Host–Bacterial Interactions: Outcomes of Antimicrobial Peptide Applications
Abstract
:1. Introduction
2. Bacterial Membrane Interaction with Host Cells
2.1. Secretion Systems in the Bacteria Membrane
2.1.1. Secretion Systems in Gram-Negative Bacteria
2.1.2. Secretion Systems in Both Gram-Negative Bacteria and Gram-Positive Bacteria
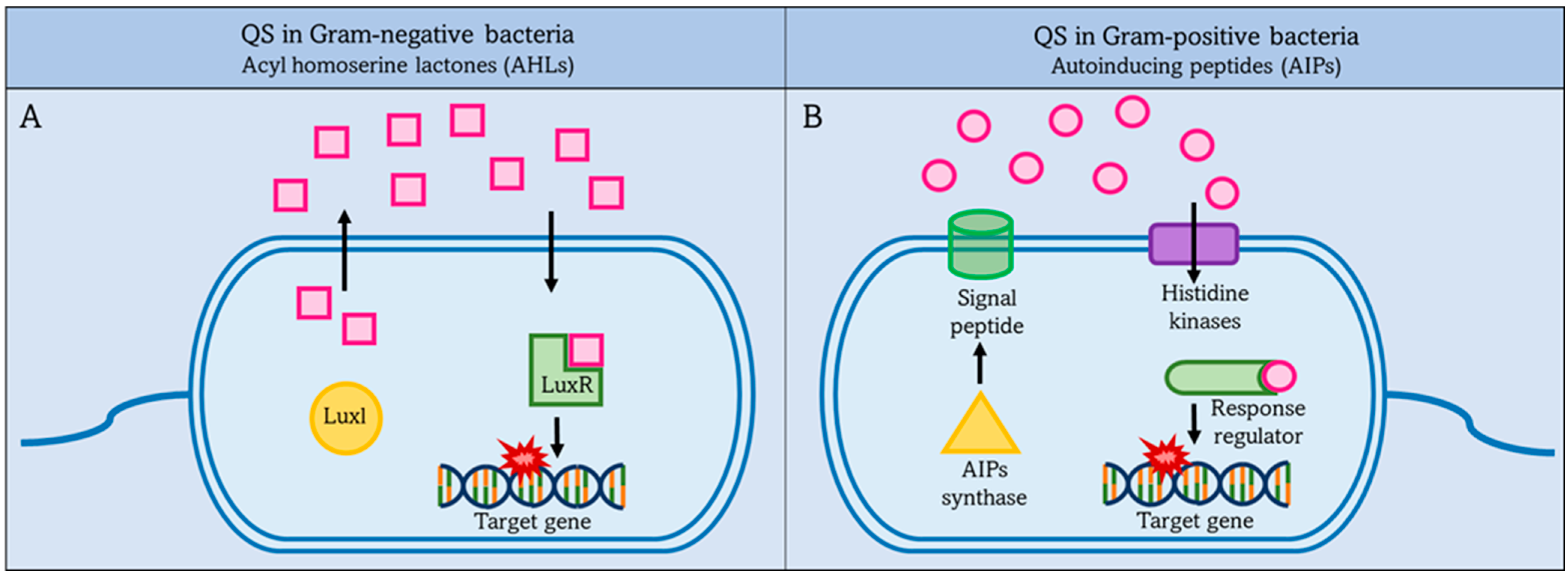
2.2. Quorum Sensing (QS)
2.2.1. QS in Gram-Negative Bacteria
2.2.2. Quorum Sensing in Gram-Positive Bacteria
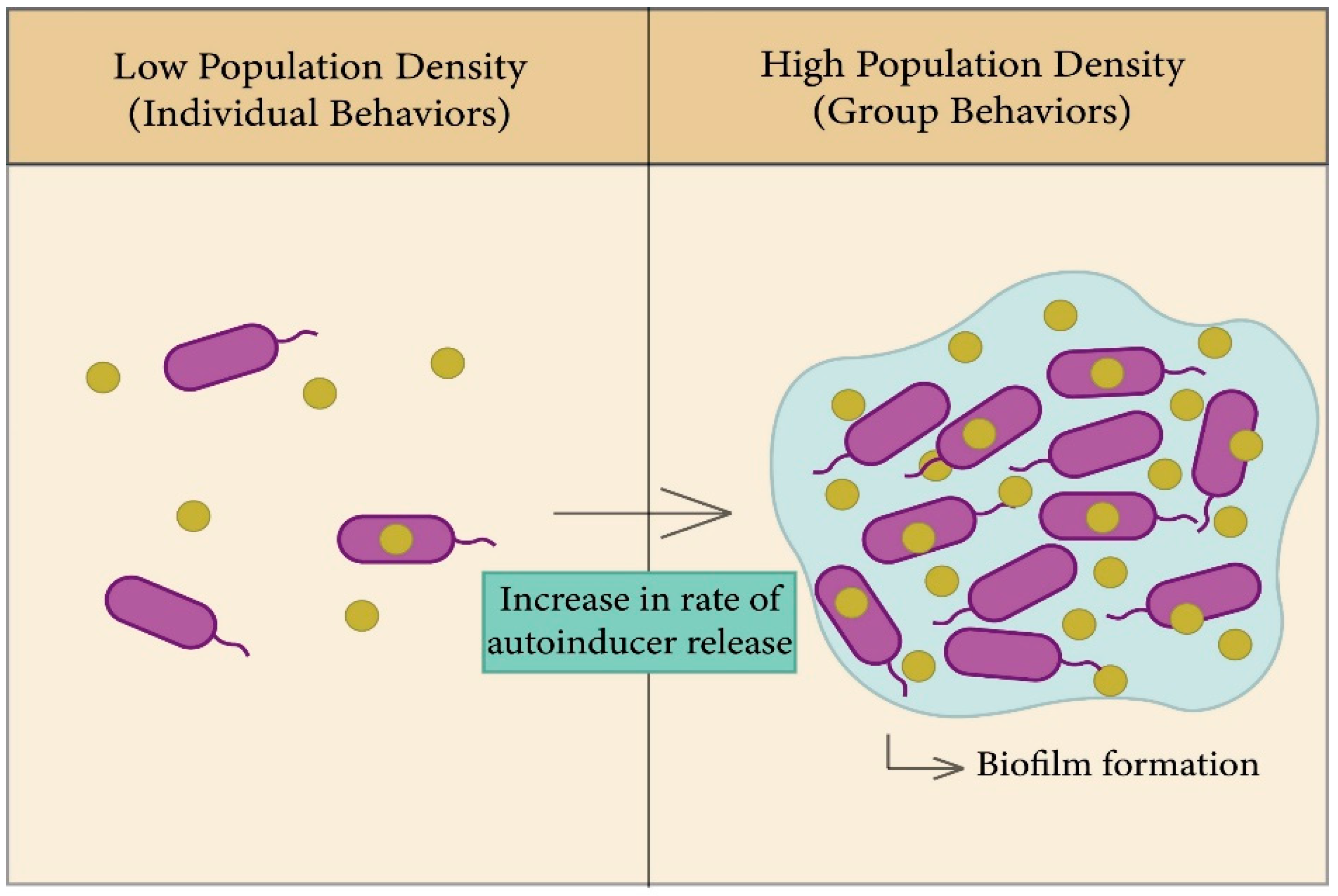
2.2.3. The Role of Quorum Sensing in Biofilm Formation
| Autoinducers | Microorganisms | Receptors | Phenotypes | References |
|---|---|---|---|---|
| Autoinducing peptides (AIPs) | Gram-positive bacteria | Response regulator | Genetic competence. | [69] |
| Autoinducer-2 (AI-2) | Many Gram-negative and Gram-positive bacteria | LuxP, LsrB, and dCACHE | Virulence, biofilm formation, and protease. | [70,71] |
| CAI-1 autoinducer | Vibrio | CqsS | Virulence, biofilm formation, and protease. | [71,72] |
| HAI-1 | Vibrio harveyi and Vibrio parahaemolyticus | LuxN | Biofilm formation, bioluminescence, TTS, and protease. | [73] |
| Acyl homoserine lactones (AHLs) | Gram-negative bacteria and commensals | LuxR | Elastase, biofilm formation, virulence factors, and exotoxins. | [74,75] |
| Competence-stimulating peptides (CSPs) | Streptococcus pneumoniae | ComD | Biofilm formation, and virulence. | [76] |
| comX-inducing peptide (XIP) | Streptococcus mutans | ComR | Antibiotic tolerance, genetic competence, and dormancy. | [77,78] |
| ComX | Bacillus subtilis | ComP | Protease and biofilm formation. | [79,80] |
3. Bacterial Membrane Interaction with Antimicrobials
Phospholipid Targeting by Antimicrobials
4. AMPs as Molecules Targeting Bacteria
4.1. Membrane Targeting Mechanism of AMPs
4.1.1. Barrel-Stave Models
4.1.2. Toroidal Pore Models
4.1.3. Carpet Model
4.2. Classification of the AMPs Based on Amino Acid-Rich Species
4.2.1. Proline-Rich Peptides
4.2.2. Tryptophan and Arginine-Rich AMPs
4.2.3. Histidine-Rich Peptides
4.2.4. Glycine-Rich AMPs
| Amino Acid-Rich Species | Description | Biological Source | Biological Activity | Peptides | Peptide Sequence | Bacteria | MIC (μg/mL) | Animal Model\In Vitro | References |
|---|---|---|---|---|---|---|---|---|---|
| Proline-Rich Peptides | Large group of small and medium in size, heterogeneous, proline-containing peptides that are arranged in peculiar sequences. | Marine sponges (aquatic), microorganisms, and mammals. | Antibacterial and antifungal. | Api88 | Gu-ONNRPVYIPRPRPPHPRL-NH2 | P. aeruginosa | 32 | In vitro | [98,120,121,122,123,124,125] |
| Pyrrhocoricin | VDKGSYLPRPTPPRPIYNRN | K. pneumonia | 4 | In vitro | |||||
| Bac7 | RRIRPRPPRLPRPRPR | E. coli | 8 | In vitro | |||||
| Arginine-Rich AMPs | Composed of small size peptides and has a short cyclic half-life as it is digested by blood proteases. | Frogs (amphibian) and insects | Antibacterial, antiviral, antifungal, antitumor, and immunomodulatory. | Cecropin D | WNPFKELEKVGQRVRDAVISAGPAVATVAQATALAK-NH2 | S. aureus | 4.55 | In vitro | [126,127,128,129] |
| Cecropin B | KWKVFKKIEKMGRNIRNGIVKAGPAIAVLGEAKAL-NH2 | H. parasuis | 2 | - | |||||
| Cecropin P1 | SWLSKTAKKLENSAKKRISEGIAIAIQGGPR-NH2 | E. coli | 1000 | - | |||||
| Histidine-Rich Peptides | Peptides with short sequences, usually H6, and are widely used in the production of recombinant protein as purification tags. | - | Antibacterial, antifungal and antivirus. | HALO | KKALLOHALHOLALLOHLAHOLKKA | P. aeruginosa | 0.168 | In vitro | [130,131,132,133] |
| LAH4 | (KKALLALALHHLAHLALHLALALKKA) | E. coli and S. aureus | 277.9 | In vitro | |||||
| Glycine-Rich AMPs | Consists of a series of peptides with a high content of glycine, which has a low profile in animals. | Aquatic and mammals | Antibacterial and antifungal. | Persulcatusin | GFGCPFNQGACHRHCRSIGRRGGYCAGLFKQTCTCYSR-NH2 | S. aureus | 0.156–1.25 | - | [129,134,135,136] |
| Attacins B | QAGALTINSDGTSGAV-VKVPITGNENHKFSALGSVDLT-NQMKL GAATAGLAYDNGNGHGATLT KTHIPGFGDKMTAAGKVNLFHN DNHDFSAKAFATKNMP-NIPQVPNFNTVGAGVDYMFKDKIGASANAAHTDFINRNDYS-LGGKLNLFKTPTTSLDFNAGWKKF DTPFFKSSWEPSTSFSFSKYF | P. maltophilia | 0.080–0.094 | Insect cells | |||||
| Attacins E | DAHGALTLNSDGTSGAVVKVPFAGNDKNIVSAIGSVDLT-DRQKL GAATAGVALDNINGHGLSLTDT HIPGFGDKMTAAGKVNVFHNDNHDITAKAFATRNMPDIANVPN FNTVGGGIDYMFKDKIG TRNMPSIPNVPNFN-TIGGGVDYMYKNKVGASLGMASTPFLDRKDYSAMG WEPNFGFSLSKYF | E. coli | 0.040–0.046 | Insect cells |
4.3. Bacterial Resistance to AMPs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and Function of Bacterial Outer Membrane Proteins: Barrels in a Nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems—An Overview. Microbiol. Spectr. 2016, 4, 4.1.13. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Bassler, B.L.; Losick, R. Bacterially Speaking. Cell 2006, 125, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting Bacterial Membrane Function: An Underexploited Mechanism for Treating Persistent Infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [Green Version]
- Herzog, I.M.; Fridman, M. Design and Synthesis of Membrane-Targeting Antibiotics: From Peptides- to Aminosugar-Based Antimicrobial Cationic Amphiphiles. Med. Chem. Commun. 2014, 5, 1014–1026. [Google Scholar] [CrossRef]
- Daugelavičius, R.; Bakienė, E.; Bamford, D.H. Stages of Polymyxin B Interaction with the Escherichia Coli Cell Envelope. Antimicrob. Agents Chemother. 2000, 44, 2969–2978. [Google Scholar] [CrossRef] [Green Version]
- Bunchorntavakul, C.; Chamroonkul, N.; Chavalitdhamrong, D. Bacterial Infections in Cirrhosis: A Critical Review and Practical Guidance. World J. Hepatol. 2016, 8, 307–321. [Google Scholar] [CrossRef]
- Dever, L.A.; Dermody, T.S. Mechanisms of Bacterial Resistance to Antibiotics. Arch. Intern. Med. 1991, 151, 886–895. [Google Scholar] [CrossRef]
- Ayoub Moubareck, C. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. [Google Scholar] [CrossRef]
- Hollmann, A.; Martinez, M.; Maturana, P.; Semorile, L.C.; Maffia, P.C. Antimicrobial Peptides: Interaction With Model and Biological Membranes and Synergism with Chemical Antibiotics. Front. Chem. 2018, 6, 204. [Google Scholar] [CrossRef] [Green Version]
- Peterson, J.W. Bacterial Pathogenesis. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Wilson, J.W.; Schurr, M.J.; LeBlanc, C.L.; Ramamurthy, R.; Buchanan, K.L.; Nickerson, C.A. Mechanisms of Bacterial Pathogenicity. Postgrad. Med. J. 2002, 78, 216–224. [Google Scholar] [CrossRef]
- Norrby, S.R. Chapter 54—Urinary Tract Infections. In Antibiotic and Chemotherapy, 9th ed.; Finch, R.G., Greenwood, D., Norrby, S.R., Whitley, R.J., Eds.; W.B. Saunders: London, UK, 2010; pp. 694–701. ISBN 978-0-7020-4064-1. [Google Scholar]
- Doron, S.; Gorbach, S.L. Bacterial Infections: Overview. Int. Encycl. Public Health 2008, 273–282. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship Between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef] [Green Version]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [Green Version]
- Depluverez, S.; Devos, S.; Devreese, B. The Role of Bacterial Secretion Systems in the Virulence of Gram-Negative Airway Pathogens Associated with Cystic Fibrosis. Front. Microbiol. 2016, 7, 1336. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Holland, I.B.; Schmitt, L. The Type 1 Secretion Pathway—The Hemolysin System and Beyond. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 1629–1641. [Google Scholar] [CrossRef] [Green Version]
- Morgan, J.L.W.; Acheson, J.F.; Zimmer, J. Structure of a Type-1 Secretion System ABC Transporter. Structure 2017, 25, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Kanonenberg, K.; Schwarz, C.K.W.; Schmitt, L. Type I Secretion Systems—A Story of Appendices. Res. Microbiol. 2013, 164, 596–604. [Google Scholar] [CrossRef]
- Delepelaire, P. Type I Secretion in Gram-Negative Bacteria. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2004, 1694, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Jun, S.-Y.; Yoon, B.-Y.; Song, S.; Lee, K.; Ha, N.-C. Membrane Fusion Proteins of Type I Secretion System and Tripartite Efflux Pumps Share a Binding Motif for TolC in Gram-Negative Bacteria. PLoS ONE 2012, 7, e40460. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G.J. The Type II Secretion System: Biogenesis, Molecular Architecture and Mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.; Bolhuis, A. Tat-Dependent Protein Targeting in Prokaryotes and Chloroplasts. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2004, 1694, 135–147. [Google Scholar] [CrossRef] [Green Version]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III Secretion Systems and Disease. Clin. Microbiol. Rev. 2007, 20, 535–549. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Reeves, A.Z.; Klein, J.A.; Twedt, D.J.; Knodler, L.A.; Lesser, C.F. The Type III Secretion System Apparatus Determines the Intracellular Niche of Bacterial Pathogens. Proc. Natl. Acad. Sci. USA 2016, 113, 4794–4799. [Google Scholar] [CrossRef] [Green Version]
- Meuskens, I.; Saragliadis, A.; Leo, J.C.; Linke, D. Type V Secretion Systems: An Overview of Passenger Domain Functions. Front. Microbiol. 2019, 10, 1163. [Google Scholar] [CrossRef] [Green Version]
- Leo, J.C.; Grin, I.; Linke, D. Type V Secretion: Mechanism(s) of Autotransport through the Bacterial Outer Membrane. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1088–1101. [Google Scholar] [CrossRef] [Green Version]
- Grijpstra, J.; Arenas, J.; Rutten, L.; Tommassen, J. Autotransporter Secretion: Varying on a Theme. Res. Microbiol. 2013, 164, 562–582. [Google Scholar] [CrossRef]
- Henderson, I.R.; Navarro-Garcia, F.; Desvaux, M.; Fernandez, R.C.; Ala’Aldeen, D. Type V Protein Secretion Pathway: The Autotransporter Story. Microbiol. Mol. Biol. Rev. 2004, 68, 692–744. [Google Scholar] [CrossRef] [Green Version]
- Lien, Y.-W.; Lai, E.-M. Type VI Secretion Effectors: Methodologies and Biology. Front. Cell. Infect. Microbiol. 2017, 7, 254. [Google Scholar] [CrossRef] [Green Version]
- Ho, B.T.; Dong, T.G.; Mekalanos, J.J. A View to a Kill: The Bacterial Type VI Secretion System. Cell Host Microbe 2014, 15, 9–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Garcia, F.; Ruiz-Perez, F.; Cataldi, Á.; Larzábal, M. Type VI Secretion System in Pathogenic Escherichia Coli: Structure, Role in Virulence, and Acquisition. Front. Microbiol. 2019, 10, 1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, Load, Fire: The Type VI Secretion System, a Bacterial Nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef]
- Wang, T.; Si, M.; Song, Y.; Zhu, W.; Gao, F.; Wang, Y.; Zhang, L.; Zhang, W.; Wei, G.; Luo, Z.-Q.; et al. Type VI Secretion System Transports Zn2+ to Combat Multiple Stresses and Host Immunity. PLoS Pathog. 2015, 11, e1005020. [Google Scholar] [CrossRef] [Green Version]
- Gorasia, D.G.; Veith, P.D.; Reynolds, E.C. The Type IX Secretion System: Advances in Structure, Function and Organisation. Microorganisms 2020, 8, 1173. [Google Scholar] [CrossRef]
- Lauber, F.; Deme, J.C.; Lea, S.M.; Berks, B.C. Type 9 Secretion System Structures Reveal a New Protein Transport Mechanism. Nature 2018, 564, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Koneru, L. Type IX Secretion System: Characterization of an Effector Protein and an Insight into the Role of c-Terminal Domain Dimeration in Outer Membrane Translocation; University of Louisville: Louisville, KY, USA, 2017. [Google Scholar]
- Cascales, E.; Christie, P.J. The Versatile Bacterial Type IV Secretion Systems. Nat. Rev. Microbiol. 2003, 1, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgro, G.G.; Oka, G.U.; Souza, D.P.; Cenens, W.; Bayer-Santos, E.; Matsuyama, B.Y.; Bueno, N.F.; dos Santos, T.R.; Alvarez-Martinez, C.E.; Salinas, R.K.; et al. Bacteria-Killing Type IV Secretion Systems. Front. Microbiol. 2019, 10, 1078. [Google Scholar] [CrossRef]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.S.; Salinas, R.K.; et al. Bacterial Killing via a Type IV Secretion System. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef] [Green Version]
- Kubori, T.; Koike, M.; Bui, X.T.; Higaki, S.; Aizawa, S.-I.; Nagai, H. Native Structure of a Type IV Secretion System Core Complex Essential for Legionella Pathogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11804–11809. [Google Scholar] [CrossRef] [Green Version]
- Houben, E.N.G.; Korotkov, K.V.; Bitter, W. Take Five—Type VII Secretion Systems of Mycobacteria. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 1707–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, B.L.; Tak, U.; Mendonça, J.C.; Nagao, P.E.; Niederweis, M.; Doran, K.S. A Type VII Secretion System in Group B Streptococcus Mediates Cytotoxicity and Virulence. PLoS Pathog. 2021, 17, e1010121. [Google Scholar] [CrossRef] [PubMed]
- Kengmo Tchoupa, A.; Watkins, K.E.; Jones, R.A.; Kuroki, A.; Alam, M.T.; Perrier, S.; Chen, Y.; Unnikrishnan, M. The Type VII Secretion System Protects Staphylococcus Aureus against Antimicrobial Host Fatty Acids. Sci. Rep. 2020, 10, 14838. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef]
- Reading, N.C.; Sperandio, V. Quorum Sensing: The Many Languages of Bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. QUORUM SENSING: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Nunan, L.; Lightner, D.; Pantoja, C.; Gomez-Jimenez, S. Detection of Acute Hepatopancreatic Necrosis Disease (AHPND) in Mexico. Dis. Aquat. Org. 2014, 111, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Tran, P.T.N.; Kumar, V.; Bossier, P. Do Acute Hepatopancreatic Necrosis Disease-Causing PirABVP Toxins Aggravate Vibriosis? Emerg. Microbes Infect. 2020, 9, 1919–1932. [Google Scholar] [CrossRef]
- Lin, S.-J.; Huang, J.-Y.; Le, P.-T.; Lee, C.-T.; Chang, C.-C.; Yang, Y.-Y.; Su, E.C.-Y.; Lo, C.-F.; Wang, H.-C. Expression of the AHPND Toxins PirAvp and PirBvp Is Regulated by Components of the Vibrio Parahaemolyticus Quorum Sensing (QS) System. Int. J. Mol. Sci. 2022, 23, 2889. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B. Quorum-Sensing Signal-Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Winans, S.C.; Glick, B.R.; Charles, T.C. Identification and Characterization of New LuxR/LuxI-Type Quorum Sensing Systems from Metagenomic Libraries. Environ. Microbiol. 2010, 12, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, G.; Zhu, Y.; Bi, J.; Hao, H.; Hou, H. Effect of the LuxI/R Gene on AHL-Signaling Molecules and QS Regulatory Mechanism in Hafnia Alvei H4. AMB Express 2019, 9, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariminik, A.; Baseri-Salehi, M.; Kheirkhah, B. Pseudomonas Aeruginosa Quorum Sensing Modulates Immune Responses: An Updated Review Article. Immunol. Lett. 2017, 190, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van de Wiele, C.; De Spiegeleer, B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In Vitro and In Vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef] [Green Version]
- Sturme, M.H.J.; Kleerebezem, M.; Nakayama, J.; Akkermans, A.D.L.; Vaughan, E.E.; de Vos, W.M. Cell to Cell Communication by Autoinducing Peptides in Gram-Positive Bacteria. Antonie Leeuwenhoek 2002, 81, 233–243. [Google Scholar] [CrossRef]
- Monnet, V.; Gardan, R. Quorum-Sensing Regulators in Gram-Positive Bacteria: ‘Cherchez Le Peptide’. Mol. Microbiol. 2015, 97, 181–184. [Google Scholar] [CrossRef]
- Perez-Pascual, D.; Monnet, V.; Gardan, R. Bacterial Cell–Cell Communication in the Host via RRNPP Peptide-Binding Regulators. Front. Microbiol. 2016, 7, 706. [Google Scholar] [CrossRef] [Green Version]
- Junges, R.; Salvadori, G.; Shekhar, S.; Åmdal, H.A.; Periselneris, J.N.; Chen, T.; Brown, J.S.; Petersen, F.C. A Quorum-Sensing System That Regulates Streptococcus Pneumoniae Biofilm Formation and Surface Polysaccharide Production. mSphere 2017, 2, e00324-17. [Google Scholar] [CrossRef] [Green Version]
- Preda, V.G.; Săndulescu, O. Communication Is the Key: Biofilms, Quorum Sensing, Formation and Prevention. Discoveries 2019, 7, e100. [Google Scholar] [CrossRef]
- Tremblay, Y.D.; Lévesque, C.; Segers, R.P.; Jacques, M. Method to Grow Actinobacillus Pleuropneumoniaebiofilm on a Biotic Surface. BMC Vet. Res. 2013, 9, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The Formation of Biofilms by Pseudomonas Aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms. BioMed Res. Int. 2015, 2015, e759348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm Formation Mechanisms and Targets for Developing Antibiofilm Agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Loo, C.Y.; Corliss, D.A.; Ganeshkumar, N. Streptococcus Gordonii Biofilm Formation: Identification of Genes That Code for Biofilm Phenotypes. J. Bacteriol. 2000, 182, 1374–1382. [Google Scholar] [CrossRef] [Green Version]
- Chopp, D.L.; Kirisits, M.J.; Moran, B.; Parsek, M.R. A Mathematical Model of Quorum Sensing in a Growing Bacterial Biofilm. J. Ind. Microbiol. Biotechnol. 2002, 29, 339–346. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- Zhang, L.; Li, S.; Liu, X.; Wang, Z.; Jiang, M.; Wang, R.; Xie, L.; Liu, Q.; Xie, X.; Shang, D.; et al. Sensing of Autoinducer-2 by Functionally Distinct Receptors in Prokaryotes. Nat. Commun. 2020, 11, 5371. [Google Scholar] [CrossRef]
- Hammer, B.K.; Bassler, B.L. Quorum Sensing Controls Biofilm Formation in Vibrio Cholerae. Mol. Microbiol. 2003, 50, 101–104. [Google Scholar] [CrossRef]
- Ng, W.-L.; Perez, L.; Wei, Y.; Kraml, C.; Semmelhack, M.; Bassler, B. Signal Production and Detection Specificity in Vibrio CqsA/CqsS Quorum-Sensing Systems. Mol. Microbiol. 2011, 79, 1407–1417. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. The Vibrio Harveyi Quorum-Sensing System Uses Shared Regulatory Components to Discriminate between Multiple Autoinducers. Genes Dev. 2006, 20, 2754–2767. [Google Scholar] [CrossRef] [Green Version]
- Coquant, G.; Grill, J.-P.; Seksik, P. Impact of N-Acyl-Homoserine Lactones, Quorum Sensing Molecules, on Gut Immunity. Front. Immunol. 2020, 11, 1827. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Schaefer, A.L.; Coutinho, B.G.; Brown, P.J.B.; Greenberg, E.P. An Aryl-Homoserine Lactone Quorum-Sensing Signal Produced by a Dimorphic Prosthecate Bacterium. Proc. Natl. Acad. Sci. USA 2018, 115, 7587–7592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Lin, J.; Harrington, A.; Cornilescu, G.; Lau, G.W.; Tal-Gan, Y. Designing Cyclic Competence-Stimulating Peptide (CSP) Analogs with Pan-Group Quorum-Sensing Inhibition Activity in Streptococcus Pneumoniae. Proc. Natl. Acad. Sci. USA 2020, 117, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Leung, V.; Dufour, D.; Levesque, C. Death and Survival in Streptococcus Mutans: Differing Outcomes of a Quorum-Sensing Signalling Peptide. Front. Microbiol. 2015, 6, 1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikash, C.R.; Tal-Gan, Y. Structure Activity Relationship Study of the XIP Quorum Sensing Pheromone in Streptococcus Mutans Reveal Inhibitors of the Competence Regulon. ACS Chem. Biol. 2020, 15, 2833–2841. [Google Scholar] [CrossRef]
- Kalamara, M.; Spacapan, M.; Mandic-Mulec, I.; Stanley-Wall, N.R. Social Behaviours by Bacillus Subtilis: Quorum Sensing, Kin Discrimination and Beyond. Mol. Microbiol. 2018, 110, 863–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Špacapan, M.; Danevčič, T.; Štefanic, P.; Porter, M.; Stanley-Wall, N.R.; Mandic-Mulec, I. The ComX Quorum Sensing Peptide of Bacillus Subtilis Affects Biofilm Formation Negatively and Sporulation Positively. Microorganisms 2020, 8, 1131. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular Mechanisms of Membrane Targeting Antibiotics. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of Lipids in the Interaction of Antimicrobial Peptides with Membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Chi, X.; Fan, Q.; Zhang, Y.; Liang, K.; Wan, L.; Zhou, Q.; Li, Y. Structural Mechanism of Phospholipids Translocation by MlaFEDB Complex. Cell Res. 2020, 30, 1127–1135. [Google Scholar] [CrossRef]
- Epand, R.M.; D’Souza, K.; Berno, B.; Schlame, M. Membrane Curvature Modulation of Protein Activity Determined by NMR. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thennarasu, S.; Huang, R.; Lee, D.-K.; Yang, P.; Maloy, L.; Chen, Z.; Ramamoorthy, A. Limiting an Antimicrobial Peptide to the Lipid-Water Interface Enhances Its Bacterial Membrane Selectivity—A Case Study on MSI-367. Biochemistry 2010, 49, 10595–10605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killian, J.A.; Salemink, I.; de Planque, M.R.R.; Lindblom, G.; Koeppe, R.E.; Greathouse, D.V. Induction of Nonbilayer Structures in Diacylphosphatidylcholine Model Membranes by Transmembrane α-Helical Peptides: Importance of Hydrophobic Mismatch and Proposed Role of Tryptophans. Biochemistry 1996, 35, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Látrová, K.; Havlová, N.; Večeřová, R.; Pinkas, D.; Bogdanová, K.; Kolář, M.; Fišer, R.; Konopásek, I.; Do Pham, D.D.; Rejman, D.; et al. Outer Membrane and Phospholipid Composition of the Target Membrane Affect the Antimicrobial Potential of First- and Second-Generation Lipophosphonoxins. Sci. Rep. 2021, 11, 10446. [Google Scholar] [CrossRef] [PubMed]
- Boto, A.; Pérez de la Lastra, J.M.; González, C.C. The Road from Host-Defense Peptides to a New Generation of Antimicrobial Drugs. Molecules 2018, 23, 311. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an Updated Data Repository of Antimicrobial Peptides. Sci. Data 2019, 6, 148. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial Peptides: Promising Alternatives in the Postfeeding Antibiotic Era. Med. Res. Rev. 2018, 39, 831–859. [Google Scholar] [CrossRef]
- Joo, H.-S.; Fu, C.-I.; Otto, M. Bacterial Strategies of Resistance to Antimicrobial Peptides. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Fleury, Y.; Dayem, M.A.; Montagne, J.J.; Chaboisseau, E.; Caer, J.P.L.; Nicolas, P.; Delfour, A. Covalent Structure, Synthesis, and Structure-Function Studies of Mesentericin Y 10537, a Defensive Peptide from Gram-Positive Bacteria Leuconostoc Mesenteroides. J. Biol. Chem. 1996, 271, 14421–14429. [Google Scholar] [CrossRef] [Green Version]
- Paiva, A.D.; Breukink, E.; Mantovani, H.C. Role of Lipid II and Membrane Thickness in the Mechanism of Action of the Lantibiotic Bovicin HC5. Antimicrob. Agents Chemother. 2011, 55, 5284–5293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh Moghaddam, M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski, L.; Silva-Pereira, I.; Kyaw, C. Antibiotic Development Challenges: The Various Mechanisms of Action of Antimicrobial Peptides and of Bacterial Resistance. Front. Microbiol. 2013, 4, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Rozek, A.; Hancock, R.E.W. Interaction of Cationic Antimicrobial Peptides with Model Membranes. J. Biol. Chem. 2001, 276, 35714–35722. [Google Scholar] [CrossRef] [Green Version]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Ehrenstein, G.; Lecar, H. Electrically Gated Ionic Channels in Lipid Bilayers. Q. Rev. Biophys. 1977, 10, 1–34. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [Green Version]
- Lipkin, R.B.; Lazaridis, T. Implicit Membrane Investigation of the Stability of Antimicrobial Peptide β-Barrels and Arcs. J. Membr. Biol. 2015, 248, 469–486. [Google Scholar] [CrossRef] [Green Version]
- Rapaport, D.; Shai, Y. Interaction of Fluorescently Labeled Pardaxin and Its Analogues with Lipid Bilayers. J. Biol. Chem. 1991, 266, 23769–23775. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, D.; Leontiadou, H.; Mark, A.E.; Marrink, S.-J. Toroidal Pores Formed by Antimicrobial Peptides Show Significant Disorder. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 2308–2317. [Google Scholar] [CrossRef] [Green Version]
- Sitaram, N.; Nagaraj, R. Interaction of Antimicrobial Peptides with Biological and Model Membranes: Structural and Charge Requirements for Activity. Biochim. Biophys. Acta (BBA)-Biomembr. 1999, 1462, 29–54. [Google Scholar] [CrossRef] [Green Version]
- Rozek, A.; Friedrich, C.L.; Hancock, R.E.W. Structure of the Bovine Antimicrobial Peptide Indolicidin Bound to Dodecylphosphocholine and Sodium Dodecyl Sulfate Micelles. Biochemistry 2000, 39, 15765–15774. [Google Scholar] [CrossRef]
- Shai, Y. Mode of Action of Membrane Active Antimicrobial Peptides. Pept. Sci. 2002, 66, 236–248. [Google Scholar] [CrossRef]
- Fernandez, D.I.; Brun, A.P.L.; Whitwell, T.C.; Sani, M.-A.; James, M.; Separovic, F. The Antimicrobial Peptide Aurein 1.2 Disrupts Model Membranes via the Carpet Mechanism. Phys. Chem. Chem. Phys. 2012, 14, 15739–15751. [Google Scholar] [CrossRef]
- Welch, N.G.; Li, W.; Hossain, M.A.; Separovic, F.; O’Brien-Simpson, N.M.; Wade, J.D. (Re)Defining the Proline-Rich Antimicrobial Peptide Family and the Identification of Putative New Members. Front. Chem. 2020, 8, 607769. [Google Scholar] [CrossRef]
- Shamova, O.; Brogden, K.A.; Zhao, C.; Nguyen, T.; Kokryakov, V.N.; Lehrer, R.I. Purification and Properties of Proline-Rich Antimicrobial Peptides from Sheep and Goat Leukocytes. Infect. Immun. 1999, 67, 4106–4111. [Google Scholar] [CrossRef] [Green Version]
- Cytryńska, M.; Rahnamaeian, M.; Zdybicka-Barabas, A.; Dobslaff, K.; Züchner, T.; Sacheau, G.; Innis, C.A.; Vilcinskas, A. Proline-Rich Antimicrobial Peptides in Medicinal Maggots of Lucilia Sericata Interact with Bacterial DnaK But Do Not Inhibit Protein Synthesis. Front. Pharmacol. 2020, 11, 532. [Google Scholar] [CrossRef]
- Yang, C.-H.; Chen, Y.-C.; Peng, S.-Y.; Tsai, A.P.-Y.; Lee, T.J.-F.; Yen, J.-H.; Liou, J.-W. An Engineered Arginine-Rich α-Helical Antimicrobial Peptide Exhibits Broad-Spectrum Bactericidal Activity against Pathogenic Bacteria and Reduces Bacterial Infections in Mice. Sci. Rep. 2018, 8, 14602. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.; Jowitt, T.A.; Harris, L.K.; Knight, C.G.; Dobson, C.B. The Lexicon of Antimicrobial Peptides: A Complete Set of Arginine and Tryptophan Sequences. Commun. Biol. 2021, 4, 605. [Google Scholar] [CrossRef]
- Kichler, A.; Leborgne, C.; März, J.; Danos, O.; Bechinger, B. Histidine-Rich Amphipathic Peptide Antibiotics Promote Efficient Delivery of DNA into Mammalian Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 1564–1568. [Google Scholar] [CrossRef] [Green Version]
- Kacprzyk, L.; Rydengård, V.; Mörgelin, M.; Davoudi, M.; Pasupuleti, M.; Malmsten, M.; Schmidtchen, A. Antimicrobial Activity of Histidine-Rich Peptides Is Dependent on Acidic Conditions. Biochim. Biophys. Acta (BBA)-Biomembr. 2007, 1768, 2667–2680. [Google Scholar] [CrossRef] [Green Version]
- de Jesus Oliveira, T.; de Oliveira, U.C.; da Silva Junior, P.I. Serrulin: A Glycine-Rich Bioactive Peptide from the Hemolymph of the Yellow Tityus Serrulatus Scorpion. Toxins 2019, 11, 517. [Google Scholar] [CrossRef] [Green Version]
- Verdon, J.; Coutos-Thevenot, P.; Rodier, M.-H.; Landon, C.; Depayras, S.; Noel, C.; La Camera, S.; Moumen, B.; Greve, P.; Bouchon, D.; et al. Armadillidin H, a Glycine-Rich Peptide from the Terrestrial Crustacean Armadillidium Vulgare, Displays an Unexpected Wide Antimicrobial Spectrum with Membranolytic Activity. Front. Microbiol. 2016, 7, 1484. [Google Scholar] [CrossRef] [Green Version]
- D’Este, F.; Benincasa, M.; Cannone, G.; Furlan, M.; Scarsini, M.; Volpatti, D.; Gennaro, R.; Tossi, A.; Skerlavaj, B.; Scocchi, M. Antimicrobial and Host Cell-Directed Activities of Gly/Ser-Rich Peptides from Salmonid Cathelicidins. Fish Shellfish. Immunol. 2016, 59, 456–468. [Google Scholar] [CrossRef]
- Vitali, A. Proline-Rich Peptides: Multifunctional Bioactive Molecules as New Potential Therapeutic Drugs. Curr. Protein Pept. Sci. 2015, 16, 147–162. [Google Scholar] [CrossRef]
- Fang, W.-Y.; Dahiya, R.; Qin, H.-L.; Mourya, R.; Maharaj, S. Natural Proline-Rich Cyclopolypeptides from Marine Organisms: Chemistry, Synthetic Methodologies and Biological Status. Mar. Drugs 2016, 14, 194. [Google Scholar] [CrossRef] [Green Version]
- Mardirossian, M.; Sola, R.; Beckert, B.; Collis, D.; Stasi, A.; Armas, F.; Hilpert, K.; Wilson, D.; Scocchi, M. Proline-Rich Peptides with Improved Antimicrobial Activity against E. Coli, K. Pneumoniae, and A. Baumannii. ChemMedChem 2019, 14, 2025–2033. [Google Scholar] [CrossRef] [Green Version]
- Rosengren, K.J.; Göransson, U.; Otvos Jr., L.; Craik, D.J. Cyclization of Pyrrhocoricin Retains Structural Elements Crucial for the Antimicrobial Activity of the Native Peptide. Pept. Sci. 2004, 76, 446–458. [Google Scholar] [CrossRef]
- Lai, P.-K.; Tresnak, D.T.; Hackel, B.J. Identification and Elucidation of Proline-Rich Antimicrobial Peptides with Enhanced Potency and Delivery. Biotechnol. Bioeng. 2019, 116, 2439–2450. [Google Scholar] [CrossRef]
- Holfeld, L.; Knappe, D.; Hoffmann, R. Proline-Rich Antimicrobial Peptides Show a Long-Lasting Post-Antibiotic Effect on Enterobacteriaceae and Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2018, 73, 933–941. [Google Scholar] [CrossRef]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 1184–1202. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Andina, D.; Nahar, S.; Leroux, J.-C.; Gauthier, M.A. Releasable and Traceless PEGylation of Arginine-Rich Antimicrobial Peptides. Chem. Sci. 2017, 8, 4082–4086. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Huang, Y.; Zheng, H.; Tang, L.; He, J.; Xiang, L.; Liu, D.; Jiang, H. Secretion and Activity of Antimicrobial Peptide Cecropin D Expressed in Pichia Pastoris. Exp. Ther. Med. 2012, 4, 1063–1068. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Lointier, M.; Dussouillez, C.; Glattard, E.; Kichler, A.; Bechinger, B. Different Biological Activities of Histidine-Rich Peptides Are Favored by Variations in Their Design. Toxins 2021, 13, 363. [Google Scholar] [CrossRef]
- Ferrer-Miralles, N.; Corchero, J.L.; Kumar, P.; Cedano, J.A.; Gupta, K.C.; Villaverde, A.; Vazquez, E. Biological Activities of Histidine-Rich Peptides; Merging Biotechnology and Nanomedicine. Microb. Cell Factories 2011, 10, 101. [Google Scholar] [CrossRef] [Green Version]
- Mason, A.J.; Moussaoui, W.; Abdelrahman, T.; Boukhari, A.; Bertani, P.; Marquette, A.; Shooshtarizaheh, P.; Moulay, G.; Boehm, N.; Guerold, B.; et al. Structural Determinants of Antimicrobial and Antiplasmodial Activity and Selectivity in Histidine-Rich Amphipathic Cationic Peptides. J. Biol. Chem. 2009, 284, 119–133. [Google Scholar] [CrossRef] [Green Version]
- Georgescu, J.; Munhoz, V.H.O.; Bechinger, B. NMR Structures of the Histidine-Rich Peptide LAH4 in Micellar Environments: Membrane Insertion, PH-Dependent Mode of Antimicrobial Action, and DNA Transfection. Biophys. J. 2010, 99, 2507–2515. [Google Scholar] [CrossRef] [Green Version]
- Baumann, T.; Kämpfer, U.; Schürch, S.; Schaller, J.; Largiadèr, C.; Nentwig, W.; Kuhn-Nentwig, L. Ctenidins: Antimicrobial Glycine-Rich Peptides from the Hemocytes of the Spider Cupiennius Salei. Cell Mol. Life Sci. 2010, 67, 2787–2798. [Google Scholar] [CrossRef] [Green Version]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of Antimicrobial Peptides. A Review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef] [Green Version]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A Promising Class of Insect Antimicrobial Peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and Consequences of Bacterial Resistance to Antimicrobial Peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Sieprawska-Lupa, M.; Mydel, P.; Krawczyk, K.; Wójcik, K.; Puklo, M.; Lupa, B.; Suder, P.; Silberring, J.; Reed, M.; Pohl, J.; et al. Degradation of Human Antimicrobial Peptide LL-37 by Staphylococcus Aureus-Derived Proteinases. Antimicrob. Agents Chemother. 2004, 48, 4673–4679. [Google Scholar] [CrossRef] [Green Version]
- Teufel, P.; Götz, F. Characterization of an Extracellular Metalloprotease with Elastase Activity from Staphylococcus Epidermidis. J. Bacteriol. 1993, 175, 4218–4224. [Google Scholar] [CrossRef] [Green Version]
- Schmidtchen, A.; Frick, I.-M.; Andersson, E.; Tapper, H.; Björck, L. Proteinases of Common Pathogenic Bacteria Degrade and Inactivate the Antibacterial Peptide LL-37. Mol. Microbiol. 2002, 46, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Frick, I.-M.; Nordin, S.L.; Baumgarten, M.; Mörgelin, M.; Sørensen, O.E.; Olin, A.I.; Egesten, A. Constitutive and Inflammation-Dependent Antimicrobial Peptides Produced by Epithelium Are Differentially Processed and Inactivated by the Commensal Finegoldia magna and the Pathogen Streptococcus pyogenes. J. Immunol. 2011, 187, 4300–4309. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.C.; Garbe, J.; Collin, M. Cysteine Proteinase SpeB from Streptococcus Pyogenes—A Potent Modifier of Immunologically Important Host and Bacterial Proteins. Gruyter 2011, 392, 1077–1088. [Google Scholar] [CrossRef]
- Barańska-Rybak, W.; Sonesson, A.; Nowicki, R.; Schmidtchen, A. Glycosaminoglycans Inhibit the Antibacterial Activity of LL-37 in Biological Fluids. J. Antimicrob. Chemother. 2006, 57, 260–265. [Google Scholar] [CrossRef]
- Stumpe, S.; Schmid, R.; Stephens, D.L.; Georgiou, G.; Bakker, E.P. Identification of OmpT as the Protease That Hydrolyzes the Antimicrobial Peptide Protamine before It Enters Growing Cells OfEscherichia Coli. J. Bacteriol. 1998, 180, 4002–4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guina, T.; Yi, E.C.; Wang, H.; Hackett, M.; Miller, S.I. A PhoP-Regulated Outer Membrane Protease of Salmonella Enterica Serovar Typhimurium Promotes Resistance to Alpha-Helical Antimicrobial Peptides. J. Bacteriol. 2000, 182, 4077–4086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galván, E.M.; Lasaro, M.A.S.; Schifferli, D.M. Capsular Antigen Fraction 1 and Pla Modulate the Susceptibility of Yersinia Pestis to Pulmonary Antimicrobial Peptides Such as Cathelicidin. Infect. Immun. 2008, 76, 1456–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Preston, J.F.; Romeo, T. The PgaABCD Locus of Escherichia Coli Promotes the Synthesis of a Polysaccharide Adhesin Required for Biofilm Formation. J. Bacteriol. 2004, 186, 2724–2734. [Google Scholar] [CrossRef] [Green Version]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide Intercellular Adhesin (PIA) Protects Staphylococcus Epidermidis against Major Components of the Human Innate Immune System. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef]
- Høiby, N. Pseudomonas aeruginosa infection in cystic fibrosis. Relationship between Mucoid Strains of Pseudomonas Aeruginosa and the Humoral Immune Response. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 1974, 82B, 551–558. [Google Scholar] [CrossRef]
- Evans, L.R.; Linker, A. Production and Characterization of the Slime Polysaccharide of Pseudomonas Aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate Overproduction Affects Pseudomonas Aeruginosa Biofilm Structure and Function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef] [Green Version]
- Foschiatti, M.; Cescutti, P.; Tossi, A.; Rizzo, R. Inhibition of Cathelicidin Activity by Bacterial Exopolysaccharides. Mol. Microbiol. 2009, 72, 1137–1146. [Google Scholar] [CrossRef]
- Herasimenka, Y.; Benincasa, M.; Mattiuzzo, M.; Cescutti, P.; Gennaro, R.; Rizzo, R. Interaction of Antimicrobial Peptides with Bacterial Polysaccharides from Lung Pathogens. Peptides 2005, 26, 1127–1132. [Google Scholar] [CrossRef]
- Bera, A.; Biswas, R.; Herbert, S.; Kulauzovic, E.; Weidenmaier, C.; Peschel, A.; Götz, F. Influence of Wall Teichoic Acid on Lysozyme Resistance in Staphylococcus Aureus. J. Bacteriol. 2007, 189, 280–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, N.; Araki, Y.; Ito, E. Structure of the Linkage Units between Ribitol Teichoic Acids and Peptidoglycan. J. Bacteriol. 1985, 161, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Götz, F. Inactivation of the Dlt Operon InStaphylococcus Aureus Confers Sensitivity to Defensins, Protegrins, and Other Antimicrobial Peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahne, D.; Leimkuhler, C.; Lu, W.; Walsh, C. Glycopeptide and Lipoglycopeptide Antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Wright, G.D.; Dutka-Malen, S.; Arthur, M.; Courvalin, P.; Walsh, C.T. Molecular Basis for Vancomycin Resistance in Enterococcus Faecium BM4147: Biosynthesis of a Depsipeptide Peptidoglycan Precursor by Vancomycin Resistance Proteins VanH and VanA. ACS Publ. 2002, 30, 10408–10415. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkatheri, A.H.; Yap, P.S.-X.; Abushelaibi, A.; Lai, K.-S.; Cheng, W.-H.; Lim, S.-H.E. Host–Bacterial Interactions: Outcomes of Antimicrobial Peptide Applications. Membranes 2022, 12, 715. https://doi.org/10.3390/membranes12070715
Alkatheri AH, Yap PS-X, Abushelaibi A, Lai K-S, Cheng W-H, Lim S-HE. Host–Bacterial Interactions: Outcomes of Antimicrobial Peptide Applications. Membranes. 2022; 12(7):715. https://doi.org/10.3390/membranes12070715
Chicago/Turabian StyleAlkatheri, Asma Hussain, Polly Soo-Xi Yap, Aisha Abushelaibi, Kok-Song Lai, Wan-Hee Cheng, and Swee-Hua Erin Lim. 2022. "Host–Bacterial Interactions: Outcomes of Antimicrobial Peptide Applications" Membranes 12, no. 7: 715. https://doi.org/10.3390/membranes12070715
APA StyleAlkatheri, A. H., Yap, P. S.-X., Abushelaibi, A., Lai, K.-S., Cheng, W.-H., & Lim, S.-H. E. (2022). Host–Bacterial Interactions: Outcomes of Antimicrobial Peptide Applications. Membranes, 12(7), 715. https://doi.org/10.3390/membranes12070715






