Extracellular Vesicles as Novel Drug-Delivery Systems through Intracellular Communications
Abstract
:1. Introduction
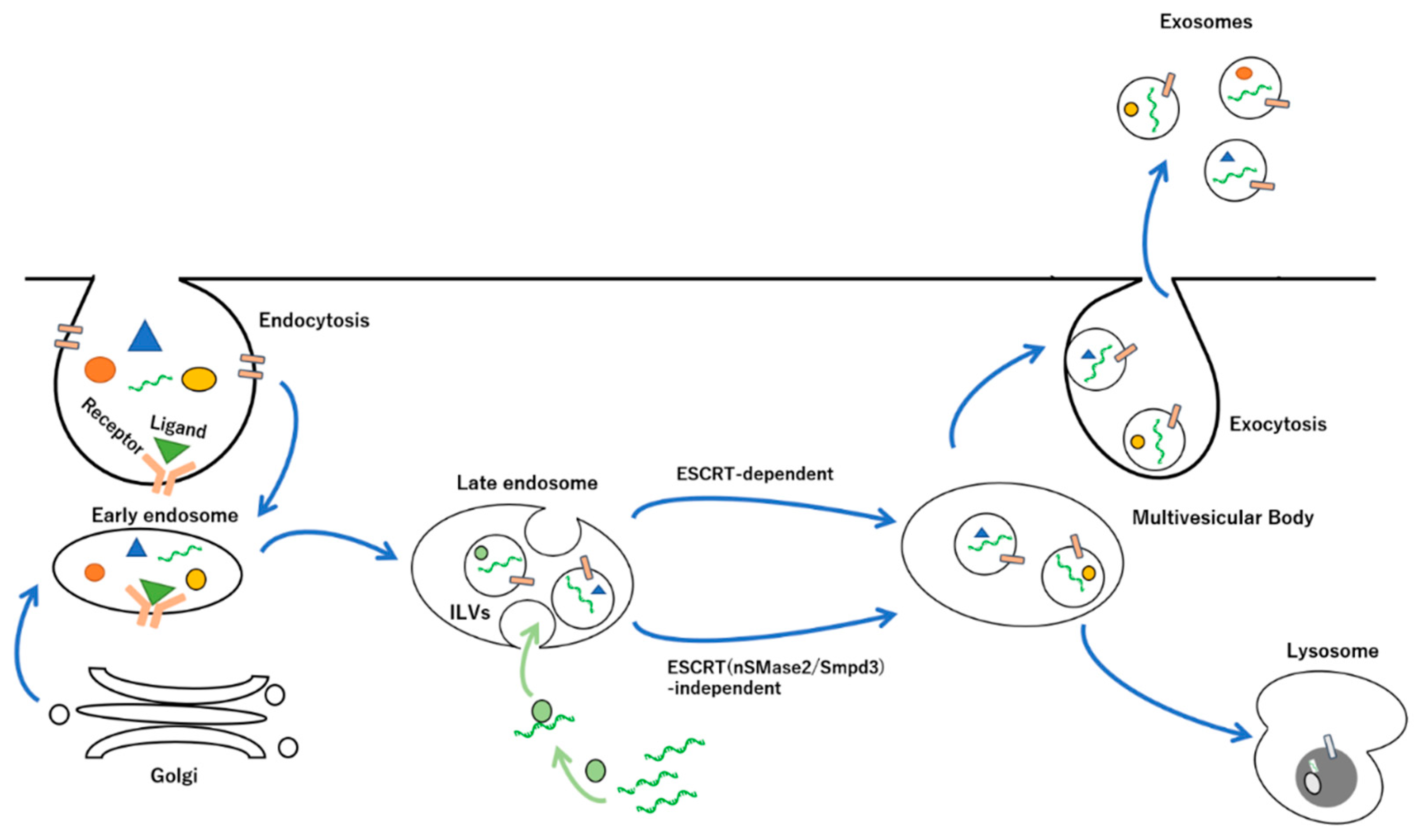
2. Biogenesis and Release of Exosomes
2.1. Endosomal Sorting Complexes Required for Transport (ESCRT)-Dependent and -Independent Pathways
2.2. Proteins, Peptides, Lipid, and Nucleic Acids in Exosomes
3. EV-Mediated DDS
3.1. microRNAs in EV-Mediated DDS
3.2. Modification of EVs as DDS Methods
3.3. Recovery of EVs as DDSs
3.4. Preparation of EVs as DDSs
4. EV-Encapsulated Adeno-Associated Virus
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Panvongsa, W.; Pegtel, D.M.; Voortman, J. More than a Bubble: Extracellular Vesicle microRNAs in Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 1160. [Google Scholar] [CrossRef] [PubMed]
- Toden, S.; Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Hosseinalizadeh, H.; Mahmoodpour, M.; Ebrahimi, A. Circulating non-coding RNAs as a diagnostic and management biomarker for breast cancer: Current insights. Mol. Biol. Rep. 2022, 49, 705–715. [Google Scholar] [CrossRef]
- Dolati, S.; Shakouri, S.K.; Dolatkhah, N.; Yousefi, M.; Jadidi-Niaragh, F.; Sanaie, S. The role of exosomal non-coding RNAs in aging-related diseases. Biofactors 2021, 47, 292–310. [Google Scholar] [CrossRef] [PubMed]
- Cannavicci, A.; Zhang, Q.; Kutryk, M.J.B. Non-Coding RNAs and Hereditary Hemorrhagic Telangiectasia. J. Clin. Med. 2020, 9, 3333. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, M.; Zhang, L.; Lou, J.; Zhou, F.; Fang, M. Non-coding RNA in thyroid cancer-Functions and mechanisms. Cancer Lett. 2021, 496, 117–126. [Google Scholar] [CrossRef]
- Hwang, S.; Yang, Y.M. Exosomal microRNAs as diagnostic and therapeutic biomarkers in non-malignant liver diseases. Arch. Pharm. Res. 2021, 44, 574–587. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, L.; Zhao, G.; Chen, D.; Song, X.; Momtazi-Borojeni, A.A.; Yuan, H. Circulating exosomal microRNAs as emerging non-invasive clinical biomarkers in heart failure: Mega bio-roles of a nano bio-particle. IUBMB Life 2020, 72, 2546–2562. [Google Scholar] [CrossRef]
- Cavallaro, S.; Pevere, F.; Stridfeldt, F.; Görgens, A.; Paba, C.; Sahu, S.S.; Mamand, D.R.; Gupta, D.; El Andaloussi, S.; Linnros, J.; et al. Multiparametric Profiling of Single Nanoscale Extracellular Vesicles by Combined Atomic Force and Fluorescence Microscopy: Correlation and Heterogeneity in Their Molecular and Biophysical Features. Small 2021, 17, e2008155. [Google Scholar] [CrossRef]
- Zhang, J.; Li, H.; Fan, B.; Xu, W.; Zhang, X. Extracellular vesicles in normal pregnancy and pregnancy-related diseases. J. Cell Mol. Med. 2020, 24, 4377–4388. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Hu, Y.; Xu, S.; Liu, F.; Gao, Y. Exosomal microRNAs as Potential Biomarkers and Therapeutic Agents for Acute Ischemic Stroke: New Expectations. Front. Neurol. 2022, 12, 747380. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, Y.; Yin, Y.; Jia, X.; Mao, L. Mechanism of cargo sorting into small extracellular vesicles. Bioengineered 2021, 12, 8186–8201. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L.; Diergaarde, B.; Hong, C.S. Tumor-Derived Exosomes (TEX) and Their Role in Immuno-Oncology. Int. J. Mol. Sci. 2021, 22, 6234. [Google Scholar] [CrossRef] [PubMed]
- Stentz, R.; Carvalho, A.L.; Jones, E.J.; Carding, S.R. Fantastic voyage: The journey of intestinal microbiota-derived microvesicles through the body. Biochem. Soc. Trans. 2018, 46, 1021–1027. [Google Scholar] [CrossRef]
- Gomez, N.; James, V.; Onion, D.; Fairclough, L.C. Extracellular vesicles and chronic obstructive pulmonary disease (COPD): A systematic review. Respir. Res. 2022, 23, 82. [Google Scholar] [CrossRef]
- Clancy, J.W.; Schmidtmann, M.; D’Souza-Schorey, C. The ins and outs of microvesicles. FASEB Bioadv. 2021, 3, 399–406. [Google Scholar] [CrossRef]
- Zhang, W.; Huang, P.; Lin, J.; Zeng, H. The Role of Extracellular Vesicles in Osteoporosis: A Scoping Review. Membranes 2022, 12, 324. [Google Scholar] [CrossRef]
- Lima, T.S.; Souza, W.; Geaquinto, L.R.; Sanches, P.L.; Stepień, E.L.; Meneses, J.; Fernández-de Gortari, E.; Meisner-Kober, N.; Himly, M.; Granjeiro, J.M.; et al. Nanomaterial Exposure, Extracellular Vesicle Biogenesis and Adverse Cellular Outcomes: A Scoping Review. Nanomaterials 2022, 12, 1231. [Google Scholar] [CrossRef]
- Jiang, X.; You, L.; Zhang, Z.; Cui, X.; Zhong, H.; Sun, X.; Ji, C.; Chi, X. Biological Properties of Milk-Derived Extracellular Vesicles and Their Physiological Functions in Infant. Front. Cell Dev. Biol. 2021, 9, 693534. [Google Scholar] [CrossRef]
- Stam, J.; Bartel, S.; Bischoff, R.; Wolters, J.C. Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1169, 122604. [Google Scholar] [CrossRef]
- Nehrbas, J.; Butler, J.T.; Chen, D.W.; Kurre, P. Extracellular Vesicles and Chemotherapy Resistance in the AML Microenvironment. Front. Oncol. 2020, 10, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, A.; Ochiya, T. Exosomes and extracellular vesicles: Rethinking the essential values in cancer biology. Semin. Cancer Biol. 2021, 74, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lyden, D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019, 14, 1027–1053. [Google Scholar] [CrossRef]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e6. [Google Scholar] [CrossRef] [Green Version]
- Clancy, J.W.; Boomgarden, A.C.; D’Souza-Schorey, C. Profiling and promise of supermeres. Nat. Cell Biol. 2021, 23, 1217–1219. [Google Scholar] [CrossRef]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Graves-Deal, R.; Trinh, V.Q.; Ramirez, M.A.; Sohn, Y.; Neininger, A.C.; Taneja, N.; McKinley, E.T.; et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021, 23, 1240–1254. [Google Scholar] [CrossRef]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Thangaraju, K.; Neerukonda, S.N.; Katneni, U.; Buehler, P.W. Extracellular Vesicles from Red Blood Cells and Their Evolving Roles in Health, Coagulopathy and Therapy. Int. J. Mol. Sci. 2020, 22, 153. [Google Scholar] [CrossRef]
- Hade, M.D.; Suire, C.N.; Suo, Z. Mesenchymal Stem Cell-Derived Exosomes: Applications in Regenerative Medicine. Cells 2021, 10, 1959. [Google Scholar] [CrossRef]
- Choi, D.; Lee, T.H.; Spinelli, C.; Chennakrishnaiah, S.; D’Asti, E.; Rak, J. Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Semin. Cell Dev. Biol. 2017, 67, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.; Simons, M. Exosomes: Vesicular carriers for intercellular communication in neurodegenerative disorders. Cell Tissue Res. 2013, 352, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Hu, Y.W.; Zheng, L.; Wang, Q. Characteristics and Roles of Exosomes in Cardiovascular Disease. DNA Cell Biol. 2017, 36, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Jia, L.; Zheng, Y.; Li, W. Salivary Exosomes: Emerging Roles in Systemic Disease. Int. J. Biol. Sci. 2018, 14, 633–643. [Google Scholar] [CrossRef]
- Guay, C.; Regazzi, R. Exosomes as new players in metabolic organ cross-talk. Diabetes Obes. Metab. 2017, 19, 137–146. [Google Scholar] [CrossRef]
- Li, S.P.; Lin, Z.X.; Jiang, X.Y.; Yu, X.Y. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta. Pharmacol. Sin. 2018, 39, 542–551. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.B.; Bellingham, S.A.; Hill, A.F. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J. Biol. Chem. 2015, 290, 3455–3467. [Google Scholar] [CrossRef] [Green Version]
- Atukorala, I.; Mathivanan, S. The Role of Post-Translational Modifications in Targeting Protein Cargo to Extracellular Vesicles. Subcell. Biochem. 2021, 97, 45–60. [Google Scholar] [CrossRef]
- Harada, Y.; Suzuki, T.; Fukushige, T.; Kizuka, Y.; Yagi, H.; Yamamoto, M.; Kondo, K.; Inoue, H.; Kato, K.; Taniguchi, N.; et al. Generation of the heterogeneity of extracellular vesicles by membrane organization and sorting machineries. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 681–691. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H.; Odorizzi, G. Get on the exosome bus with ALIX. Nat. Cell Biol. 2012, 14, 654–655. [Google Scholar] [CrossRef] [PubMed]
- Stoorvogel, W. Resolving sorting mechanisms into exosomes. Cell Res. 2015, 25, 531–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, G.; Zimmermann, P. Heparanase Involvement in Exosome Formation. Adv. Exp. Med. Biol. 2020, 1221, 285–307. [Google Scholar] [CrossRef]
- Addi, C.; Presle, A.; Frémont, S.; Cuvelier, F.; Rocancourt, M.; Milin, F.; Schmutz, S.; Chamot-Rooke, J.; Douche, T.; Duchateau, M.; et al. The Flemmingsome reveals an ESCRT-to-membrane coupling via ALIX/syntenin/syndecan-4 required for completion of cytokinesis. Nat. Commun. 2020, 11, 1941. [Google Scholar] [CrossRef]
- Cone, A.S.; Hurwitz, S.N.; Lee, G.S.; Yuan, X.; Zhou, Y.; Li, Y.; Meckes, D.G., Jr. Alix and Syntenin-1 direct amyloid precursor protein trafficking into extracellular vesicles. BMC Mol. Cell Biol. 2020, 21, 58. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Tanno, H.; Komada, M. The ubiquitin code and its decoding machinery in the endocytic pathway. J. Biochem. 2013, 153, 497–504. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Muallem, S.; Kim, S.H.; Kwon, K.B.; Kim, M.S. Exosomal release through TRPML1-mediated lysosomal exocytosis is required for adipogenesis. Biochem. Biophys. Res. Commun. 2019, 510, 409–415. [Google Scholar] [CrossRef]
- Nag, S.; Rani, S.; Mahanty, S.; Bissig, C.; Arora, P.; Azevedo, C.; Saiardi, A.; van der Sluijs, P.; Delevoye, C.; van Niel, G.; et al. Rab4A organizes endosomal domains for sorting cargo to lysosome-related organelles. J. Cell Sci. 2018, 131, jcs216226. [Google Scholar] [CrossRef] [Green Version]
- Rome, S.; Forterre, A.; Mizgier, M.L.; Bouzakri, K. Skeletal Muscle-Released Extracellular Vesicles: State of the Art. Skeletal Muscle-Released Extracellular Vesicles: State of the Art. Front. Physiol. 2019, 10, 929. [Google Scholar] [CrossRef]
- Aheget, H.; Tristán-Manzano, M.; Mazini, L.; Cortijo-Gutierrez, M.; Galindo-Moreno, P.; Herrera, C.; Martin, F.; Marchal, J.A.; Benabdellah, K. Exosome: A New Player in Translational Nanomedicine. J. Clin. Med. 2020, 9, 2380. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T.; Rahman, S.; Khan, S.; Tasduq, S.A.; Choi, I. Biology, Pathophysiological Role, and Clinical Implications of Exosomes: A Critical Appraisal. Cells 2019, 8, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.N.; Chaudhuri, A.; Aqil, F.; Dehari, D.; Munagala, R.; Singh, S.; Gupta, R.C.; Agrawal, A.K. Exosomes as Emerging Drug Delivery and Diagnostic Modality for Breast Cancer: Recent Advances in Isolation and Application. Cancers 2022, 14, 1435. [Google Scholar] [CrossRef] [PubMed]
- Jayaseelan, V.P. Emerging role of exosomes as promising diagnostic tool for cancer. Cancer Gene. Ther. 2020, 27, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Ghafourian, M.; Mahdavi, R.; Akbari Jonoush, Z.; Sadeghi, M.; Ghadiri, N.; Farzaneh, M.; Mousavi Salehi, A. The implications of exosomes in pregnancy: Emerging as new diagnostic markers and therapeutics targets. Cell Commun. Signal 2022, 20, 51. [Google Scholar] [CrossRef]
- Aleksejeva, E.; Zarovni, N.; Dissanayake, K.; Godakumara, K.; Vigano, P.; Fazeli, A.; Jaakma, Ü.; Salumets, A. Extracellular vesicle research in reproductive science: Paving the way for clinical achievements. Biol. Reprod. 2022, 106, 408–424. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, N.; Yan, T.; Shi, Y.N.; Chen, J.; Zhang, C.J.; Xie, X.J.; Liao, D.F.; Qin, L. The crosstalk: Exosomes and lipid metabolism. Cell Commun. Signal. 2020, 18, 119. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sundaram, K.; Mu, J.; Dryden, G.W.; Sriwastva, M.K.; Lei, C.; Zhang, L.; Qiu, X.; Xu, F.; Yan, J.; et al. High-fat diet-induced upregulation of exosomal phosphatidylcholine contributes to insulin resistance. Nat. Commun. 2021, 12, 213. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ren, Y.; Sundaram, K.; Mu, J.; Sriwastva, M.K.; Dryden, G.W.; Lei, C.; Zhang, L.; Yan, J.; Zhang, X.; et al. miR-375 prevents high-fat diet-induced insulin resistance and obesity by targeting the aryl hydrocarbon receptor and bacterial tryptophanase (tnaA) gene. Theranostics 2021, 11, 4061–4077. [Google Scholar] [CrossRef]
- Rodrigues, A.C.; Spagnol, A.R.; Frias, F.T.; de Mendonça, M.; Araújo, H.N.; Guimarães, D.; Silva, W.J.; Bolin, A.P.; Murata, G.M.; Silveira, L. Intramuscular Injection of miR-1 Reduces Insulin Resistance in Obese Mice. Front. Physiol. 2021, 12, 676265. [Google Scholar] [CrossRef]
- Jung, M.K.; Mun, J.Y. Sample Preparation and Imaging of Exosomes by Transmission Electron Microscopy. J. Vis. Exp. 2018, 131, 56482. [Google Scholar] [CrossRef]
- Corrado, C.; Barreca, M.M.; Zichittella, C.; Alessandro, R.; Conigliaro, A. Molecular Mediators of RNA Loading into Extracellular Vesicles. Cells 2021, 10, 3355. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Geng, X.; Lin, X.; Zhang, Y.; Li, Q.; Guo, Y.; Fang, C.; Wang, H. Exosomal circular RNA sorting mechanisms and their function in promoting or inhibiting cancer. Oncol. Lett. 2020, 19, 3369–3380. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Asano, Y.; Ui-Tei, K. Modulation of MicroRNA Processing by Dicer via Its Associated dsRNA Binding Proteins. Noncoding RNA 2021, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, B.N.; Hata, A. Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun. Signal 2009, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Sheng, P.; Fields, C.; Aadland, K.; Wei, T.; Kolaczkowski, O.; Gu, T.; Kolaczkowski, B.; Xie, M. Dicer cleaves 5’-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res. 2018, 46, 5737–5752. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, J.S.; Langlois, R.A.; Pham, A.M.; Tenoever, B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 2012, 18, 1338–1346. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Tan, P.P.S.; Hall, D.; Chilian, W.M.; Chia, Y.C.; Mohd Zain, S.; Lim, H.M.; Kumar, D.N.; Ching, S.M.; Low, T.Y.; Md Noh, M.F.; et al. Exosomal microRNAs in the development of essential hypertension and its potential as biomarkers. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1486–H1497. [Google Scholar] [CrossRef]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Akgül, B.; Erdoğan, İ. Intracytoplasmic Re-localization of miRISC Complexes. Front. Genet. 2018, 9, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizutani, A.; Fukuda, M.; Ibata, K.; Shiraishi, Y.; Mikoshiba, K. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 2000, 275, 9823–9831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, D.J.; Franklin, J.L.; Dou, Y.; Liu, Q.; Higginbotham, J.N.; Demory Beckler, M.; Weaver, A.M.; Vickers, K.; Prasad, N.; Levy, S.; et al. KRAS-dependent sorting of miRNA to exosomes. Elife 2015, 4, e07197. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Life 2016, 5, e19276. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Yamada, Y. Nucleic Acid Drugs-Current Status, Issues, and Expectations for Exosomes. Cancers 2021, 13, 5002. [Google Scholar] [CrossRef]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef]
- Neumeier, J.; Meister, G. siRNA Specificity: RNAi Mechanisms and Strategies to Reduce Off-Target Effects. Front. Plant Sci. 2021, 11, 526455. [Google Scholar] [CrossRef]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar]
- Choudhary, S.; Lee, H.C.; Maiti, M.; He, Q.; Cheng, P.; Liu, Q.; Liu, Y. A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell Biol. 2007, 27, 3995–4005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, M.P.; Langer, R.; Jensen, K.F. Intracellular Delivery by Membrane Disruption: Mechanisms, Strategies, and Concepts. Chem. Rev. 2018, 118, 7409–7531. [Google Scholar] [CrossRef] [PubMed]
- Durymanov, M.; Reineke, J. Non-viral Delivery of Nucleic Acids: Insight Into Mechanisms of Overcoming Intracellular Barriers. Front. Pharmacol. 2018, 9, 971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.Z.; Xu, Y.; Xie, J.F.; Jiang, Z.H.; Peng, L.H. Ginsenoside as a new stabilizer enhances the transfection efficiency and biocompatibility of cationic liposome. Biomater. Sci. 2021, 9, 8373–8385. [Google Scholar] [CrossRef]
- Deshayes, S.; Simeoni, F.; Morris, M.C.; Divita, G.; Heitz, F. Peptide-mediated delivery of nucleic acids into mammalian cells. Methods Mol. Biol. 2007, 386, 299–308. [Google Scholar] [CrossRef]
- Della Pelle, G.; Kostevšek, N. Nucleic Acid Delivery with Red-Blood-Cell-Based Carriers. Int. J. Mol. Sci. 2021, 22, 5264. [Google Scholar] [CrossRef]
- Hammond, S.M.; Aartsma-Rus, A.; Alves, S.; Borgos, S.E.; Buijsen, R.A.M.; Collin, R.W.J.; Covello, G.; Denti, M.A.; Desviat, L.R.; Echevarría, L.; et al. Delivery of oligonucleotide-based therapeutics: Challenges and opportunities. EMBO Mol. Med. 2021, 13, e13243. [Google Scholar] [CrossRef]
- Kimura, N.; Maeki, M.; Sato, Y.; Note, Y.; Ishida, A.; Tani, H.; Harashima, H.; Tokeshi, M. Development of the iLiNP Device: Fine Tuning the Lipid Nanoparticle Size within 10 nm for Drug Delivery. ACS Omega 2018, 3, 5044–5051. [Google Scholar] [CrossRef]
- Ewert, K.K.; Scodeller, P.; Simón-Gracia, L.; Steffes, V.M.; Wonder, E.A.; Teesalu, T.; Safinya, C.R. Cationic Liposomes as Vectors for Nucleic Acid and Hydrophobic Drug Therapeutics. Pharmaceutics 2021, 13, 1365. [Google Scholar] [CrossRef]
- Fan, Y.; Marioli, M.; Zhang, K. Analytical characterization of liposomes and other lipid nanoparticles for drug delivery. J. Pharm. Biomed. Anal. 2021, 192, 113642. [Google Scholar] [CrossRef]
- Filipczak, N.; Pan, J.; Yalamarty, S.S.K.; Torchilin, V.P. Recent advancements in liposome technology. Adv. Drug Deliv. Rev. 2020, 156, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Goutas, D.; Pergaris, A.; Goutas, N.; Theocharis, S. Utilizing Exosomal-EPHs/Ephrins as Biomarkers and as a Potential Platform for Targeted Delivery of Therapeutic Exosomes. Int. J. Mol. Sci. 2022, 23, 3551. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.; Huang, D.; Sang, C.; Zhong, T.; Zhang, Z.; Tang, Z. Advances in Mesenchymal Stem Cell-Derived Exosomes as Drug Delivery Vehicles. Front. Bioeng. Biotechnol. 2022, 9, 797359. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.C.; Li, C.; Zhang, W.; Pi, W.; Han, N. Potential Effects of Exosomes and Their MicroRNA Carrier on Osteoporosis. Curr. Pharm. Des. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Chen, W.; Ye, J.; Wang, Y. Potential Role of Exosomes in Ischemic Stroke Treatment. Biomolecules 2022, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yue, C.; Gao, S.; Li, S.; Zhou, J.; Chen, J.; Fu, J.; Sun, W.; Hua, C. Promising Roles of Exosomal microRNAs in Systemic Lupus Erythematosus. Front. Immunol. 2021, 12, 757096. [Google Scholar] [CrossRef]
- Jurgielewicz, B.; Stice, S.; Yao, Y. Therapeutic Potential of Nucleic Acids when Combined with Extracellular Vesicles. Aging Dis. 2021, 12, 1476–1493. [Google Scholar] [CrossRef]
- Xu, M.; Feng, T.; Liu, B.; Qiu, F.; Xu, Y.; Zhao, Y.; Zheng, Y. Engineered exosomes: Desirable target-tracking characteristics for cerebrovascular and neurodegenerative disease therapies. Theranostics 2021, 11, 8926–8944. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, P.; Tan, H.; Chen, X.; Wang, Q.; Chen, T. Exosomes as Smart Nanoplatforms for Diagnosis and Therapy of Cancer. Front. Oncol. 2021, 11, 743189. [Google Scholar] [CrossRef]
- Jiang, L.; Gu, Y.; Du, Y.; Tang, X.; Wu, X.; Liu, J. Engineering Exosomes Endowed with Targeted Delivery of Triptolide for Malignant Melanoma Therapy. ACS Appl. Mater. Interfaces 2021, 13, 42411–42428. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Gnanasampanthapandian, D.; Rajasingh, J.; Palaniyandi, K. Stem Cell-Derived Exosomes Potential Therapeutic Roles in Cardiovascular Diseases. Front. Cardiovasc. Med. 2021, 8, 723236. [Google Scholar] [CrossRef] [PubMed]
- Moghadasi, S.; Elveny, M.; Rahman, H.S.; Suksatan, W.; Jalil, A.T.; Abdelbasset, W.K.; Yumashev, A.V.; Shariatzadeh, S.; Motavalli, R.; Behzad, F.; et al. A paradigm shift in cell-free approach: The emerging role of MSCs-derived exosomes in regenerative medicine. J. Transl. Med. 2021, 19, 302. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhao, Y.C.; Niu, Z.F.; Fan, H.J.; Hou, S.K.; Guo, X.Q.; Sang, L.; Lv, Q. Mesenchymal stem cell-derived small extracellular vesicles in the treatment of human diseases: Progress and prospect. World J. Stem Cells 2021, 13, 49–63. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Huang, Y.; Li, R.; Ye, S.; Lin, S.; Yin, G.; Xie, Q. Recent Advances in the Use of Exosomes in Sjögren’s Syndrome. Front. Immunol. 2020, 11, 1509. [Google Scholar] [CrossRef]
- Markov, O.; Oshchepkova, A.; Mironova, N. Immunotherapy Based on Dendritic Cell-Targeted/-Derived Extracellular Vesicles-A Novel Strategy for Enhancement of the Anti-tumor Immune Response. Front. Pharmacol. 2019, 10, 1152. [Google Scholar] [CrossRef] [Green Version]
- Yang, P.; Peng, Y.; Feng, Y.; Xu, Z.; Feng, P.; Cao, J.; Chen, Y.; Chen, X.; Cao, X.; Yang, Y.; et al. Immune Cell-Derived Extracellular Vesicles-New Strategies in Cancer Immunotherapy. Front. Immunol. 2021, 12, 771551. [Google Scholar] [CrossRef]
- Hosseini, R.; Asef-Kabiri, L.; Yousefi, H.; Sarvnaz, H.; Salehi, M.; Akbari, M.E.; Eskandari, N. The roles of tumor-derived exosomes in altered differentiation, maturation and function of dendritic cells. Mol. Cancer 2021, 20, 83. [Google Scholar] [CrossRef]
- Wang, Y.; Xiang, Y.; Xin, V.W.; Wang, X.W.; Peng, X.C.; Liu, X.Q.; Wang, D.; Li, N.; Cheng, J.T.; Lyv, Y.N.; et al. Dendritic cell biology and its role in tumor immunotherapy. J. Hematol. Oncol. 2020, 13, 107. [Google Scholar] [CrossRef]
- Han, Y.; Jones, T.W.; Dutta, S.; Zhu, Y.; Wang, X.; Narayanan, S.P.; Fagan, S.C.; Zhang, D. Overview and Update on Methods for Cargo Loading into Extracellular Vesicles. Processes 2021, 9, 356. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, M.; Wang, J.; Su, L.; Lin, J.; Yan, X. Active cargo loading into extracellular vesicles: Highlights the heterogeneous encapsulation behaviour. J. Extracell. Vesicles 2021, 10, e12163. [Google Scholar] [CrossRef] [PubMed]
- Rankin-Turner, S.; Vader, P.; O’Driscoll, L.; Giebel, B.; Heaney, L.M.; Davies, O.G. A call for the standardised reporting of factors affecting the exogenous loading of extracellular vesicles with therapeutic cargos. Adv. Drug Deliv. Rev. 2021, 173, 479–491. [Google Scholar] [CrossRef]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Raghav, A.; Jeong, G.B. A systematic review on the modifications of extracellular vesicles: A revolutionized tool of nano-biotechnology. J. Nanobiotechnol. 2021, 19, 459. [Google Scholar] [CrossRef] [PubMed]
- N’Diaye, E.R.; Orefice, N.S.; Ghezzi, C.; Boumendjel, A. Chemically Modified Extracellular Vesicles and Applications in Radiolabeling and Drug Delivery. Pharmaceutics 2022, 14, 653. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.H.; Liang, M.X.; Wu, Y.; Yang, K.; Tang, J.H.; Zhang, W. Extracellular vesicles as drug vectors for precise cancer treatment. Nanomedicine 2021, 16, 1519–1537. [Google Scholar] [CrossRef]
- Urzì, O.; Gasparro, R.; Ganji, N.R.; Alessandro, R.; Raimondo, S. Plant-RNA in Extracellular Vesicles: The Secret of Cross-Kingdom Communication. Membranes 2022, 12, 352. [Google Scholar] [CrossRef]
- He, B.; Hamby, R.; Jin, H. Plant extracellular vesicles: Trojan horses of cross-kingdom warfare. FASEB Bioadv. 2021, 3, 657–664. [Google Scholar] [CrossRef]
- Cuesta, C.M.; Guerri, C.; Ureña, J.; Pacual, M. Role of Microbiota-Derived Extracellular Vesicles in Gut-Brain Communication. Int. J. Mol. Sci. 2021, 22, 4235. [Google Scholar] [CrossRef]
- Pérez-Cabezas, B.; Santarém, N.; Cecílio, P.; Silva, C.; Silvestre, R.; AM Catita, J.; Cordeiro da Silva, A. More than just exosomes: Distinct Leishmania infantum extracellular products potentiate the establishment of infection. J. Extracell. Vesicles 2018, 8, 1541708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morishita, M.; Horita, M.; Higuchi, A.; Marui, M.; Katsumi, H.; Yamamoto, A. Characterizing Different Probiotic-Derived Extracellular Vesicles as a Novel Adjuvant for Immunotherapy. Mol. Pharm. 2021, 18, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Sagayama, R.; Yamawaki, Y.; Yamaguchi, M.; Katsumi, H.; Yamamoto, A. Activation of Host Immune Cells by Probiotic-Derived Extracellular Vesicles via TLR2-Mediated Signaling Pathways. Biol. Pharm. Bull. 2022, 45, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, J.H.; Kim, E.H.; Reaney, M.J.T.; Shim, Y.Y.; Chung, M.J. Immunomodulatory Activity of Extracellular Vesicles of Kimchi-Derived Lactic Acid Bacteria (Leuconostoc mesenteroides, Latilactobacillus curvatus, and Lactiplantibacillus plantarum). Foods 2022, 11, 313. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Lin, H.; Wang, M.; Zhao, Y.; Liu, H.; Min, Y.; Yang, X.; Gao, Y.; Yang, M. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J. Anim. Sci. Biotechnol. 2021, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Cerione, R.A.; Antonyak, M.A. Isolation and characterization of extracellular vesicles produced by cell lines. STAR Protoc. 2021, 2, 100295. [Google Scholar] [CrossRef]
- Tang, Y.T.; Huang, Y.Y.; Zheng, L.; Qin, S.H.; Xu, X.P.; An, T.X.; Xu, Y.; Wu, Y.S.; Hu, X.M.; Ping, B.H.; et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucl. Acids 2021, 2, 80–103. [Google Scholar] [CrossRef]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating Methods for Isolation and Quantification of Exosomes: A Review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef]
- Clos-Sansalvador, M.; Monguió-Tortajada, M.; Roura, S.; Franquesa, M.; Borràs, F.E. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur. J. Cell Biol. 2022, 101, 151227. [Google Scholar] [CrossRef]
- Tiwari, S.; Kumar, V.; Randhawa, S.; Verma, S.K. Preparation and characterization of extracellular vesicles. Preparation and characterization of extracellular vesicles. Am. J. Reprod. Immunol. 2021, 85, e13367. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; McNamara, R.P.; Dittmer, D.P. Purification Methods and the Presence of RNA in Virus Particles and Extracellular Vesicles. Viruses 2020, 12, 917. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Zhou, H.; Qian, X. Syntaxin 2 promotes colorectal cancer growth by increasing the secretion of exosomes. J. Cancer 2021, 12, 2050–2058. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.Y.; Sung, C.W.; Chen, C.; Cheng, C.M.; Lin, D.P.; Huang, C.T.; Hsu, M.Y. Advances in exosomes technology. Clin. Chim. Acta 2019, 493, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Guo, J.; Zhang, Z.; Duan, M.; Wang, G.; Qian, Y.; Zhao, H.; Yang, Z.; Jiang, X. Application of engineered extracellular vesicles for targeted tumor therapy. J. Biomed. Sci. 2022, 29, 14. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yang, Q.; Sun, X.; Wang, Y. Recent Advancements in the Loading and Modification of Therapeutic Exosomes. Front. Bioeng. Biotechnol. 2020, 8, 586130. [Google Scholar] [CrossRef] [PubMed]
- Busatto, S.; Iannotta, D.; Walker, S.A.; Di Marzio, L.; Wolfram, J. A Simple and Quick Method for Loading Proteins in Extracellular Vesicles. Pharmaceuticals 2021, 14, 356. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Luan, X.; Sansanaphongpricha, K.; Myers, I.; Chen, H.; Yuan, H.; Sun, D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta. Pharmacol. Sin. 2017, 38, 754–763. [Google Scholar] [CrossRef] [Green Version]
- Gangadaran, P.; Ahn, B.C. Extracellular Vesicle- and Extracellular Vesicle Mimetics-Based Drug Delivery Systems: New Perspectives, Challenges, and Clinical Developments. Pharmaceutics 2020, 12, 442. [Google Scholar] [CrossRef]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.W.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F.; et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef] [PubMed]
- Soma, E.; Yamayoshi, A.; Toda, Y.; Mishima, Y.; Hosogi, S.; Ashihara, E. Successful Incorporation of Exosome-Capturing Antibody-siRNA Complexes into Multiple Myeloma Cells and Suppression of Targeted mRNA Transcripts. Cancers 2022, 14, 566. [Google Scholar] [CrossRef] [PubMed]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, V.; Berglund, M.; Skogberg, G.; Lindgren, S.; Lundqvist, C.; Gudmundsdottir, J.; Thörn, K.; Telemo, E.; Ekwall, O. Thymic exosomes promote the final maturation of thymocytes. Sci. Rep. 2016, 6, 36479. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Bunggulawa, E.J.; Wang, W.; Yin, T.; Wang, N.; Durkan, C.; Wang, Y.; Wang, G. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 2018, 16, 81. [Google Scholar] [CrossRef] [Green Version]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [Green Version]
- Clayton, A.; Turkes, A.; Dewitt, S.; Steadman, R.; Mason, M.D.; Hallett, M.B. Adhesion and signaling by B cell-derived exosomes: The role of integrins. FASEB J. 2004, 18, 977–979. [Google Scholar] [CrossRef]
- Bastos-Amador, P.; Pérez-Cabezas, B.; Izquierdo-Useros, N.; Puertas, M.C.; Martinez-Picado, J.; Pujol-Borrell, R.; Naranjo-Gómez, M.; Borràs, F.E. Capture of cell-derived microvesicles (exosomes and apoptotic bodies) by human plasmacytoid dendritic cells. J. Leukoc. Biol. 2012, 91, 751–758. [Google Scholar] [CrossRef]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, X.; Liu, C.; Wang, H.; Wang, L.; Xiao, F.; Guo, Z.; Zhang, H. Surface Phosphatidylserine Is Responsible for the Internalization on Microvesicles Derived from Hypoxia-Induced Human Bone Marrow Mesenchymal Stem Cells into Human Endothelial Cells. PLoS ONE 2016, 11, e0147360. [Google Scholar] [CrossRef] [Green Version]
- Bhatta, M.; Shenoy, G.N.; Loyall, J.L.; Gray, B.D.; Bapardekar, M.; Conway, A.; Minderman, H.; Kelleher, R.J., Jr.; Carreno, B.M.; Linette, G.; et al. Novel phosphatidylserine-binding molecule enhances antitumor T-cell responses by targeting immunosuppressive exosomes in human tumor microenvironments. J. Immunother. Cancer 2021, 9, e003148. [Google Scholar] [CrossRef]
- Meier, A.F.; Fraefel, C.; Seyffert, M. The Interplay between Adeno-Associated Virus and its Helper Viruses. Viruses 2020, 12, 662. [Google Scholar] [CrossRef]
- Zinn, E.; Vandenberghe, L.H. Adeno-associated virus: Fit to serve. Curr. Opin. Virol. 2014, 8, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Asokan, A.; Samulski, R.J. Adeno-associated virus serotypes: Vector toolkit for human gene therapy. Mol. Ther. 2006, 14, 316–327. [Google Scholar] [CrossRef]
- Wörner, T.P.; Bennett, A.; Habka, S.; Snijder, J.; Friese, O.; Powers, T.; Agbandje-McKenna, M.; Heck, A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021, 12, 1642. [Google Scholar] [CrossRef]
- Gonçalves, M.A. Adeno-associated virus: From defective virus to effective vector. Virol. J. 2005, 2, 43. [Google Scholar] [CrossRef] [Green Version]
- Owens, R.A. Second generation adeno-associated virus type 2-based gene therapy systems with the potential for preferential integration into AAVS1. Curr. Gene Ther. 2002, 2, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Rivière, C.; Danos, O.; Douar, A.M. Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Ther. 2006, 13, 1300–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blackburn, S.D.; Steadman, R.A.; Johnson, F.B. Attachment of adeno-associated virus type 3H to fibroblast growth factor receptor 1. Arch. Virol. 2006, 151, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Asokan, A.; Hamra, J.B.; Govindasamy, L.; Agbandje-McKenna, M.; Samulski, R.J. Adeno-associated virus type 2 contains an integrin alpha5beta1 binding domain essential for viral cell entry. J. Virol. 2006, 80, 8961–8969. [Google Scholar] [CrossRef] [Green Version]
- Woodard, K.T.; Liang, K.J.; Bennett, W.C.; Samulski, R.J. Heparan Sulfate Binding Promotes Accumulation of Intravitreally Delivered Adeno-associated Viral Vectors at the Retina for Enhanced Transduction but Weakly Influences Tropism. J. Virol. 2016, 90, 9878–9888. [Google Scholar] [CrossRef] [Green Version]
- Walters, R.W.; Pilewski, J.M.; Chiorini, J.A.; Zabner, J. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 2002, 277, 23709–23713. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Li, Z.; Huang, S.; Yan, B.; He, S.; Chen, F.; Liang, Y. AAV-Containing Exosomes as a Novel Vector for Improved Gene Delivery to Lung Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 707607. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Yang, M.; Qiu, R.; Li, Y.; Bian, S.; Hao, B.; Lei, B. Intravitreal Injection of an Exosome-Associated Adeno-Associated Viral Vector Enhances Retinoschisin 1 Gene Transduction in the Mouse Retina. Hum. Gene Ther. 2021, 32, 707–716. [Google Scholar] [CrossRef]
- Maurya, S.; Jayandharan, G.R. Exosome-associated SUMOylation mutant AAV demonstrates improved ocular gene transfer efficiency in vivo. Virus Res. 2020, 283, 197966. [Google Scholar] [CrossRef]
- Khan, N.; Maurya, S.; Bammidi, S.; Jayandharan, G.R. AAV6 Vexosomes Mediate Robust Suicide Gene Delivery in a Murine Model of Hepatocellular Carcinoma. Mol. Ther. Methods Clin. Dev. 2020, 17, 497–504. [Google Scholar] [CrossRef]
- Meliani, A.; Boisgerault, F.; Fitzpatrick, Z.; Marmier, S.; Leborgne, C.; Collaud, F.; Simon Sola, M.; Charles, S.; Ronzitti, G.; Vignaud, A.; et al. Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors. Blood Adv. 2017, 1, 2019–2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wassmer, S.J.; Carvalho, L.S.; György, B.; Vandenberghe, L.H.; Maguire, C.A. Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection. Sci. Rep. 2017, 7, 45329. [Google Scholar] [CrossRef] [PubMed]
- Hudry, E.; Martin, C.; Gandhi, S.; György, B.; Scheffer, D.I.; Mu, D.; Merkel, S.F.; Mingozzi, F.; Fitzpatrick, Z.; Dimant, H.; et al. Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016, 23, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maguire, C.A.; Balaj, L.; Sivaraman, S.; Crommentuijn, M.H.; Ericsson, M.; Mincheva-Nilsson, L.; Baranov, V.; Gianni, D.; Tannous, B.A.; Sena-Esteves, M.; et al. Microvesicle-associated AAV vector as a novel gene delivery system. Mol. Ther. 2012, 20, 960–971. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.; Dietz, L.; Gong, Y.; Eichler, F.; Nammour, J.; Ng, C.; Grimm, D.; Maguire, C.A. Neutralizing Antibody Evasion and Transduction with Purified Extracellular Vesicle-Enveloped Adeno-Associated Virus Vectors. Hum. Gene Ther. 2021, 32, 1457–1470. [Google Scholar] [CrossRef]
- György, B.; Fitzpatrick, Z.; Crommentuijn, M.H.; Mu, D.; Maguire, C.A. Naturally enveloped AAV vectors for shielding neutralizing antibodies and robust gene delivery in vivo. Biomaterials 2014, 35, 7598–7609. [Google Scholar] [CrossRef] [Green Version]



| Pharmaceutical Technology | Advantage |
|---|---|
| Liposome | Improved transduction and safety |
| Microcapsule | Extension in dosing interval |
| PEG modification * | Extension in dosing interval |
| Transdermal administration | Improved convenience and safety |
| Sublingual administration | Immediate effect improvement |
| Antibody drug conjugate | Improved transduction and safety |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsuzaka, Y.; Yashiro, R. Extracellular Vesicles as Novel Drug-Delivery Systems through Intracellular Communications. Membranes 2022, 12, 550. https://doi.org/10.3390/membranes12060550
Matsuzaka Y, Yashiro R. Extracellular Vesicles as Novel Drug-Delivery Systems through Intracellular Communications. Membranes. 2022; 12(6):550. https://doi.org/10.3390/membranes12060550
Chicago/Turabian StyleMatsuzaka, Yasunari, and Ryu Yashiro. 2022. "Extracellular Vesicles as Novel Drug-Delivery Systems through Intracellular Communications" Membranes 12, no. 6: 550. https://doi.org/10.3390/membranes12060550
APA StyleMatsuzaka, Y., & Yashiro, R. (2022). Extracellular Vesicles as Novel Drug-Delivery Systems through Intracellular Communications. Membranes, 12(6), 550. https://doi.org/10.3390/membranes12060550






