix Packing. PLoS ONE 2012, 7, e44263. [Google Scholar] [CrossRef] [PubMed]
- Yusof, R.; Clum, S.; Wetzel, M.; Murthy, H.M.K.; Padmanabhan, R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J. Biol. Chem. 2000, 275, 9963–9969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clum, S.; Ebner, K.E. Padmanabhan, Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 1997, 272, 30715–30723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robin, G.; Chappell, K.; Stoermer, M.J.; Hu, S.H.; Young, P.R.; Fairlie, D.P.; Martin, J.L. Structure of West Nile virus NS3 protease: Ligand stabilization of the catalytic conformation. J. Mol. Biol. 2009, 385, 1568–1577. [Google Scholar] [CrossRef]
- Noble, C.G.; Seh, C.C.; Chao, A.T.; Shi, P.Y. Ligand-bound structures of the dengue virus protease reveal the active conformation. J. Virol. 2011, 86, 438–446. [Google Scholar] [CrossRef] [Green Version]
- Radichev, I.; Shiryaev, S.A.; Aleshin, A.E.; Ratnikov, B.I.; Smith, J.W.; Liddington, R.C.; Strongin, A.Y. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B-NS3 proteinase. J. Gen. Virol. 2008, 89, 636–641. [Google Scholar] [CrossRef]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef]
- de la Cruz, L.; Nguyen, T.H.; Ozawa, K.; Shin, J.; Graham, B.; Huber, T.; Otting, G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: Fold analysis by pseudocontact shifts. J. Am. Chem. Soc. 2011, 133, 19205–19215. [Google Scholar] [CrossRef]
- Kim, Y.M.; Gayen, S.; Kang, C.; Joy, J.; Huang, Q.; Chen, A.S.; Wee, J.L.; Ang, M.J.; Lim, H.A.; Hung, A.W.; et al. NMR analysis of a novel enzymatically active unlinked dengue NS2B-NS3 protease complex. J. Biol. Chem. 2013, 288, 12891–12900. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, C. Structure and Dynamics of Zika Virus Protease and Its Insights into Inhibitor Design. Biomedicines 2021, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, C. Mechanisms of Action for Small Molecules Revealed by Structural Biology in Drug Discovery. Int. J. Mol. Sci. 2020, 21, 5262. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kang, C. Insights into Structures and Dynamics of Flavivirus Proteases from NMR Studies. Int. J. Mol. Sci. 2020, 21, 2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cruz, L.; Chen, W.N.; Graham, B.; Otting, G. Binding mode of the activity-modulating C-terminal segment of NS2B to NS3 in the dengue virus NS2B-NS3 protease. FEBS J. 2014, 281, 1517–1533. [Google Scholar] [CrossRef]
- Chen, W.N.; Loscha, K.V.; Nitsche, C.; Graham, B.; Otting, G. The dengue virus NS2B-NS3 protease retains the closed conformation in the complex with BPTI. FEBS Lett. 2014, 588, 2206–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.C.; Ozawa, K.; Qi, R.; Vasudevan, S.G.; Lim, S.P.; Otting, G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLOS Negl. Trop. Dis. 2009, 3, e561. [Google Scholar] [CrossRef]
- Choksupmanee, O.; Hodge, K.; Katzenmeier, G.; Chimnaronk, S. Structural platform for the autolytic activity of an intact NS2B-NS3 protease complex from dengue virus. Biochemistry 2012, 51, 2840–2851. [Google Scholar] [CrossRef]
- Wu, R.-H.; Tsai, M.-H.; Tsai, K.-N.; Tian, J.N.; Wu, J.-S.; Wu, S.-Y.; Chern, J.-H.; Chen, C.-H.; Yueh, A.; Diamond, M.S. Mutagenesis of Dengue Virus Protein NS2A Revealed a Novel Domain Responsible for Virus-Induced Cytopathic Effect and Interactions between NS2A and NS2B Transmembrane Segments. J. Virol. 2017, 91, e01836-16. [Google Scholar] [CrossRef] [Green Version]
- Cordero, J.G.; Juárez, M.L.; González-Y-Merchand, J.A.; Barrón, L.C.; Castañeda, B.G. Caveolin-1 in Lipid Rafts Interacts with Dengue Virus NS3 during Polyprotein Processing and Replication in HMEC-1 Cells. PLoS ONE 2014, 9, e90704. [Google Scholar]
- Li, X.D.; Deng, C.L.; Ye, H.Q.; Zhang, H.L.; Zhang, Q.Y.; Chen, D.D.; Zhang, P.T.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Transmembrane Domains of NS2B Contribute to both Viral RNA Replication and Particle Formation in Japanese Encephalitis Virus. J. Virol. 2016, 90, 5735–5749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- León-Juárez, M.; Martínez-Castillo, M.; Shrivastava, G.; García-Cordero, J.; Villegas-Sepulveda, N.; Mondragón-Castelán, M.; Mondragón-Flores, R.; Cedillo-Barrón, L. Recombinant Dengue virus protein NS2B alters membrane permeability in different membrane models. Virol. J. 2016, 13, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, S.; Luthra, P.; Sanchez-Aparicio, M.T.; Maestre, A.M.; Patel, J.; Lamothe, F.; Fredericks, A.C.; Tripathi, S.; Zhu, T.; Pintado-Silva, J.; et al. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat. Microbiol. 2017, 2, 17037. [Google Scholar] [CrossRef]
- Yu, L.; Liu, P. Cytosolic DNA sensing by cGAS: Regulation, function, and human diseases. Signal Transduct. Target. Ther. 2021, 6, 170. [Google Scholar] [CrossRef] [PubMed]
- Gayen, S.; Chen, A.S.; Huang, Q.; Kang, C. West Nile Virus (WNV) protease and membrane interactions revealed by NMR spectroscopy. Biochem. Biophys. Res. Commun. 2012, 423, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Su, X.C.; Ozawa, K.; Yagi, H.; Lim, S.P.; Wen, D.; Ekonomiuk, D.; Huang, D.; Keller, T.H.; Sonntag, S.; Caflisch, A.; et al. NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease. FEBS J. 2009, 276, 4244–4255. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Wei, N.; Doan, D.N.; Paradkar, P.N.; Chong, Y.; Davidson, A.D.; Kotaka, M.; Lescar, J.; Vasudevan, S.G. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 2010, 285, 18817–18827. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Li, Q.; Chen, A.S.; Kang, C. West Nile virus protease activity in detergent solutions and application for affinity tag removal. Anal. Biochem. 2013, 435, 44–46. [Google Scholar] [CrossRef]
- Liew, L.S.; Lee, M.Y.; Wong, Y.L.; Cheng, J.; Li, Q.; Kang, C. Selection of suitable detergents for obtaining an active dengue protease in its natural form from E. coli. Protein Expr. Purif. 2016, 121, 141–148. [Google Scholar] [CrossRef]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Bühler, S.; Bartenschlager, R. The Non-structural Protein 4A of Dengue Virus Is an Integral Membrane Protein Inducing Membrane Alterations in a 2K-regulated Manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Amberg, S.M.; Chambers, T.J.; Rice, C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 1993, 67, 2327–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Kumar, P.; Giri, R. Zika virus NS4A cytosolic region (residues 1–48) is an intrinsically disordered domain and folds upon binding to lipids. Virology 2020, 550, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.F.; Schwarten, M.; Hoffmann, S.; Willbold, D.; Sklan, E.H.; Koenig, B. Amino Terminal Region of Dengue Virus NS4A Cytosolic Domain Binds to Highly Curved Liposomes. Viruses 2015, 7, 4119–4130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, Y.F.; Schwarten, M.; Schunke, S.; Thiagarajan-Rosenkranz, P.; Hoffmann, S.; Sklan, E.H.; Willbold, D.; Koenig, B.W. Dengue virus NS4A cytoplasmic domain binding to liposomes is sensitive to membrane curvature. Biochim. Biophys. Acta 2015, 1848, 1119–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, J.; Torres, J. Trimerization of the N-Terminal Tail of Zika Virus NS4A Protein: A Potential In Vitro Antiviral Screening Assay. Membranes 2021, 11, 335. [Google Scholar] [CrossRef]
- Tan, M.J.A.; Brown, N.G.; Chan, K.W.K.; Jin, J.Y.; Kong, S.Y.Z.; Vasudevan, S.G. Mutations in the cytoplasmic domain of dengue virus NS4A affect virus fitness and interactions with other non-structural proteins. J. Gen. Virol. 2020, 101, 941–953. [Google Scholar] [CrossRef]
- Klaitong, P.; Smith, D.R. Roles of Non-Structural Protein 4A in Flavivirus Infection. Viruses 2021, 13, 2077. [Google Scholar] [CrossRef]
- Mikulasova, A.; Gillespie, L.K.; Ambrose, R.L.; Aktepe, T.E.; Trenerry, A.M.; Liebscher, S.; Mackenzie, J.M. A Putative Lipid-Associating Motif in the West Nile Virus NS4A Protein Is Required for Efficient Virus Replication. Front. Cell Dev. Biol. 2021, 9, 655606. [Google Scholar] [CrossRef]
- Ambrose, R.L.; Mackenzie, J.M. A Conserved Peptide in West Nile Virus NS4A Protein Contributes to Proteolytic Processing and Is Essential for Replication. J. Virol. 2011, 85, 11274–11282. [Google Scholar] [CrossRef] [Green Version]
- Ambrose, R.L.; Mackenzie, J.M. Conserved amino acids within the N-terminus of the West Nile virus NS4A protein contribute to virus replication, protein stability and membrane proliferation. Virology 2015, 481, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Teo, C.S.H.; Chu, J.J.H. Cellular Vimentin Regulates Construction of Dengue Virus Replication Complexes through Interaction with NS4A Protein. J. Virol. 2014, 88, 1897–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The Endoplasmic Reticulum Provides the Membrane Platform for Biogenesis of the Flavivirus Replication Complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junjhon, J.; Pennington, J.G.; Edwards, T.J.; Perera, R.; Lanman, J.; Kuhn, R.J.; Doms, R.W. Ultrastructural Characterization and Three-Dimensional Architecture of Replication Sites in Dengue Virus-Infected Mosquito Cells. J. Virol. 2014, 88, 4687–4697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.E.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, A.M.; Shurtleff, M.J.; Popova, K.D.; Kulsuptrakul, J.; Weissman, J.S.; Puschnik, A.S. The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. eLife 2019, 8, 48469. [Google Scholar] [CrossRef]
- Lin, M.H.; Hsu, H.J.; Bartenschlager, R.; Fischer, W.B. Membrane undulation induced by NS4A of Dengue virus: A molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2013, 32, 1552–1562. [Google Scholar] [CrossRef]
- Stern, O.; Hung, Y.-F.; Valdau, O.; Yaffe, Y.; Harris, E.; Hoffmann, S.; Willbold, D.; Sklan, E.H. An N-Terminal Amphipathic Helix in Dengue Virus Nonstructural Protein 4A Mediates Oligomerization and Is Essential for Replication. J. Virol. 2013, 87, 4080–4085. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.M.; Xie, X.; Zou, J.; Li, S.H.; Lee, M.Y.; Dong, H.; Qin, C.F.; Kang, C.; Shi, P.Y. Determinants of Dengue Virus NS4A Protein Oligomerization. J. Virol. 2015, 89, 6171–6183. [Google Scholar] [CrossRef] [Green Version]
- Riva, L.; Goellner, S.; Biering, S.B.; Huang, C.-T.; Rubanov, A.N.; Haselmann, U.; Warnes, C.M.; Jesus, P.D.D.; Martin-Sancho, L.; Terskikh, A.V.; et al. The Compound SBI-0090799 Inhibits Zika Virus Infection by Blocking De Novo Formation of the Membranous Replication Compartment. J. Virol. 2021, 95, e00996-21. [Google Scholar] [CrossRef]
- Preugschat, F.; Yao, C.W.; Strauss, J.H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J. Virol. 1990, 64, 4364–4374. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Mohan, P.M.; Padmanabhan, R. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J. Virol. 1992, 66, 7549–7554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiryaev, S.A.; Chernov, A.V.; Aleshin, A.E.; Shiryaeva, T.N.; Strongin, A.Y. NS4A regulates the ATPase activity of the NS3 helicase: A novel cofactor role of the non-structural protein NS4A from West Nile virus. J. Gen. Virol. 2009, 90, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Genetic Interaction of Flavivirus Nonstructural Proteins NS1 and NS4A as a Determinant of Replicase Function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Płaszczyca, A.; Scaturro, P.; Neufeldt, C.J.; Cortese, M.; Cerikan, B.; Ferla, S.; Brancale, A.; Pichlmair, A.; Bartenschlager, R. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLOS Pathog. 2019, 15, e1007736. [Google Scholar] [CrossRef]
- Zou, J.; Xie, X.; Wang, Q.Y.; Dong, H.; Lee, M.Y.; Kang, C.; Yuan, Z.; Shi, P.Y. Characterization of dengue virus NS4A and NS4B protein interaction. J. Virol. 2015, 89, 3455–3470. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.-S.; Lee, S.-A.; Ge, J.; Wang, S.; Goldman, S.-A.; Zlokovic, B.-V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Ketkar, H.; Geng, T.; Lo, E.; Wang, L.; Xi, J.; Sun, Q.; Zhu, Z.; Cui, Y.; Yang, L.; et al. Zika Virus Non-structural Protein 4A Blocks the RLR-MAVS Signaling. Front. Microbiol. 2018, 9, 1350. [Google Scholar] [CrossRef] [Green Version]
- Ngueyen, T.T.N.; Kim, S.J.; Lee, J.Y.; Myoung, J. Zika Virus Proteins NS2A and NS4A Are Major Antagonists that Reduce IFN-β Promoter Activity Induced by the MDA5/RIG-I Signaling Pathway. J. Microbiol. Biotechnol. 2019, 29, 1665–1674. [Google Scholar]
- McLean, J.E.; Wudzinska, A.; Datan, E.; Quaglino, D.; Zakeri, Z. Flavivirus NS4A-induced Autophagy Protects Cells against Death and Enhances Virus Replication. J. Biol. Chem. 2011, 286, 22147–22159. [Google Scholar] [CrossRef] [Green Version]
- Aktepe, T.E.; Liebscher, S.; Prier, J.E.; Simmons, C.P.; Mackenzie, J.M. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017, 21, 1639–1654. [Google Scholar] [CrossRef] [Green Version]
- Hung, Y.F.; Valdau, O.; Schunke, S.; Stern, O.; Koenig, B.W.; Willbold, D.; Hoffmann, S. Recombinant production of the amino terminal cytoplasmic region of dengue virus non-structural protein 4A for structural studies. PLoS ONE 2014, 9, e86482. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Xie, X.; Le, T.L.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of flavivirus NS4B protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef] [Green Version]
- Westaway, E.G.; Khromykh, A.A.; Kenney, M.T.; Mackenzie, J.M.; Jones, M.K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 1997, 234, 31–41. [Google Scholar] [CrossRef]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated Cleavages at the West Nile Virus NS4A-2K-NS4B Junctions Play a Major Role in Rearranging Cytoplasmic Membranes and Golgi Trafficking of the NS4A Protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [Green Version]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Zhan, Y.; Li, X.; Bai, X.; Yuan, F.; Ma, L.; Wang, X.; Xie, M.; Wu, W.; Chen, Z. Novel insights into the function of an N-terminal region of DENV2 NS4B for the optimal helicase activity of NS3. Virus Res. 2021, 295, 198318. [Google Scholar] [CrossRef]
- Fanunza, E.; Grandi, N.; Quartu, M.; Carletti, F.; Ermellino, L.; Milia, J.; Corona, A.; Capobianchi, M.R.; Ippolito, G.; Tramontano, E. INMI1 Zika Virus NS4B Antagonizes the Interferon Signaling by Suppressing STAT1 Phosphorylation. Viruses 2021, 13, 2448. [Google Scholar] [CrossRef]
- Tian, J.-N.; Yang, C.-C.; Chuang, C.-K.; Tsai, M.-H.; Wu, R.-H.; Chen, C.-T.; Yueh, A. A Dengue Virus Type 2 (DENV-2) NS4B-Interacting Host Factor, SERP1, Reduces DENV-2 Production by Suppressing Viral RNA Replication. Viruses 2019, 11, 787. [Google Scholar] [CrossRef] [Green Version]
- Youn, S.; Li, T.; McCune, B.T.; Edeling, M.A.; Fremont, D.H.; Cristea, I.M.; Diamond, M.S. Evidence for a Genetic and Physical Interaction between Nonstructural Proteins NS1 and NS4B That Modulates Replication of West Nile Virus. J. Virol. 2012, 86, 7360–7371. [Google Scholar] [CrossRef] [Green Version]
- Barrett, P.J.; Chen, J.; Cho, M.-K.; Kim, J.-H.; Lu, Z.; Mathew, S.; Peng, D.; Song, Y.; van Horn, W.D.; Zhuang, T.; et al. The Quiet Renaissance of Protein Nuclear Magnetic Resonance. Biochemistry 2013, 52, 1303–1320. [Google Scholar] [CrossRef]
- Zou, J.; Lee, T.L.; Wang, Q.Y.; Xie, X.; Lu, S.; Yau, Y.H.; Yuan, Z.; Shochat, S.G.; Kang, C.; Lescar, J.; et al. Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. J. Virol. 2015, 89, 3471–3483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz-Jordan, J.L.; Laurent-Rolle, M.; Ashour, J.; Martinez-Sobrido, L.; Ashok, M.; Lipkin, W.I.; Garcia-Sastre, A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005, 79, 8004–8013. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.B. 19F-NMR in target-based drug discovery. Curr. Med. Chem. 2019, 26, 4964–4983. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Carbajal, P.; Soto-Acosta, R.; Angel-Ambrocio, A.H.; Cervantes-Salazar, M.; Loranca-Vega, C.I.; Herrera-Martínez, M.; del Angel, R.M. The calmodulin antagonist W-7 (N-(6-aminohexyl)-5-chloro-1-naphthalenesulfonamide hydrochloride) inhibits DENV infection in Huh-7 cells. Virology 2017, 501, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Nemésio, H.; Villalaín, J. Membrane Interacting Regions of Dengue Virus NS2A Protein. J. Phys. Chem. B 2014, 118, 10142–10155. [Google Scholar] [CrossRef] [PubMed]
- Fajardo-Sánchez, E.; Galiano, V.; Villalaín, J. Spontaneous membrane insertion of a dengue virus NS2A peptide. Arch. Biochem. Biophys. 2017, 627, 56–66. [Google Scholar] [CrossRef]
- Soto-Acosta, R.; Bautista-Carbajal, P.; Syed, G.H.; Siddiqui, A.; del Angel, R.M. Nordihydroguaiaretic acid (NDGA) inhibits replication and viral morphogenesis of dengue virus. Antivir. Res. 2014, 109, 132–140. [Google Scholar] [CrossRef]
- Li, Y.; Loh, Y.R.; Hung, A.W.; Kang, C. Characterization of molecular interactions between Zika virus protease and peptides derived from the C-terminus of NS2B. Biochem. Biophys. Res. Commun. 2018, 503, 691–696. [Google Scholar] [CrossRef]
- Yin, Z.; Patel, S.J.; Wang, W.L.; Wang, G.; Chan, W.L.; Rao, K.R.; Alam, J.; Jeyaraj, D.A.; Ngew, X.; Patel, V.; et al. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorganic Med. Chem. Lett. 2006, 16, 36–39. [Google Scholar] [CrossRef]
- Tomlinson, S.M.; Malmstrom, R.D.; Russo, A.; Mueller, N.; Pang, Y.P.; Watowich, S.J. Structure-based discovery of dengue virus protease inhibitors. Antivir. Res. 2009, 82, 110–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, D.; Abbenante, G.; Fairlie, D.P. Protease inhibitors: Current status and future prospects. J. Med. Chem. 2000, 43, 305–341. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, M.; Ghosh, S.; Bell, J.A.; Sherman, W.; Hardy, J.A. Allosteric Inhibition of the NS2B-NS3 Protease from Dengue Virus. ACS Chem. Biol. 2013, 8, 2744–2752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, L.; Dang, M.; Roy, A.; Kang, J.; Song, J. Curcumin Allosterically Inhibits the Dengue NS2B-NS3 Protease by Disrupting Its Active Conformation. ACS Omega 2020, 5, 25677–25686. [Google Scholar] [CrossRef]
- Shiryaev, S.A.; Farhy, C.; Pinto, A.; Huang, C.-T.; Simonetti, N.; Ngono, A.E.; Dewing, A.; Shresta, S.; Pinkerton, A.B.; Cieplak, P.; et al. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir. Res. 2017, 143, 218–229. [Google Scholar] [CrossRef]
- Yao, Y.; Huo, T.; Lin, Y.L.; Nie, S.; Wu, F.; Hua, Y.; Wu, J.; Kneubehl, A.R.; Vogt, M.B.; Rico-Hesse, R.; et al. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 2019, 141, 6832–6836. [Google Scholar] [CrossRef]
- Li, Q.; Kang, C. Perspectives on Fragment-based Drug Discovery: A Strategy Applicable to Diverse Targets. Curr. Top. Med. Chem. 2021, 21, 1099–1112. [Google Scholar]
- Quek, J.P.; Liu, S.; Zhang, Z.; Li, Y.; Ng, E.Y.; Loh, Y.R.; Hung, A.W.; Luo, D.; Kang, C. Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease. Antivir. Res. 2020, 175, 104707. [Google Scholar] [CrossRef]
- Chang, Y.S.; Graves, B.; Guerlavais, V.; Tovar, C.; Packman, K.; To, K.-H.; Olson, K.A.; Kesavan, K.; Gangurde, P.; Mukherjee, A.; et al. Stapled α−helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA 2013, 110, E3445–E3454. [Google Scholar] [CrossRef] [Green Version]
- Mabonga, L.; Kappo, A.P. Peptidomimetics: A Synthetic Tool for Inhibiting Protein–Protein Interactions in Cancer. Int. J. Pept. Res. Ther. 2020, 26, 225–241. [Google Scholar] [CrossRef] [Green Version]
- Zou, B.; Chan, W.L.; Ding, M.; Leong, S.Y.; Nilar, S.; Seah, P.G.; Liu, W.; Karuna, R.; Blasco, F.; Yip, A.; et al. Lead Optimization of Spiropyrazolopyridones: A New and Potent Class of Dengue Virus Inhibitors. ACS Med. Chem. Lett. 2015, 6, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A.M.; Klein, C.D. On track to tackle dengue: History and future of NS4B ligands. Cell Host Microbe 2021, 29, 1735–1737. [Google Scholar] [CrossRef] [PubMed]
- Low, J.G.; Gatsinga, R.; Vasudevan, S.G.; Sampath, A. Dengue Antiviral Development: A Continuing Journey. In Dengue and Zika: Control and Antiviral Treatment Strategies; Hilgenfeld, R., Vasudevan, S.G., Eds.; Springer: Singapore, 2018; pp. 319–332. [Google Scholar]
- Knyazhanskaya, E.; Morais, M.C.; Choi, K.H. Chapter Ten—Flavivirus enzymes and their inhibitors. In The Enzymes; Cameron, C.E., Arnold, J.J., Kaguni, L.S., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 265–303. [Google Scholar]
- Kang, C.; Keller, T.H.; Luo, D. Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol. 2017, 25, 797–808. [Google Scholar] [CrossRef]
- Obi, J.O.; Gutiérrez-Barbosa, H.; Chua, J.V.; Deredge, D.J. Current Trends and Limitations in Dengue Antiviral Research. Trop. Med. Infect. Dis. 2021, 6, 180. [Google Scholar] [CrossRef]
- Lescar, J.; Soh, S.; Lee, L.T.; Vasudevan, S.G.; Kang, C.; Lim, S.P. The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. Adv. Exp. Med. Biol. 2018, 1062, 115–129. [Google Scholar] [PubMed]
- Biering, S.B.; Harris, E. A step towards therapeutics for dengue. Nature 2021, 598, 420–421. [Google Scholar] [CrossRef]
- Li, Y.; Kang, C. Solution NMR Spectroscopy in Target-Based Drug Discovery. Molecules 2017, 22, 1399. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Kang, C. Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins. Membranes 2022, 12, 231. https://doi.org/10.3390/membranes12020231
Li Q, Kang C. Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins. Membranes. 2022; 12(2):231. https://doi.org/10.3390/membranes12020231
Chicago/Turabian StyleLi, Qingxin, and Congbao Kang. 2022. "Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins" Membranes 12, no. 2: 231. https://doi.org/10.3390/membranes12020231
APA StyleLi, Q., & Kang, C. (2022). Structures and Dynamics of Dengue Virus Nonstructural Membrane Proteins. Membranes, 12(2), 231. https://doi.org/10.3390/membranes12020231





