Disulfiram Oxy-Derivatives Suppress Protein Retrotranslocation across the ER Membrane to the Cytosol and Initiate Paraptosis-like Cell Death
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cell Culture and Treatment
2.3. Immunoblotting
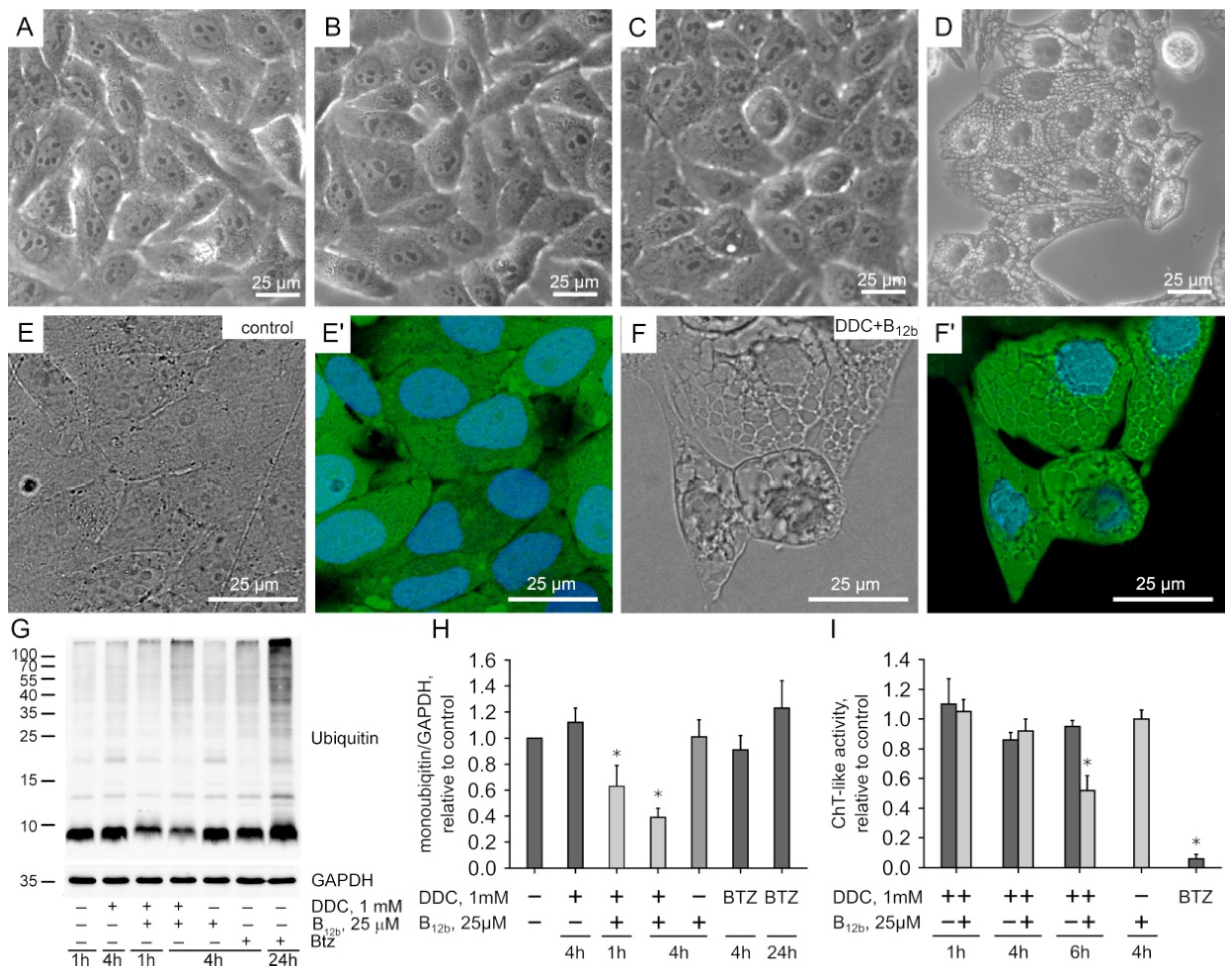
2.4. Analysis of the Chymotrypsin-like Activity of Proteasome
2.5. CFSE Staining
2.6. Sample Preparation for Mass Spectrometry
2.7. Mass Spectrometry and Liquid Chromatography
2.8. Bioinformatics Analysis of Ubiquitome
2.9. Statistical Analysis
3. Results
3.1. Effect of Exogenous DSFoxy on Protein Ubiquitination
3.2. Functional Analysis of Ubiquitome in PANTHER
3.3. Individual Analysis of Ub-Proteins in Groups Formed According to the Classification of UniProtKB
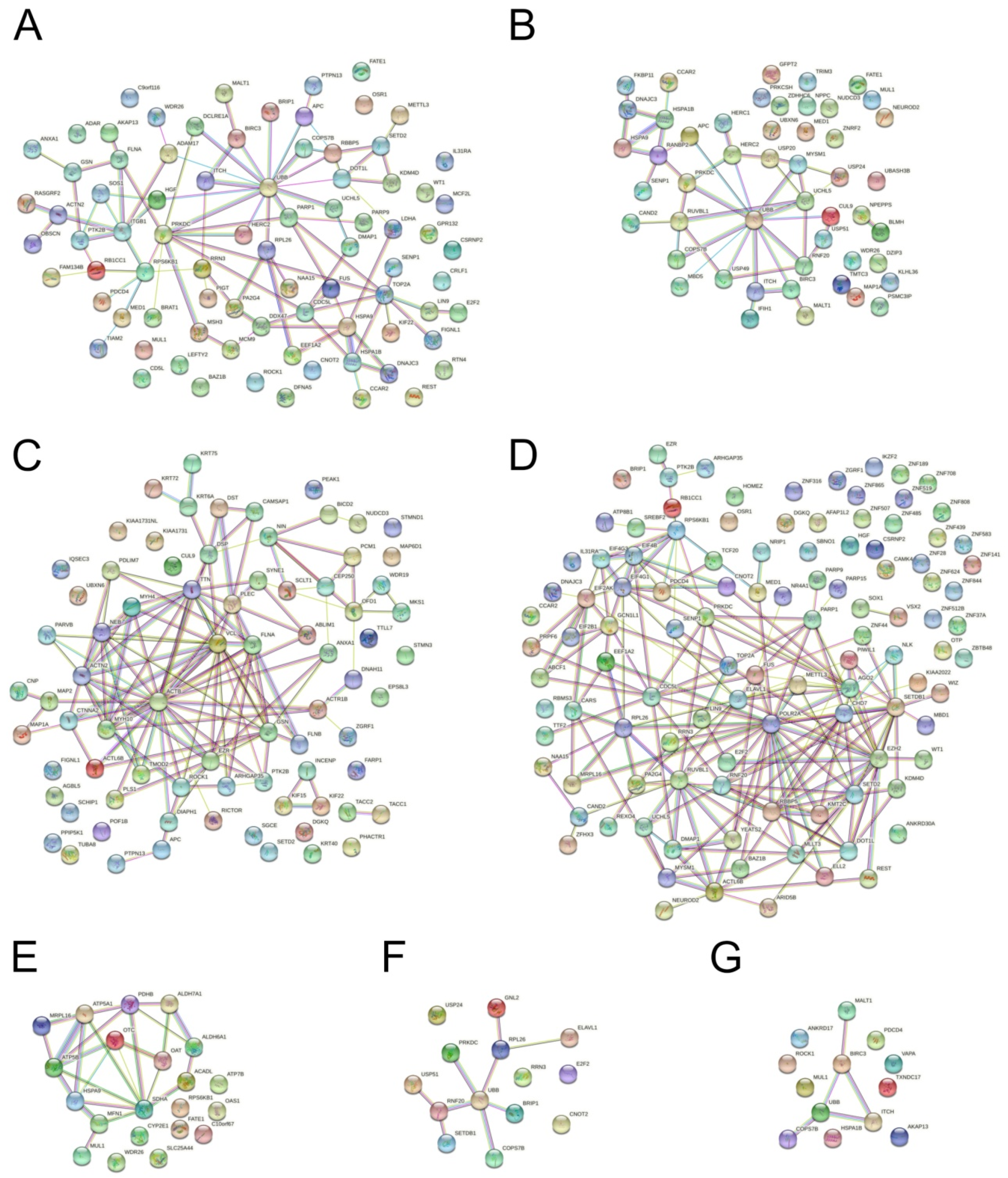
3.4. Protein Interaction Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burger, A.; Amemiya, Y.; Kitching, R.; Seth, A.K. Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia 2006, 8, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Jaynes, P.W.; Saei, A.; Iyengar, P.V.; Richard, J.L.C.; Eichhorn, P.J.A. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 456–483. [Google Scholar] [CrossRef] [PubMed]
- Erl, W.; Weber, C.; Hansson, G.K. Pyrrolidine dithiocarbamate-induced apoptosis depends on cell type, density, and the presence of Cu2+ and Zn2+. Am. J. Physiol. Cell Physiol. 2000, 278, C1116–C1125. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.; Matsukawa, E.; Miura, A.; Shouji, A.; Asou, K.; Ishikawa, M. Diethyldithiocarbamate-induced cytotoxicity and apoptosis in leukemia cell lines. Biol. Pharm. Bull. 2003, 26, 964–968. [Google Scholar] [CrossRef]
- Kona, F.R.; Buac, D.; Burger, A.M. Disulfiram, and disulfiram derivatives as novel potential anticancer drugs targeting the ubiquitin-proteasome system in both preclinical and clinical studies. Curr. Cancer Drug Targets 2011, 11, 338–346. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.S.; Xi, Y.; Miller, J.R.; Brownell, A.L.; Zeng, Q.; Yoo, G.H.; Garshott, D.M.; O’Brien, M.B.; Galinato, A.E.; Cai, P.; et al. Disulfiram (antabuse) activates ROS-dependent ER stress and apoptosis in oral cavity squamous cell carcinoma. J. Clin. Med. 2019, 8, 611. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xue, X.; Wang, L.; Wang, W.; Han, J.; Sun, X.; Zhang, H.; Liu, Y.; Che, X.; Yang, J.; et al. Suppressing autophagy enhances disulfiram/copper-induced apoptosis in non-small cell lung cancer. Eur. J. Pharmacol. 2018, 827, 1–12. [Google Scholar] [CrossRef]
- Mays, D.C.; Nelson, A.N.; Fauq, A.H.; Shriver, Z.H.; Veverka, K.A.; Naylor, S.; Lipsky, J.J. S-Methyl N,N-diethylthiocarbamate sulfone, a potential metabolite of disulfiram and potent inhibitor of low Km mitochondrial aldehyde dehydrogenase. Biochem. Pharmacol. 1995, 49, 693–700. [Google Scholar] [CrossRef]
- Nobel, C.S.; Kimland, M.; Nicholson, D.W.; Orrenius, S.; Slater, A.F. Disulfiram is a potent inhibitor of proteases of the caspase family. Chem. Res. Toxicol. 1997, 10, 1319–1324. [Google Scholar] [CrossRef]
- Solovieva, M.E.; Shatalin, Y.V.; Solovyev, V.V.; Sazonov, A.V.; Kutyshenko, V.P.; Akatov, V.S. Hydroxycobalamin catalyzes the oxidation of diethyldithiocarbamate and increases its cytotoxicity independently of copper ions. Redox Biol. 2019, 20, 28–37. [Google Scholar] [CrossRef]
- Solovieva, M.; Shatalin, Y.; Fadeev, R.; Krestinina, O.; Baburina, Y.; Kruglov, A.; Kharechkina, E.; Kobyakova, M.; Rogachevsky, V.; Shishkova, E.; et al. Vitamin B12b enhances the cytotoxicity of diethyldithiocarbamate in a synergistic manner, inducing the paraptosis-like death of human larynx carcinoma cells. Biomolecules 2020, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Solovieva, M.; Shatalin, Y.; Odinokova, I.; Krestinina, O.; Baburina, Y.; Mishukov, A.; Lomovskaya, Y.; Pavlik, L.; Mikheeva, I.; Holmuhamedov, E.; et al. Disulfiram oxy-derivatives induce entosis or paraptosis-like death in breast cancer MCF-7 cells depending on the duration of treatment. Biochim. Biophys. Acta Gen. Subj. 2022, 1866, 130184. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa-West, N.; Dunlop, R.A.; Rodgers, K.J. Using an in vitro model to study oxidised protein accumulation in ageing fibroblasts. Biochim. Biophys. Acta 2015, 1850, 2177–2184. [Google Scholar] [CrossRef]
- Buneeva, O.; Kopylov, A.; Kapitsa, I.; Ivanova, E.; Zgoda, V.; Medvedev, A. The effect of neurotoxin MPTP and neuroprotector isatin on the profile of ubiquitinated brain mitochondrial proteins. Cells 2018, 7, 91. [Google Scholar] [CrossRef]
- Wis’niewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.N.; Perlman, D.H.; Costello, C.E.; McComb, M.E. Software tool for researching annotations of proteins: Open-source protein annotation software with data visualization. Anal. Chem. 2009, 81, 9819–9823. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-F.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Ninagawa, S.; George, G.; Mori, K. Mechanisms of productive folding and endoplasmic reticulum-associated degradation of glycoproteins and non-glycoproteins. Biochim. Biophys. Acta Gen. Subj. 2021, 1865, 129812. [Google Scholar] [CrossRef]
- de Bie, P.; Ciechanover, A. Ubiquitination of E3 ligases: Self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011, 1, 1393–1402. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, A.L.; Ciechanover, A. Targeting Proteins for Destruction by the Ubiquitin System: Implications for Human Pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 73–96. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Hahn, A.A.; Hu, S.; Yang, X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 2011, 286, 35380–35387. [Google Scholar] [CrossRef]
- Wei, Y.; Xu, X. UFMylation: A Unique & Fashionable Modification for Life. Genom. Proteom. Bioinf. 2016, 14, 140–146. [Google Scholar] [CrossRef]
- Pratt-Hyatt, M.; Lin, H.-L.; Hollenberg, P.F. Mechanism-Based Inactivation of Human CYP2E1 by Diethyldithocarbamate. Drug Metab. Dispos. 2010, 38, 2286–2292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guan, S.; Acharya, P.; Koop, D.R.; Liu, Y.; Liao, M.; Burlingame, A.L.; Correia, M.A. Ubiquitin-dependent proteasomal degradation of human liver cytochrome P450 2E1: Identification of sites targeted for phosphorylation and ubiquitination. J. Biol. Chem. 2011, 286, 9443–9456. [Google Scholar] [CrossRef]
- Müller, E.; Salcan, S.; Bongardt, S.; Barbosa, D.M.; Krüger, M.; Kötter, S. E3-ligase knock down revealed differential titin degradation by autopagy and the ubiquitin proteasome system. Sci. Rep. 2021, 11, 21134. [Google Scholar] [CrossRef]
- Li, J.; Li, P.F.; Dietz, R.; von Harsdorf, R. Intracellular superoxide induces apoptosis in VSMCs: Role of mitochondrial membrane potential, cytochrome C and caspases. Apoptosis 2002, 7, 511–517. [Google Scholar] [CrossRef]
- Dumay, A.; Rincheval, V.; Trotot, P.; Mignotte, B.; Vayssière, J.L. The superoxide dismutase inhibitor diethyldithiocarbamate has antagonistic effects on apoptosis by triggering both cytochrome c release and caspase inhibition. Free Radic. Biol. Med. 2006, 40, 1377–1390. [Google Scholar] [CrossRef]
- Jivan, R.; Damelin, L.H.; Birkhead, M.; Rousseau, A.L.; Veale, R.B.; Mavri-Damelin, D. Disulfiram/copper-disulfiram damages multiple protein degradation and turnover pathways and cytotoxicity is enhanced by metformin in oesophageal squamous cell carcinoma cell lines. J. Cell. Biochem. 2015, 116, 2334–2343. [Google Scholar] [CrossRef]
- Brüning, A.; Kast, R.E. Oxidizing to death: Disulfiram for cancer cell killing. Cell Cycle 2014, 13, 1513–1514. [Google Scholar] [CrossRef]
- Wang, L.; Gundelach, J.; Bram, R. Cycloheximide promotes paraptosis induced by inhibition of cyclophilins in glioblastoma multiforme. Cell Death Dis. 2017, 8, e2807. [Google Scholar] [CrossRef] [PubMed]
- Gharibani, P.; Modi, J.; Menzie, J.; Alexandrescu, A.; Ma, Z.; Tao, R.; Prentice, H.; Wu, J.Y. Comparison between single and combined post-treatment with S-Methyl-N,N-diethylthiolcarbamate sulfoxide and taurine following transient focal cerebral ischemia in rat brain. Neuroscience 2015, 300, 460–473. [Google Scholar] [CrossRef]
- Sicari, D.; Centonze, F.G.; Pineau, R.; Le Reste, P.J.; Negroni, L.; Chat, S.; Mohtar, M.A.; Thomas, D.; Gillet, R.; Hupp, T.; et al. Reflux of Endoplasmic Reticulum proteins to the cytosol inactivates tumor suppressors. EMBO Rep. 2021, 22, e51412. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Reed, J.C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 2013, 1833, 3460–3470. [Google Scholar] [CrossRef]
- Foot, N.; Henshall, T.; Kumar, S. Ubiquitination and the Regulation of Membrane Proteins. Physiol. Rev. 2017, 97, 253–281. [Google Scholar] [CrossRef] [PubMed]
- Spang, A. Retrograde traffic from the Golgi to the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013391. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Bailly-Maitre, B.; Reed, J.C. Endoplasmic reticulum stress: Cell life and death decisions. J. Clin. Investig. 2005, 115, 2656–2664. [Google Scholar] [CrossRef]
- Brahemi, G.; Kona, F.R.; Fiasella, A.; Buac, D.; Soukupová, J.; Brancale, A.; Burger, A.M.; Westwell, A.D. Exploring the structural requirements for inhibition of the Ubiquitin E3 Ligase Breast Cancer Associated Protein 2 (BCA2) as a treatment for breast cancer. J. Med. Chem. 2010, 53, 2757–2765. [Google Scholar] [CrossRef]
- Wang, Z.; Nie, Z.; Chen, W.; Zhou, Z.; Kong, Q.; Seth, A.K.; Liu, R.; Chen, C. RNF115/BCA2 E3 ubiquitin ligase promotes breast cancer cell proliferation through targeting p21Waf1/Cip1 for ubiquitin-mediated degradation. Neoplasia 2013, 15, 1028–1035. [Google Scholar] [CrossRef]
- Carvalho, P.; Stanley, A.M.; Rapoport, T.A. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell 2010, 143, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Scialpi, F.; Malatesta, M.; Peschiaroli, A.; Rossi, M.; Melino, G.; Bernassola, F. Itch self-polyubiquitylation occurs through lysine-63 linkages. Biochem. Pharmacol. 2008, 76, 1515–1521. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, N.O.; Rapoport, T.A. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 2017, 169, 722–735.e9. [Google Scholar] [CrossRef]
- Tepedelen, B.E.; Kirmizibayrak, P.B. Endoplasmic Reticulum-Associated Degradation (ERAD). In Endoplasmic Reticulum; Català, A., Ed.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Han, J.J.W.; Ho, D.V.; Kim, H.M.; Lee, J.Y.; Jeon, Y.S.; Chan, J.Y. The deubiquitinating enzyme USP7 regulates the transcription factor Nrf1 by modulating its stability in response to toxic metal exposure. J. Biol. Chem. 2021, 296, 100732. [Google Scholar] [CrossRef]
- Hu, S.; Xiang, Y.; Qiu, L.; Wang, M.; Zhang, Y. Activation of the membrane-bound Nrf1 transcription factor by USP19, a ubiquitin-specific protease C-terminally anchored in the endoplasmic reticulum. Biochim. Biophys. Acta. Mol. Cell Res. 2022, 1869, 119299. [Google Scholar] [CrossRef]
- Peterson, B.G.; Glaser, M.L.; Rapoport, T.A.; Baldridge, R.D. Cycles of autoubiquitination and deubiquitination regulate the ERAD ubiquitin ligase Hrd1. eLife 2019, 8, e50903. [Google Scholar] [CrossRef]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef]
- Frani´c, D.; Zubčić, K.; Boban, M. Nuclear Ubiquitin-Proteasome Pathways in Proteostasis Maintenance. Biomolecules 2021, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Mehrtash, A.B.; Hochstrasser, M. Ubiquitin-dependent protein degradation at the endoplasmic reticulum and nuclear envelope. Semin. Cell Dev. Biol. 2019, 93, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Allensworth, J.L.; Evans, M.K.; Bertucci, F.; Aldrich, A.J.; Festa, R.A.; Finetti, P.; Ueno, N.T.; Safi, R.; McDonnell, D.P.; Thiele, D.J.; et al. Disulfiram (DSF) acts as a copper ionophore to induce copper dependent oxidative stress and mediate anti-tumor efficacy in inflammatory breast cancer. Mol. Oncol. 2015, 9, 1155–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, L.H.; Zhang, H.T.; Wang, Y.T.; Liu, S.; Zhou, W.L.; Yuan, X.Z.; Li, T.Y.; Wu, C.F.; Yang, J.Y. Disulfiram combined with copper inhibits metastasis and epithelial-mesenchymal transition in hepatocellular carcinoma through the NF-κB and TGF-β pathways. J. Cell Mol. Med. 2018, 22, 439–451. [Google Scholar] [CrossRef]
- Lin, J.; Haffner, M.C.; Zhang, Y.; Lee, B.H.; Brennen, W.N.; Britton, J.; Kachhap, S.K.; Shim, J.S.; Liu, J.O.; Nelson, W.G.; et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate 2011, 71, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Paranjpe, A.; Srivenugopal, K.S. Degradation of NF-kappaB, p53 and other regulatory redox-sensitive proteins by thiol-conjugating and -nitrosylating drugs in human tumor cells. Carcinogenesis 2013, 34, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Iwakuma, T. Non-Canonical Cell Death Induced by p53. Int. J. Mol. Sci. 2016, 17, 2068. [Google Scholar] [CrossRef] [PubMed]
- Rizzotto, D.; Englmaier, L.; Villunger, A. At a Crossroads to Cancer: How p53-Induced Cell Fate Decisions Secure Genome Integrity. Int. J. Mol. Sci. 2021, 22, 10883. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.M.; Kim, I.Y.; Seo, M.J.; Kwon, M.R.; Choi, K.S. Nutlin-3 enhances the bortezomib sensitivity of p53-defective cancer cells by inducing paraptosis. Exp. Mol. Med. 2017, 49, e365. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Domizio, A.D.; Limonta, P. The emerging role of paraptosis in tumor cell biology: Perspectives for cancer prevention and therapy with compounds. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188338. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solovieva, M.; Shatalin, Y.; Odinokova, I.; Krestinina, O.; Baburina, Y.; Lomovskaya, Y.; Pankratov, A.; Pankratova, N.; Buneeva, O.; Kopylov, A.; et al. Disulfiram Oxy-Derivatives Suppress Protein Retrotranslocation across the ER Membrane to the Cytosol and Initiate Paraptosis-like Cell Death. Membranes 2022, 12, 845. https://doi.org/10.3390/membranes12090845
Solovieva M, Shatalin Y, Odinokova I, Krestinina O, Baburina Y, Lomovskaya Y, Pankratov A, Pankratova N, Buneeva O, Kopylov A, et al. Disulfiram Oxy-Derivatives Suppress Protein Retrotranslocation across the ER Membrane to the Cytosol and Initiate Paraptosis-like Cell Death. Membranes. 2022; 12(9):845. https://doi.org/10.3390/membranes12090845
Chicago/Turabian StyleSolovieva, Marina, Yuri Shatalin, Irina Odinokova, Olga Krestinina, Yulia Baburina, Yana Lomovskaya, Anton Pankratov, Natalia Pankratova, Olga Buneeva, Arthur Kopylov, and et al. 2022. "Disulfiram Oxy-Derivatives Suppress Protein Retrotranslocation across the ER Membrane to the Cytosol and Initiate Paraptosis-like Cell Death" Membranes 12, no. 9: 845. https://doi.org/10.3390/membranes12090845
APA StyleSolovieva, M., Shatalin, Y., Odinokova, I., Krestinina, O., Baburina, Y., Lomovskaya, Y., Pankratov, A., Pankratova, N., Buneeva, O., Kopylov, A., Medvedev, A., & Akatov, V. (2022). Disulfiram Oxy-Derivatives Suppress Protein Retrotranslocation across the ER Membrane to the Cytosol and Initiate Paraptosis-like Cell Death. Membranes, 12(9), 845. https://doi.org/10.3390/membranes12090845







