Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines
Abstract
:1. Introduction
2. Analysis of Compounds in Kampo Medicines
2.1. Low-Molecular-Weight Compounds in Kampo Medicines, Crude Drugs, and Other Natural Products
2.2. Proteins in Crude Drugs
3. Previous and Potential Technologies for the Identification and Quantification of Proteins in Kampo Medicines
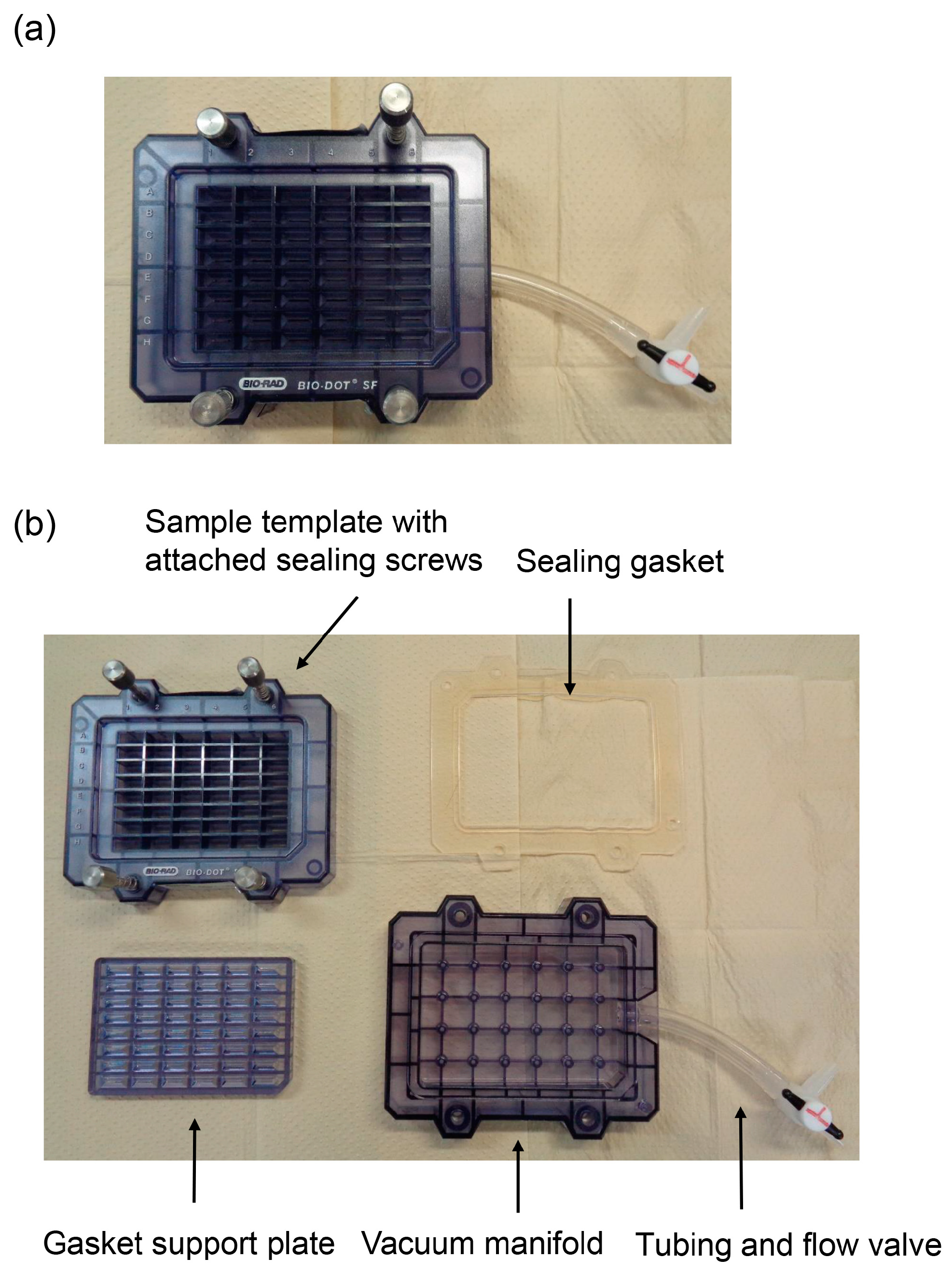
4. Equipment, Characteristics, and Methodology of the Novel Slot Blot
4.1. Equipment
4.2. PVDF Membrane
4.3. Lysis Buffer
4.4. Application of Standard and Sample Solutions and Vacuum with Water Aspirator
4.5. Protein Quantification in Standard and Sample Solutions
5. Comparing the Novel Slot Blot with Other Slot Blots
6. Potential for Identifying and Quantifying Various Rare Proteins in Kampo Medicines Using the Novel Slot Blot Method
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AGEs | Advanced glycation end products |
| CHAPS | 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate |
| PBS | Phosphate-buffered saline |
| PVDF | Polyvinylidene difluoride |
| Tris | Tris-(hydroxymethyl)-aminomethane |
| LC | Liquid chromatography |
References
- Chung, H.; Yuasa, M.; Chen, F.; Yukawa, K.; Motoo, Y.; Arai, I. The status of education for integrative medicine in Japanese medical universities with special reference to Kapo medicines. Tradit. Kampo Med. 2023, 10, 123–131. [Google Scholar] [CrossRef]
- Motoo, Y.; Seki, T.; Tsutani, K. Traditional Japanese medicine, Kampo: Its history and current status. Clin. J. Integr. Med. 2011, 17, 85–87. [Google Scholar] [CrossRef] [PubMed]
- Arai, I.; Kawabata, N. Kampo pharmaceutical products in the Japanese health-care system: Legal status and quality assurance. Tradit. Kampo. Med. 2019, 6, 3–11. [Google Scholar] [CrossRef]
- Arai, I. Clinical studies of traditional Japanese herbal medicines (Kampo): Need for evidence by modern scientific methodology. Integr. Med. Res. 2021, 10, 100722. [Google Scholar] [CrossRef] [PubMed]
- Motoo, Y.; Hakamatsuka, T.; Kawahara, N.; Arai, I.; Tsutani, K. Standards of Reporting Kampo Products (STORK) in research articles. J. Integr. Med. 2017, 15, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Motoo, Y.; Arai, I.; Tsutani, K. Use of Kampo Diagnosis in Randomized Controlled Trials of Kampo Products in Japan: A Systematic Review. PLoS ONE 2014, 9, e104422. [Google Scholar] [CrossRef] [PubMed]
- Motoo, Y.; Arai, I.; Kogure, T.; Tsutani, K. Review of the first 20 years Evidence-Based Medicine Committee of the Japan Society for Oriental Medicine. Tradit. Kampo Med. 2021, 8, 123–139. [Google Scholar] [CrossRef]
- Motoo, Y. Role of Kampo Medicine in Modern Cancer Therapy: Towards Completion of Standard Treatment. J. Nippon Med. Sch. 2022, 89, 139–144. [Google Scholar] [CrossRef]
- Motoo, Y.; Cameron, S. Kampo medicines for supportive care of patients with cancer: A brief review. Integr. Med. Res. 2022, 11, 100839. [Google Scholar] [CrossRef]
- Suzuki, T.; Yamamoto, A.; Ohsawa, M.; Motoo, Y.; Mizukami, H.; Makino, T. Effect of ninjin’yoeito and ginseng extracts on oxliplation-induced neuropathies in micie. J. Nat. Med. 2017, 71, 757–764. [Google Scholar] [CrossRef]
- Sameshima-Uto, N.; Amitani, H.; Atobe, Y.; Sameshima, Y.; Sakaki, M.; Rokot, N.; Ataka, K.; Amitani, M.; Inui, A. Herbal Medicine Ninjin’yoeito in the Treatment of Sarcopenia and Frailty. Front. Nutr. 2018, 5, 126. [Google Scholar]
- Ohnishi, Y.; Fujii, H.; Hayakawa, Y.; Sakukawa, R.; Yamamura, T.; Sakamoto, T.; Tsukada, K.; Fujimaki, M.; Nunome, S.; Komatsu, Y.; et al. Oral Administration of Kampo (Japanese Herbal) Medicine Juzen-taiho-to Inhibits Liver Metastastics of Colon 26-L5 Carcinoma Cells. Jpn. J. Cancer Res. 1998, 89, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Takagi, K.; Sugihira, T.; Kitamura, M.; Kawai, M.; Mitsuguchi, Y.; Tsukamoto, K.; Nakanishi, H.; Makino, T. Inhibitory effect of Bofutushosan (Fangfengtongshengsan) extract on the absorption of fructose in rats and mice. J. Nat. Med. 2023, 77, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Kishida, Y.; Kagawa, S.; Arimitsu, J.; Nakanishi, M.; Sakashita, N.; Otsuka, S.; Yoshikawa, H.; Hagihara, K. Go-sha-jinki-Gan (GJG), a traditional Japanese herbal medicine, protects against in senescence-accelerated mice. Phytomedicine 2015, 22, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Nakae, A.; Kishida, Y.; Baba, K.; Sakashita, N.; Shibata, M.; Yoshikawa, H.; Hagihara, K. Go-sha-jinki-Gan (GJG) ameliorates allodynia in chronic constriction injury model mice via suppression of TNF-α expression in the spinal cord. Mol. Pain. 2016, 12, 1744806916656382. [Google Scholar] [CrossRef] [PubMed]
- Hosogi, S.; Ohsawa, M.; Kato, I.; Kuwahara, A.; Inui, T.; Marunaka, Y. Improvement of Diabetes Mellitus Symptoms by Intake of Ninjin’yoeito. Front. Nutr. 2018, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Uchiyama, M.; Zhang, Q.; Harada, T.; Otsuka, K.; Shimokawa, T.; Niimi, M. Effect of 34 Kinds of Traditional Japanese Herbal Medicines on Prolongation of Cardiac Allograft Survival. Transplant. Proc. 2014, 46, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Qia, B.L.; Li, Q.L.; Zhang, Y.; Li, K.; Wang, L.; Qiao, Y. Novel Antihypertensive Peptides Derived from Adlay (Coix larchryma-jobi L. var. ma-yuen Stapf) Glutelin. Molecules 2017, 22, 123. [Google Scholar]
- Lio, B.; Ma, S.; Zhang, S.; Li, X.; Quan, R.; Wan, S. Fructus cannabis protein powder as a green and high effective corrosion inhibitor for Q235 carbon steel in 1 M HCl solution. Int. J. Biol. Macromol. 2023, 239, 124358. [Google Scholar] [CrossRef]
- Quazi, R.M.; Sajid, Z.; Zhao, C.; Hussain, I.; Ifikhar, F.; Jameel, M.; Rehman, F.U.; Ali, A. Lyophilization Based Isolation of Exosomes. Int. J. Mol. Sci. 2023, 24, 10477. [Google Scholar] [CrossRef]
- Wang, X.; Ma, Y.; Qi, X.; Ruan, X.; Zhao, F. Practically of non-invasive glucagon-loaded dissolving microneedle for life–saving treatment of severe hypoglycemia in a diabetic rat model. Int. J. Pharm. 2023, 644, 123340. [Google Scholar] [CrossRef] [PubMed]
- Armin, V.; Farnaz, M. Practical Techniques for Improving the Performance of Polymeric Membranes and Processes for Protein Separation and Purification. Iran J. Chem. Chem. Eng. 2018, 37, 1–23. [Google Scholar]
- Soxena, A.; Tripathi, B.P.; Kumar, M.; Shahi, V.K. Membrane-based techniques for the separation and purification of proteins: An overview. Adv. Colloid. Interface Sci. 2009, 145, 1–22. [Google Scholar] [CrossRef]
- Zeng, X.; Ruckenstein, E. Membranse Chromatography: Preparation and Applications to Protein Separation. Biotechnol. Prog. 1999, 15, 1003–1019. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Hasegawa, T.; Tatsuno, T.; Date, J.; Ishigaki, Y.; Nakamura, N.; Takano, F.; Ohta, T. Isolation of N-acetylneuraminic Acid and N-glycolylneuraminic Acid from Pleurocybella porrigens. J. Health Sci. 2009, 55, 373–379. [Google Scholar] [CrossRef]
- Takata, T.; Ishigaki, Y.; Shimasaki, T.; Tsuchida, H.; Motoo, Y.; Hayashi, A.; Tomosugi, N. Characterization of proteins secreted by pancreatic cancer cells with anticancer drug treatment in vitro. Oncol. Rep. 2012, 28, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Mahony, C.B.; Torres, A.; Murillo-Saich, J.; Kembel, S.; Cedeno, M.; John, P.; Bhatti, A.; Croft, A.P.; Guma, M. Dual inhibition of glycolysis and glutaminolysis for synergistic therapy of reheumatoid arthritis. Arthritis Res. Ther. 2023, 25, 176. [Google Scholar] [CrossRef]
- Liu, P.; Tang, W.; Zhao, D.; Zhou, P.; Hu, K. Active metabolites and potential mechanisms of Notopterygium incisum against obstructive sleep apanea Syndrome (OSAS): Neteork analysis and experimental assessment. Front. Pharmacol. 2023, 14, 1185100. [Google Scholar] [CrossRef]
- Lin, Y.; O’Reilly, M.A.; Hynynen, K. A PVDF Receiver for A coustic Monitoring of Microbubble-Mediated Ultrasound Brain Therapy. Sensors 2023, 23, 1369. [Google Scholar] [CrossRef]
- Vierstraete, M.; Beckers, R.; Vangeel, L.; Foriers, B.; Pletinckx, P.; Muysoms, F. Prospective cohort study on mesh shrinkage measured with MRI after robot-assisted minimal invasive retrorectus ventral hernia repair using an iron-oxide-loadef polyvinylidene fluoride mesh. Surg. Endosc. 2023, 37, 4604–4612. [Google Scholar] [CrossRef]
- Sebastian, L.; Alina, J.; Fabinshy, T.; Dominik, R.; Axel, S.; Jens, H.; Kilian, W.; Cludia, R.; Leonidas, K.; Julia, R.; et al. AbsorbaTackTM vs ProTackTM vs. sutures: A biomerchanical analysis of cervical fixation methods for laparoscopic fixations in the porcine model. Arch. Gynecol. Obstet. 2023, 307, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cadet, E.R.; King, M.W.; Cole, J.H. Comparison of the mechanical properties and anchoring performance of polyvinylidene fluoride and polypropylene barbed sutures for tendon repair. J. Biomed. Mater. Res. 2022, 110, 2258–2265. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhu, Y.; Yan, J.; Wu, W.; Wang, B. Micromechanism Study of Molecular Compatibility of PVDF/PEI Blend Membrane. Membranes 2022, 12, 809. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Han, Q.; Wu, S.; Xio, H.; Zhang, L.; Lin, Y.; Meng, F.; Zhao, S. Unveiling the Impacts of Sodium Hypochlorite on the Characteristics and Fouling Behaviors of Different Commercial Polyvinylidene Fluoride Hollw Fiber Membranes. Membranes 2022, 12, 965. [Google Scholar] [CrossRef] [PubMed]
- Sisay, E.J.; Fazekas, Á.F.; Gyulári, T.; Kopniczky, J.; Hopp, B.; Veréb, G.; Lászó, Z. Investigation of Photocatalytic PVDF Membranes Containing Inorganic Nanoparticles for Model Dairy Wastwater Treatment. Membranes 2023, 13, 656. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Wang, S.; Chen, N.; Wen, X.; Tian, G.; Zhang, L.; Cheng, P.; Zhang, J.; Tang, N. Study on Low Therminal-Conductivity of PVDF@SiAG/PET Membranes for Direct Contact Membrane Distillation Application. Membranes 2023, 13, 773. [Google Scholar] [CrossRef] [PubMed]
- Ogino, N.; Ogino, K.; Eitoku, M.; Suganuma, N.; Nagaoka, K. Filter blot method: A simple method for measuring 3-nitrotyrosine in proteins of atmospheric particulate matter. Environ. Pollut. 2023, 329, 121677. [Google Scholar] [CrossRef]
- Bickner, A.N.; Chmpion, M.M.; Hummon, A.B.; Bruening, M.L. Electroblotting through a tryptic membrane for LC-MS/MS analysis of proteins separated in electrophoretic gels. Analyst 2020, 145, 7724–7735. [Google Scholar] [CrossRef]
- Takata, T.; Ueda, T.; Sakasai-sakai, A.; Takeuchi, M. Generation of glyceraldehyde-derived advanced glycation end-products in pancreatic cancer cells and the potential of tumor promotion. World J. Gastroenterol. 2017, 23, 4910–4919. [Google Scholar] [CrossRef]
- Takata, T. Is the Novel Slot Blot a Useful Method for Quantification of Intracellular Advanced Glycation End-Products? Metabolites 2023, 13, 564. [Google Scholar] [CrossRef]
- Takata, T.; Motoo, Y. Novel In Vitro Assay of the Effects of Kampo Medicines against Intra/Extracellular Advanced Glycation End-Products in Oral, Esophageal, and Gastric Epithelial Cells. Metabolites 2023, 13, 878. [Google Scholar] [CrossRef] [PubMed]
- Phoung-Nguyen, K.; McNeill, B.A.; Aston-Mourney, K.; Rivera, L.R. Advanced Glycation End-Products and Their Effects on Gut Health. Nutrients 2023, 15, 405. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Radjiabzadeh, D.; Midina-Gomez, C.; Voortman, T.; van Merus, J.B.J.; Ikram, M.A.; Uittelinden, A.G.; Kraaij, R.; Zillekens, M.C. Advanced Glycation End Products (AGEs) in Diet and Skin in Relation to Stool Microbiota: The Rotterdam Study. Nutrients 2023, 15, 2567. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Schaffiner, S.H.; Davies, J.P.; Benton, M.L.; Plate, L.; Nordman, J.T. BRWD3 promotes KDM5 degradation to maintain H3K4 methylation levels. Proc. Natl. Acad. Sci. USA 2023, 120, e2305092120. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Fang, Z.; Wang, K.; Ye, M. Hydrophobic Derivatization Strategy Facilitates Comprehensive Profiling of Protein Methylation. J. Proteome Res. 2023, 22, 3275–3281. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, M.; Jang, M.; Chen, X.; Song, G.; Ji, H.; Wang, Z.; Zhu, X. Methylation of BRD4 by PRMT1 regulated BRD4 phophorylation and promotes ovarian cancer invasion. Cell Death Dis. 2023, 14, 624. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gai, X.; Liang, R.; Zhang, E.; Liang, X.; Liang, H.; Fu, C.; Zhou, A.; Shi, Y.; Xu, F.; et al. SIRT1-dependent deacetylation of Txnip H3K9ac is critical for exenatide-improved diabetic kidney disease. Biomed. Pharmacother. 2023, 167, 115515. [Google Scholar] [CrossRef]
- Sun, Q.; Zou, Y.; Feng, Q.; Gong, Z.; Li, M.; Chen, Z. The acetylation of pknH is linked to the ethambutol resistance of Mycobacterium tuberculosis. Arch. Microbiol. 2023, 205, 337. [Google Scholar]
- Huang, Z.; Ito, M.; Zhang, S.; Toda, T.; Takeda, J.; Ogi, T.; Ohno, K. Extremely low-frequency electromagnetic field induces acetylation of heat shock proteins and enhances protein folding. Ecotoxicol. Environ. Saf. 2023, 264, 115482. [Google Scholar] [CrossRef]
- Xiong, H.; Zheng, Z.; Zhao, C.; Zhao, M.; Wang, Q.; Zhang, P.; Li, Y.; Zhu, Y.; Zhu, S.; Li, J. Insight into the underlying molecular mechanisms of dilated cardiomyopathy through integrative analysis of data mining, iTRAQ-PRM proteomics and bioinformatics. Proteome 2023, 21, 13. [Google Scholar] [CrossRef]
- Toney, N.J.; Schlom, J.; Donahue, R.N. Phosphoflow cytometry to assess cytokine signaling pathways in peripheral immune cell function and treatment response in patients with solid tumors. J. Exp. Clin. Res. 2023, 42, 247. [Google Scholar]
- Chen, D.; Dong, X.; Chen, D.; Lin, J.; Lu, T.; Shen, J.; Ye, H. Chd1 plays a protective role in nonalcoholic fatty liver disease by regulating PPAR/PGC-1α signaling pathway. Biochem. Biophys. Res. Commun. 2023, 681, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Zappi, J.; Tong, Q.; Van der Cruyssen, R.; Cornlis, F.M.F.; Lambert, C.; Coelho, T.P.; Grisart, J.; Kague, E.; Kague, E.; Lories, R.J.; et al. Osteomodulin downregulation is associated with osteoarthritis development. Bone Res. 2023, 11, 49. [Google Scholar] [CrossRef]
- Mapunda, J.A.; Parejia, J.; Vladymyrov, M.; Bouilet, E.; Hélie, P.; Pleskač, P.; Barcos, S.; Andree, J.; Vesweber, D.; McDonald, D.M.; et al. VE-cadherin in arachoid and pia mater cells serves as a suitable landmark for in vivo imaging of CNS immune surveillance and inflammation. Nat. Commun. 2023, 14, 5837. [Google Scholar] [CrossRef] [PubMed]
- Decloquement, M.; Venuto, M.T.; Cogez, V.; Steinmetz, A.; Schulz, C.; Lion, C.; Noel, M.; Rigolot, V.; Teppa, R.E.; Biot, C.; et al. Salmonid polusialyltranseferases to generate a variety of sialic acid polymers. Sci. Rep. 2023, 13, 15610. [Google Scholar] [CrossRef] [PubMed]
- Maeder, C.; Speer, T.; Wirth, A.; Boeckel, J.; Fatima, S.; Shazad, K.; Freichet, M.; Laufs, U.; Gaul, S. Membrane-bound Interleukin-1α mediated leukocyte adhesion during atherogenesis. Front. Immunol. 2023, 14, 1252384. [Google Scholar] [CrossRef]
- Tang, L.; Ye, P.; Yao, L.; Luo, Y.; Tan, W.; Xiang, W.; Liu, Z.; Tan, L.; Xiao, J. LINC01268 promotes epithelial-mesenchymal transition, invasion and metastasis of gastric cancer via the PI3K/Akt singling pathway and targeting MARCKS. World J. Gastrointest. Oncol. 2023, 15, 1366–1383. [Google Scholar] [CrossRef]
- Leal, C.S.; Carvalho, C.A.M. In Silico Physicohemical Characterization of Fusion Proteins from Emerging Amazonian Arboviruses. Life 2023, 13, 1687. [Google Scholar] [CrossRef]
- Jobaer, M.A.; Ashrafi, S.; Asan, M.; Hassan, C.M.; Rashid, M.A.; Ismam, S.N.; Masud, M.M. Phystochemical and Bioligical Investigation of an Indigenous Plant of Bangladesh, Gynura procumbens (Lour.) Merr; Drug Discovery from Nature. Molecules 2023, 28, 4186. [Google Scholar] [CrossRef]
- Masota, N.; Ohlsen, K.; Schollmayer, C.; Meinei, C.; Holzagrabe, U. Isolation and Characterization of Galloyglucoses Effective against Multidrug-Resistant Strains of Escherichia coli and Klebsiella pneumoniae. Molecules 2022, 17, 5045. [Google Scholar] [CrossRef]
- Sananboonudom, S.; Kaewnoi, A.; Pompimon, W.; Narakaew, S.; Jiajaroen, S.; Chainok, K.; Nuntasaen, N.; Suksen, K.; Chairoungdua, A.; Limthongkul, J.; et al. Study on the absolute configuration and biological activity of rotenoids, from the leaves and twigs of Millettia pyrrhocarpa Mattapha, Forest & Hakins, sp. Nov. BMC Complement. Med. Ther. 2023, 23, 147. [Google Scholar]
- Miyano, K.; Hasegawa, S.; Asai, N.; Uzu, N.; Yatsuoka, W.; Ueno, T.; Nonaka, M.; Uezono, Y. The Japanese Herbal Medicine Hangesshashinto Induce Oral Keratinocyte Migration by Mediating the Expression of CXCL12 Through the Activation of Extracellular Signal-Regulated Kinase. Front. Pharmacol. 2022, 12, 695039. [Google Scholar] [CrossRef]
- Chung, C.; Lee, M.; Hsia, S.; Chiang, W.; Kuo, Y.; Hsu, H.; Lin, Y. Suppression on allergic inflammation of dehulled adlay (Coix lachrymal-jobi L. var. ma-yuen Stapf) in mice and anti-degranulation phytosterols from adlay bran. Food Funct. 2021, 12, 12788–12799. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, F.; Zhang, Y.; Wang, S.; Yuan, Y.; Lu, C.; Nie, J.; Nan, T.; Yang, B.; Huang, L.; et al. Application of hyperspectral imaging assistant with integrated deep learning approaches in identifiying geographical origins and predicting nutrient contents of Coix seeds. Food Chem. 2023, 404 Pt A, 134503. [Google Scholar] [CrossRef]
- Sui, Y.; Xu, D. Isolation and identification of anti-inflammatory and analgesic polysaccharides from Coix deed (Coix lacryma-jobi L. var. Ma-yuen (Roman.) Stapf). Nat. Prod. Res. 2022, 30, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Kim, Y.; Lee, J.; Kim, Y.; Han, K.; Yoon, Y.; Cho, B.; Park, K.; Lee, H.; Cho, J. Comparison of Quality, Antioxidant Capacity, and Anti-Inflammatory Activity of Adlay [Coix lacryma-jobi L. var. ma-yuen (Rom. Caill.) Stapf.] Sprout at Several Harvest Time. Plants 2023, 12, 2975. [Google Scholar] [PubMed]
- Tang, X.; Wang, Z.; Zheng, J.; Kan, J.; Chen, G.; Du, M. Physicochemical, Structure properties and in vitro hypoglycemic activity of soluble dietary fiber from adlay (Coix lachyma-jobi L. var. ma-yuen Stapf) bran treated by steam explosion. Front Nutr. 2023, 10, 1124012. [Google Scholar]
- Huang, Y.; Chen, Y.; Chen, H.; Chiang, Y.; Ali, M.; Chiang, W.; Chung, C.; Hsia, S. Ethanolic Extracts of Adlay Testa Hull and Their Active Biomolecules Exert Relaxing Effects on Ulterine Muscle Contraction through Blocking Extracellular Calcium Influx in Ex Vivo and In Vivo Studies. Biomolecules 2021, 11, 887. [Google Scholar] [CrossRef]
- Chiang, Y.; Chung, C.; Lin, J.; Chiang, W.; Chen, H.; Ali, M.; Shih, Y.; Wang, K.; Huang, T.; Chang, H. Adlay, Seed (Coix lacryma-jobi L. var. Ma-yuen Stapf.) Ethanolic Extract Fractions and Subfractions Induce Cell Cycle Arrest and Apoptosis in Human Breast and Cervical Cancer Cell Lines. Molecules 2022, 27, 3984. [Google Scholar] [CrossRef]
- Farinon, B.; Molianari, R.; Costantini, L.; Mereodino, N. The Seed of Industrial Hemp (Cannabis sativa L.): Nutritional Quality and Potential Functionality for Human Health and Nutrition. Nutrients 2020, 12, 1935. [Google Scholar] [CrossRef]
- Chen, N.; Liu, C.; Lin, W.; Ding, Y.; Bian, Z.; Huang, L.; Huang, H.; Yu, K.; Chen, S.; Sun, Y.; et al. Extract of Fructus Cannabis Ameliorates Learning and Memory Impairment Induced by D-Galactose in an Aging Rats Model. Evid.-Based Complement. Altern. Med. 2017, 2017, 4757520. [Google Scholar] [CrossRef] [PubMed]
- Iftikhar, A.; Zafar, U.; Ahmed, W.; Shabbir, M.A.; Sameen, A.; Sahar, A.; Bhat, Z.B.; Kowalczewski, P.Ł.; Jarzębski, M. Applications of Cannabis sativa L. in Food and Its Therapeutic Potential: From a Prohibited Drug to a Nutritional Supplement. Molecules 2021, 26, 7699. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Na, C.; Wang, Y. Angelica decursiva exerts antihypertensive activity by inhibiting L-type calcium channel. J. Ethnopharmacol. 2023, 313, 116527. [Google Scholar] [CrossRef]
- Mihm, A.C.P.; Bonet, L.F.S.; Aiub, C.A.F. Biochemical characterization and phytotoxic activity or protein extract from Euphoribia tirucalli L. J. Ethnopharmacol. 2022, 285, 114903. [Google Scholar]
- Moghanloo, S.A.; Forouzanfar, M.; Jafarinia, M.; Fazlollahi, M.R.; Kardar, G.A. Allergen-specific immunotherapy by recombinant Der P1 allergen-derived peptide-based vaccine in an allergic mouse model. Immun. Inflamm. Dis. 2023, 11, e878. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Stum, R.; Henrich, D.; Marzi, I.; Leppik, L. CD44+ and CD31+ extracellular vesicles (EVs) are significantly reduced in polytraumatized patients with hemorrhagic shock—Evaluation of their diagnostic and prognostic potential. Front. Immunol. 2023, 14, 1196241. [Google Scholar] [CrossRef] [PubMed]
- Yong, J.; Hakobyan, K.; Xu, J.; Mellick, A.; Wthitelock, J.; Liang, K. Comparison of protein quantification method for protein encapsulation with ZIF-8 metal-organic frameworks. Biotechnol. J. 2023, 12, e2300015. [Google Scholar] [CrossRef]
- Shegefti, S.; Bolori, S.; Nabavi-Rad, A.; Dabri, H.; Yadegar, A.; Bagaei, K. Helicobacter pylori-derived outer membrane vesicles suppress liver autophagy: A novel mechanism for H. pylori-mediated hepatic disorder. Microb. Pathog. 2023, 183, 106319. [Google Scholar] [CrossRef]
- Dai, Y.; Duan, K.; Huang, G.; Yang, X.; Jang, X.; Chen, J.; Liu, P. Inhalation of electronic cigarettes slightly affects lung function and inflammation in mice. Front. Toxicol. 2023, 5, 1232040. [Google Scholar] [CrossRef]
- Yang, C.; Liu, M.; Peng, Y.; Xu, Z.; Liu, Y.; Guo, Z.; Li, B.; Yang, X. 17β-estradiol inhibits TGF-β-induced collagen gen contraction mediated by human Tenon fibroblasts via Smads and MAPK signaling pathways. Int. J. Ophtalmol. 2023, 16, 1441–1449. [Google Scholar] [CrossRef]
- Xing, X.; Wang, H. Correlation of serum HMGB1 and HMGB2 levels with clinical symptoms in allergic rhinitis children. Medicines 2023, 102, e34921. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Zheng, H.; Deng, B.; Mahajan, B.; Grabias, B.; Kozakai, Y.; Morin, M.J.; Locke, E.; Birkett, A.; Miura, K.; et al. A Slot Blot Immunoassay for Quantitative Detection of Plasmodium falciparum Circumsporozoite Protein in Mosquito Midgut Oocyst. PLoS ONE 2014, 9, e115807. [Google Scholar] [CrossRef] [PubMed]
- Grabias, B.; Verma, N.; Zheng, H.; Tripathi, A.K.; Mlambo, G.; Morin, M.J.; Locke, E.; Kumar, S. A no film slot blot for the detection of developing P. falciparum oocysts in mosquitoes. PLoS ONE 2017, 12, e0174229. [Google Scholar] [CrossRef] [PubMed]
- Ghiani, A.; Ania, R.; Asero, R.; Bellotto, E.; Citterio, S. Ragweed pollen collected along high-traffic roads shows a higher allergenicity than pollen sampled in vegetated areas. Allergy 2012, 67, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Nimmo, J.T.; Verma, A.; Dodart, J.-C.; Wang, C.Y.; Savistchenko, J.; Melki, R.; Carare, R.O.; Nicoll, J.A.R. Novel antibodies detect additional α-synuclein pahaology in synucleinopathies: Potential development for immunotherapy. Alzheimer’s Res. Ther. 2020, 12, 159. [Google Scholar] [CrossRef]
- Barandalla, M.; Haucke, E.; Fischer, B.; Santos, A.N.; Colleoni, S.; Galli, C.; Santos, A.N.; Lazzari, G. Comparative Analysis of AGE and RAGE Levels in Human Somatic and Embryonic Stem Cell under H2O2-Induced Noncytotoxic Oxidative Stress Conditions. Oxid. Med. Cell. Longev. 2017, 2017, 4240136. [Google Scholar] [CrossRef]
- Koriyama, Y.; Furukawa, A.; Muramatsu, M.; Takino, J.; Takeuchi, M. Glyceraldehyde caused Alzheimer’s disease-like alterations in diagnostic marker levels in SH-SY5Y human neuroblastoma cells. Sci. Rep. 2015, 5, 13313. [Google Scholar] [CrossRef]
- Nasu, R.; Furukawa, A.; Suzuki, K.; Takeuchi, M.; Koriyama, Y. The Effect of Glyceraldehyde-Derived Advanced Glycation End Products on β-Tubulin-Inhibited Neurite Outgrowth in SH-SY5Y Human Neuroblastoma Cells. Nutrients 2020, 12, 2958. [Google Scholar] [CrossRef]
- Kumar, S.T.; Jagannath, S.; Francois, C.; Vanderstichele, H.; Stoops, E.; Lashuel, H.A. How specific are the conformation-specific α-synuclein antibodies? Characterization and validation of 16 α-synuclein conformation-specific antibodies using well-characterized preparations of α-synuclein monomers, fibris and oligomers with distinct structures and morphology. Neutobiol. Dis. 2020, 146, 10586. [Google Scholar]
- Takata, T.; Sakasa-Sakai, A.; Ueda, T.; Takeuchi, M. Intracellular toxic advanced glycation end-products in cardiomyocytes may cause cardiovascular disease. Sci. Rep. 2019, 9, 2121. [Google Scholar] [CrossRef]
- Takata, T.; Sakasa-Sakai, A.; Takino, J.; Takeuchi, M. Evidence for Toxic Advanced Glycation End-Poducts Generated in the Normal Rat Liver. Nutrients 2019, 11, 1612. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Sakasai-Sakai, A.; Takeuchi, M. Impact of intracellular toxic advanced glycation end-products (TAGE) on murine myoblast cell death. Diabetol. Metab. Syndr. 2020, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Takata, T.; Nakazawa, Y.; Nakamura, Y.; Guo, X.; Yamada, S.; Ishigaki, Y.; Takeuchi, M.; Miyazawa, K. Potential of an Interorgan Network Mediated by Toxic Advanced Glycation End-Products in a Rat Model. Nutrients 2021, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, C.; Sakasa-Sakai, A.; Okimura, R.; Tanaka, H.; Takata, T.; Takeuchi, M.; Matsunaga, T. Accumulation of Toxic Advanced Glycation End-Products Induces Cytotoxicity and Inflammation in Hepatocyte-Like Cells Differentiated from Human Induced Pluripotent Stem Cells. Biol. Pharm. Bull. 2021, 44, 1399–1402. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Sakasai-Sakai, A.; Takeuchi, M. Intracellular Toxic Advanced Glycation End-Products in 1.4E7 Cell Line Induce Death with Reduction of Microtubule-Associated Protein 1 Light Chain 3 and p62. Nutrients 2022, 14, 332. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Sakasa-Sakai, A.; Takeuchi, M. Intracellular Toxic Advanced Glycation End-Products May Induce Cell Death and Suppress Cardiac Fibroblasts. Metabolites 2022, 12, 615. [Google Scholar] [CrossRef] [PubMed]
- Sakasai-Sakai, A.; Takata, T.; Suzuki, H.; Maruyama, I.; Motomiya, Y.; Takeuchi, M. Immunological evidence for in vivo production of novel advanced glycation end-products from 1,5-anhydro-D-fructose, a glycogen metabolite. Sci. Rep. 2019, 9, 10194. [Google Scholar] [CrossRef] [PubMed]
- Sakasai-Sakai, A.; Takata, T.; Takeuchi, M. Intracellular Toxic Advanced Glycation End-Products Promote the Production of Reactive Oxygens Species in HepG2 Cells. Int. J. Mol. Sci. 2020, 21, 4861. [Google Scholar] [CrossRef]
- Sakasai-Sakai, A.; Takata, T.; Takeuchi, M. The Association between Accumulation of Toxic Advanced Glycation End-Products and Cytotoxic Effect in MC3T3-E1 Cells. Nutrients 2022, 14, 990. [Google Scholar] [CrossRef]
- Sakasai-Sakai, A.; Takata, T.; Takino, J.; Takeuchi, M. Impact of intracellular glyceraldehyde-derived advanced glycation end-products on human hepatocyte cell death. Sci. Rep. 2017, 7, 14282. [Google Scholar] [CrossRef]
- Browicka-Szydełko, A.; Krzystek-Korpacka, M.; Kuzan, A.; Gostomaska-Pampuch, K.; Gacka, M.; Jakobsche-Policht, U.; Adamiec, R.; Gamian, A. Non-standard AGE4 epitopes that predict polyneuropathy independently of obesity can be detected by slot dot-blot immunoassay. Adv. Clin. Exp. Med. 2020, 29, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Takino, J.; Kobayashi, Y.; Takeuchi, M. The formation of intracellular glyceraldehyde-derived advanced glycation end-products and cytotoxicity. J. Gastroenterol. 2010, 45, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Gravel, C.; Li, C.; Wang, J.; Hashem, A.M.; Jaentschke, B.; van Domselaar, G.; He, R.; Li, X. Quantitative Analyses of all Influenza Type A Viral Hemagglultinins and Neuraminidases using Universal Antibodies in Simple Slot Blot Assays. J. Vis. Exp. 2011, 4, 2784. [Google Scholar]
- Papadaki, M.; Holewinski, R.J.; Previs, S.B.; Martin, T.G.; Stachowski, M.J.; Li, A.; Blair, C.A.; Morave, C.S.; Van Eyk, J.E.; Campbell, K.S.; et al. Diabetes with heart failure increases methylglyoxal modifications in the sarcomere, which inhibit function. JCI Insight 2018, 3, e121264. [Google Scholar] [CrossRef] [PubMed]
- Gil-Agusti, M.T.; Campostrini, N.; Zolla, L.; Ciambella, C.; Invernizzi, C.; Righetti, G. Two-dimensional mapping as a tool for classification of green coffee bean species. Proteomics 2005, 5, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Tian, M.; Zong, M.; Teng, M.; Chen, Y.; Lu, J.; Jiang, J.; Liu, X.; Han, J. Proteomics Analysis of Pancreatic Ductal Adenocarcinoma Compared with Normal Adjacent Pancreatic Tissue and Pancreatic Benign Cystadenoma. Pancreatology 2009, 9, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Sakolvaree, Y.; Maneewatch, S.; Jiemsup, S.; Klaysing, B.; Tongtawe, P.; Srimanote, P.; Saengjaruk, P.; Banyen, S.; Tapchaisri, P.; Chonsa-nguan, M.; et al. Proteome and Immunome of Pathogenic Leptospira spp. Revealed by 2DE and 2DE-Immunoblotting with Immune Serum. Asian Pac. J. Allergy Immunol. 2007, 25, 53–73. [Google Scholar]
- Twine, S.M.; Mykytczuk, N.C.S.; Petit, M.; Tremblay, T.; Conlan, J.W.; Kelly, J.F. Francisella tularensis Proteome: Low Levels of ASB-14 Facilitate the Visualization of Membrane Proteins in Total Protein Extracts. J. Proteome Res. 2005, 4, 1848–1854. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, J.; Zhang, X.; Zheng, X.; Jiang, X.; Shi, L.; Yin, W.; Wang, J. Optimized samples preparation for two-dimensional gel electrophoresis of soluble proteins from chicken bursa of Fabricus. Proteome Sci. 2009, 7, 38. [Google Scholar] [CrossRef]
- Campos, A.; Puetro, M.; Prieto, A.; Cameán, A.; Almeida, A.M.; Coelho, A.; Vasconcelos, V.J. Protein extraction and two-dimensional gel electrophoresis of proteins in the marine mussel Mytilus galloprovincialis: An important tool for protein expression studies, food quality and safety assessment. Sci. Food Agric. 2013, 93, 1779–1787. [Google Scholar] [CrossRef]
- Pedroso, A.P.; Watanabe, R.L.H.; Albuquerque, K.T.; Telles, M.M.; Andrade, M.C.C.; Perez, J.D.; Sakata, M.M.; Lima, M.L.; Estadella, D.; Nascimento, C.M.O.; et al. Proteomic profiling of the rat hypothalamus. Proteome Sci. 2012, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Farinazzo, A.; Fasoli, E.; Kravchuk, A.V.; Candiano, G.; Aldini, G.; Regazzoni, L.; Righetti, P.G. En bloc elution of proteomes from combinatorial peptide ligand libraries. J. Proteom. 2009, 72, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Herbert, B. Advances in protein solubilization for two-dimensional electrophoresis. Electrophoresis 1999, 20, 660–663. [Google Scholar] [CrossRef]
- McCarthy, J.; Hopwood, F.; Oxley, D.; Laver, M.; Castagna, A.; Righetti, P.G.; Williams, K.; Herbert, B. Carbamylation of Protein in 2-D Electrophoresis-Myth or Reality? J. Proteome Res. 2003, 2, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Tit-Oon, P.; Chokchaichamnankit, D.; Khongmanee, A.; Sawangreetrakul, P.; Svasti, J.; Srisomsap, C. Comparative secretome analysis of cholanagiocaricinoma cell line in three-dimensional culture. Int. J. Oncol. 2014, 45, 2108–2116. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, Y.; Michhiro, M. Analysis of trypsin inhibition activity in human plasma proteins after separation by non-denaturing two-dimensional electrophoresis. Clin. Chim. Acta 2013, 425, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Okayama, A.; Iwafune, Y.; Yahagi, S.; Arakawa, N.; Hirano, H. Multiplex detection and identification of proteins on a PVDF membrane blocked with a synthetic polymer-based reagent. Electrophoresis 2008, 29, 4377–4380. [Google Scholar] [CrossRef]
- Nie, X.; Li, C.; Hu, S.; Xue, F.; Kang, J.; Zhang, W. An appropriate loading control for western blot analysis in animal model of myocardial ischemic infarction. Biochem. Biophys. Res. 2017, 12, 108–113. [Google Scholar] [CrossRef]
- Kurien, B.; Scefield, R.H. A brief review of other notable protein blotting methods. Methods Mol. Biol. 2009, 536, 367–384. [Google Scholar]
- Taoerdalhong, H.; Zhou, K.; Yang, F.; Dong, C. Structure, immunostimulatory activity, and the effect of ameliorating airway inflammation of polysaccharides from Pyrus sinkiangesis Yu. Int. J. Biol. Macromol. 2022, 195, 246–254. [Google Scholar] [CrossRef]
- Huang, C.; Peng, X.; Pang, D.; Li, J.; Paulsen, B.S.; Eise, F.; Chen, Y.; Chen, Z.; Jia, R.; Li, L. Pectic polysaccharide from Nelumbo nucifera leaves promotes intestinal antioxidant defense in vitro and in vivo. Food Funct. 2021, 12, 10828–10841. [Google Scholar] [CrossRef] [PubMed]






| Solution A | Solution B | Solution C | Solution D |
|---|---|---|---|
| 30 mM Tris 7 M Urea 2 M Thiourea 4% CHAPS (Ultrapure water) (pH 8.5) | 1 Protease inhibitor cocktail tablet/2 mL (ultrapure water) | 27 mM Tris 6.3 M Urea 1.8 M Thiourea 3.6% CHAPS 10% Solution B (Ultrapure water) (pH 8.5) | 30 mM Tris 7 M Urea 2 M Thiourea 4% CHAPS 4% Solution B (Ultrapure water) (pH 8.5) |
| Solution | References |
|---|---|
| C | [39,90,91,92,93,94,95,96] |
| D | [97,98,99,100] |
| No. | Sample | Membrane Type | Lysis Buffer | Error Bars | Statistically Significant Difference | References |
|---|---|---|---|---|---|---|
| 1 | Cell lysate | Nitrocellulose | RIPA | Yes | Yes | [87,88] |
| 2 | Cell lysate | PVDF | RIPA | No | No | [102] |
| 3 | Protein in virus | PVDF | 4 M Urea | Yes | No | [103] |
| 4 | Cell lysate | PVDF | Solution C | Yes | Yes | [39,90,92,94,95,96] |
| 5 | Tissue lysate | PVDF | Solution C | Yes | Yes | [91,93] |
| 6 | Cell lysate | PVDF | Solution D | Yes | Yes | [97,98,99,100] |
| 7 | Tissue lysate | PVDF | 8 M Urea, 0.1%SDS | Yes | Yes | [104] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takata, T.; Masauji, T.; Motoo, Y. Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines. Membranes 2023, 13, 896. https://doi.org/10.3390/membranes13120896
Takata T, Masauji T, Motoo Y. Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines. Membranes. 2023; 13(12):896. https://doi.org/10.3390/membranes13120896
Chicago/Turabian StyleTakata, Takanobu, Togen Masauji, and Yoshiharu Motoo. 2023. "Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines" Membranes 13, no. 12: 896. https://doi.org/10.3390/membranes13120896
APA StyleTakata, T., Masauji, T., & Motoo, Y. (2023). Potential of the Novel Slot Blot Method with a PVDF Membrane for Protein Identification and Quantification in Kampo Medicines. Membranes, 13(12), 896. https://doi.org/10.3390/membranes13120896






