Analysis of Microbial Communities in Membrane Biofilm Reactors Using a High-Density Microarray
Abstract
1. Introduction
2. Materials and Methods
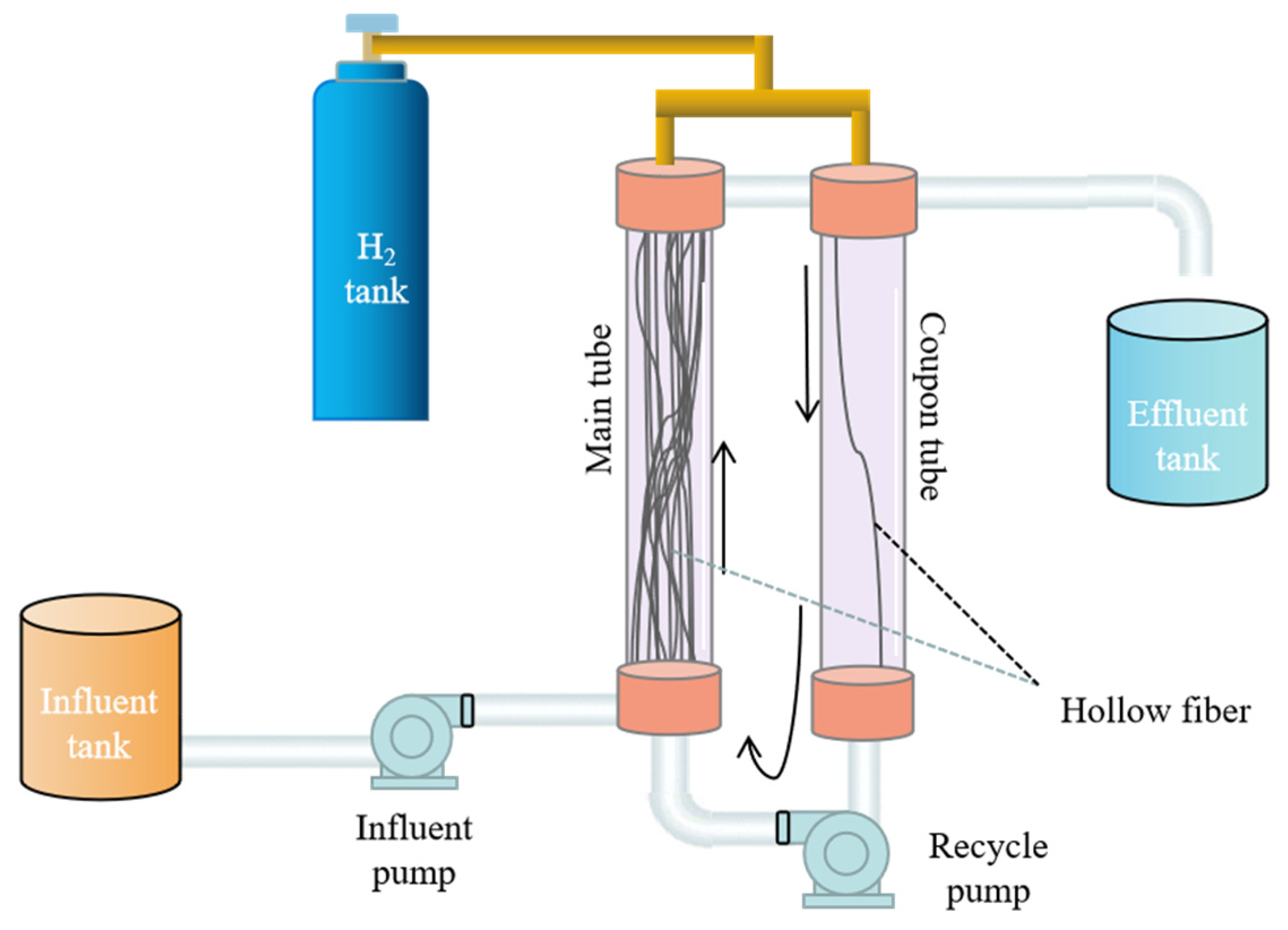
2.1. Membrane Biofilm Reactor (MBfR) Operation
2.2. Analytical Methods for Water Quality Parameters
2.3. High-Density Microarray
2.4. Phylogenetic Analysis
3. Results and Discussion
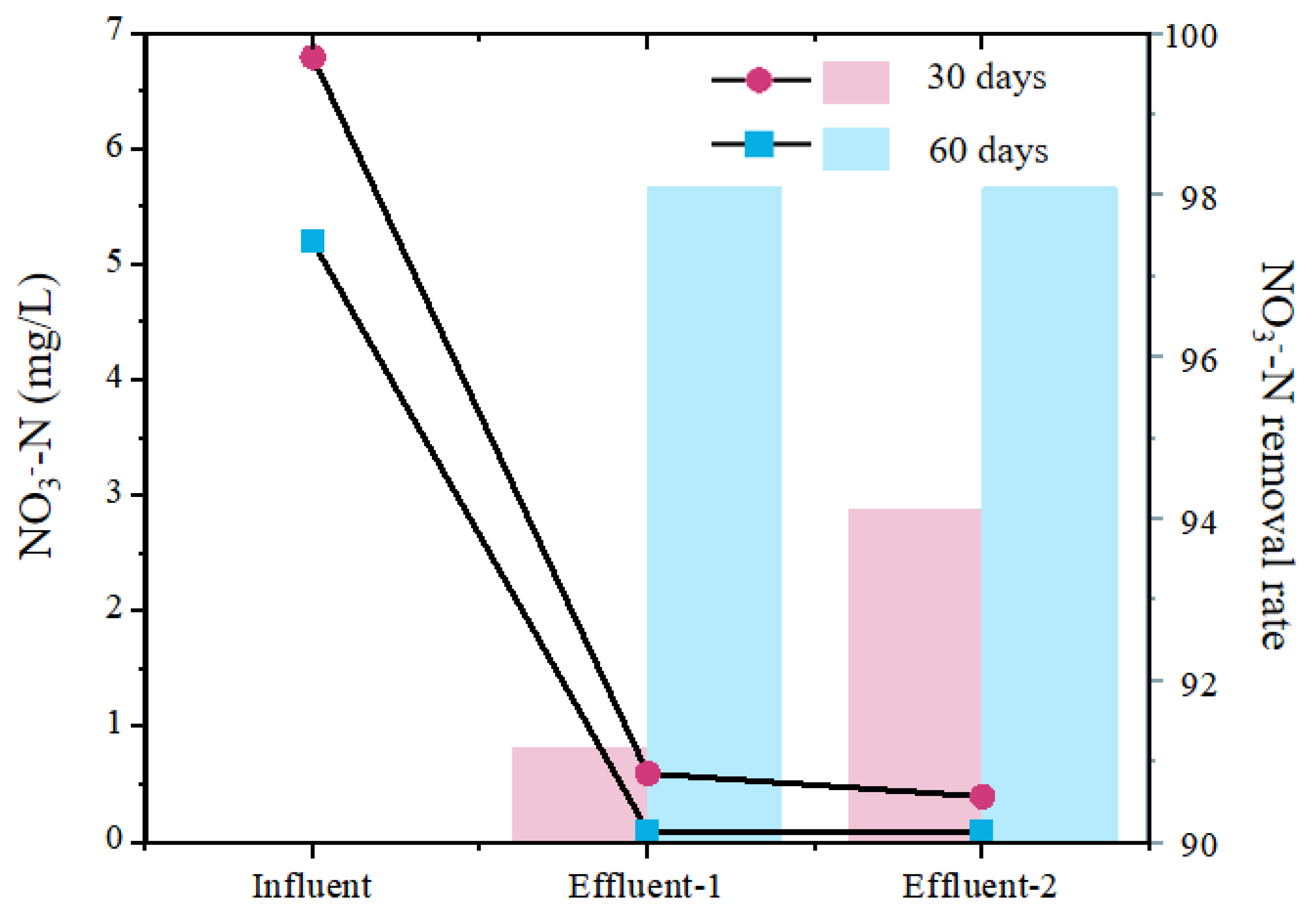
3.1. The Process Performance
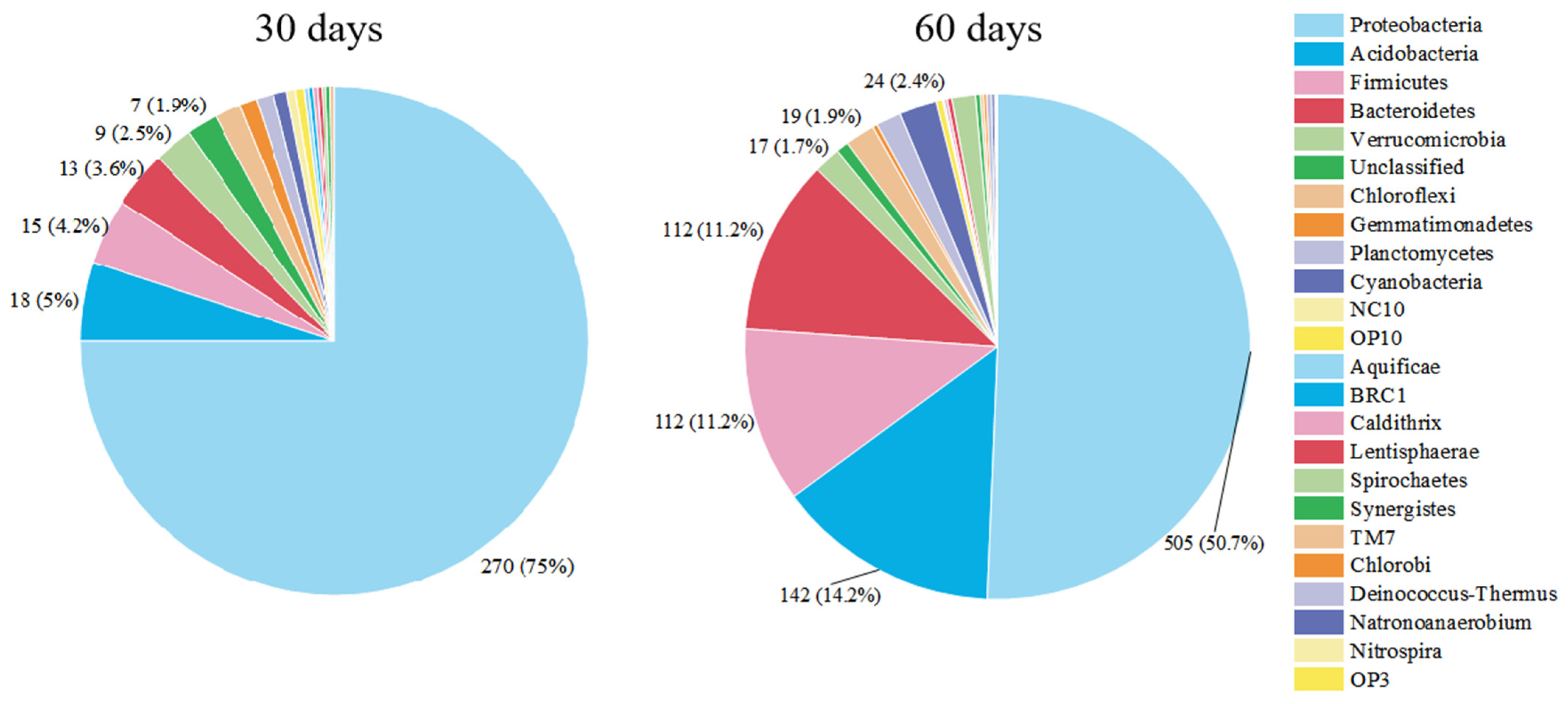
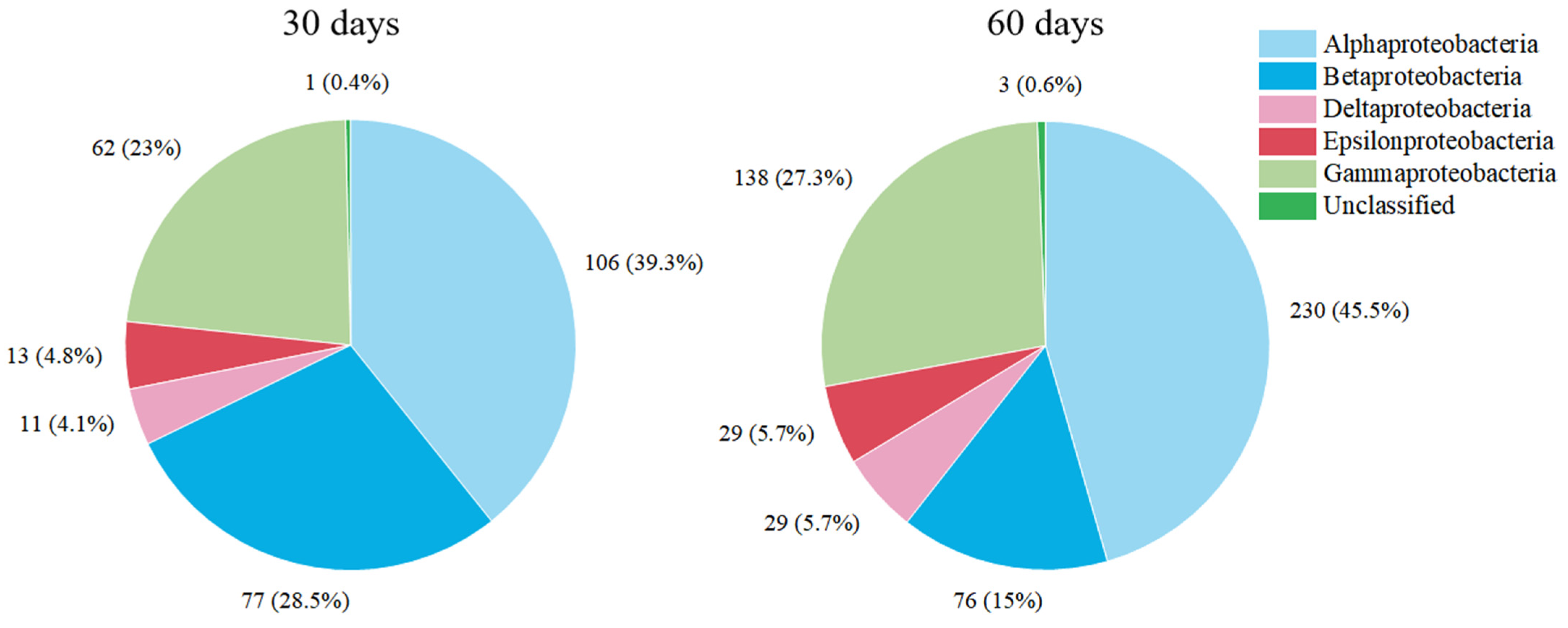
3.2. Succession of Relative Abundance
3.3. The Concentration of Microbial Communities
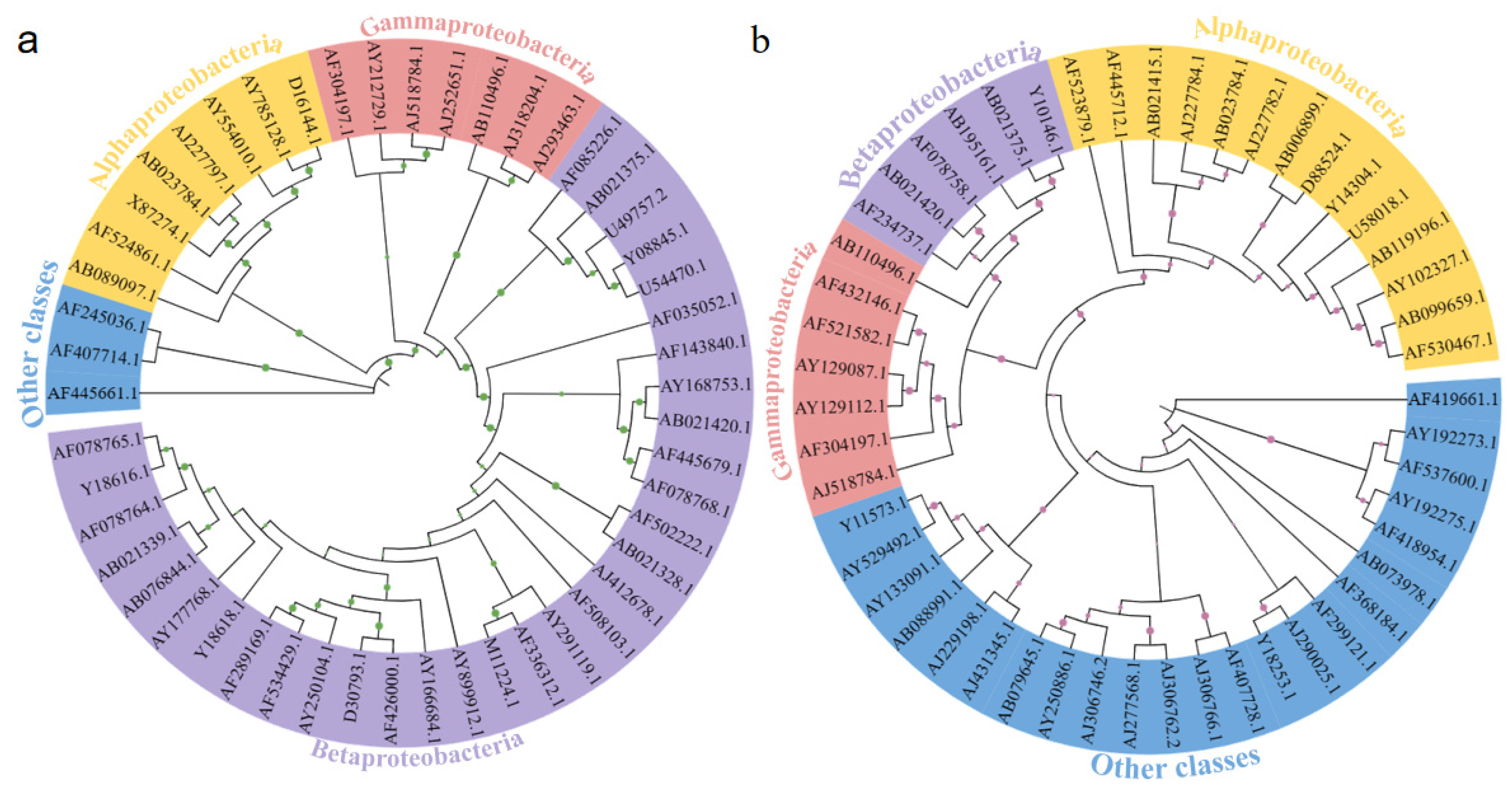
3.4. The Phylogenetic Relationships of Microbial Communities
3.5. The Functional Microbial Communities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Duan, L.; Liu, X.; Song, Y. Study on the Changes in the Microcosmic Environment in Forward Osmosis Membranes to Reduce Membrane Resistance. Membranes 2022, 12, 1203. [Google Scholar] [CrossRef]
- Zhao, Y.; Duan, L.; Liu, X.; Song, Y. Forward Osmosis Technology and Its Application on Microbial Fuel Cells: A Review. Membranes 2022, 12, 1254. [Google Scholar] [CrossRef]
- Zhao, Y.; Duan, L.; Liu, X.; Song, Y. Influence of Membrane Fouling and Reverse Salt Flux on Membrane Impedance of Forward Osmosis Microbial Fuel Cell. Membranes 2022, 12, 1165. [Google Scholar] [CrossRef]
- Hou, H.; Mengting, Z.; Duan, L.; Zhao, Y.; Zhang, Z.; Yao, M.; Zhou, B.; Zhang, H.; Hermanowicz, S.W. Removal performance and biodegradation mechanism of sulfonamides antibiotic contained wastewater by IFAS-MBR bioreactor. J. Mol. Liq. 2022, 367, 120572. [Google Scholar] [CrossRef]
- Yao, M.; Duan, L.; Song, Y.; Hermanowicz, S.W. Degradation mechanism of Ibuprofen via a forward osmosis membrane bioreactor. Bioresour. Technol. 2021, 321, 124448. [Google Scholar] [CrossRef]
- Hasar, H. Simultaneous removal of organic matter and nitrogen compounds by combining a membrane bioreactor and a membrane biofilm reactor. Bioresour. Technol. 2009, 100, 2699–2705. [Google Scholar] [CrossRef] [PubMed]
- Van Ginkel, S.W.; Yang, Z.; Kim, B.O.; Sholin, M.; Rittmann, B.E. The removal of selenate to low ppb levels from flue gas desulfurization brine using the H-2-based membrane biofilm reactor (MBfR). Bioresour. Technol. 2011, 102, 6360–6364. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yin, Y.; Wang, J. Hydrogen-based membrane biofilm reactors for nitrate removal from water and wastewater. Int. J. Hydrogen Energy 2018, 43, 1–15. [Google Scholar] [CrossRef]
- Lu, P.; Li, Z.; Zhao, H. Advances in water pollution control and resource recovery by membrane-supported biofilm reactor. Microbiol. China 2020, 47, 3287–3304. [Google Scholar]
- Duan, L.; Song, Y.; Xia, S.; Hermanowicz, S.W.; Jin, F.; Zhou, Q.; Wu, B. Detection of microbial communities in continuous and discontinuous membrane bioreactor using high-density oligonucleotide Microarray. In AIP Conference Proceedings; American Institute of Physics: College Park, MD, USA, 2010; Volume 1251. [Google Scholar]
- Duan, L.; Li, S.; Han, L.; Song, Y.; Zhou, B.; Zhang, J. Comparison between moving bed-membrane bioreactor and conventional membrane bioreactor systems. Part I: Membrane fouling. Environ. Earth Sci. 2015, 73, 4881–4890. [Google Scholar] [CrossRef]
- Tang, B.; Chen, Q.; Bin, L.; Huang, S.; Zhang, W.; Fu, F.; Li, P. Insight into the microbial community and its succession of a coupling anaerobic-aerobic biofilm on semi-suspended bio-carriers. Bioresour. Technol. 2018, 247, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Lv, P.; Zhang, J.; Fane, A.G.; McDougald, D.; Rice, S.A. Succession of biofilm communities responsible for biofouling of membrane bio-reactors (MBRs). PLoS ONE 2017, 12, e0179855. [Google Scholar] [CrossRef]
- Zhou, J.H. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 2003, 6, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, A.; Francois, P.; Charbonnier, Y.; Tangomo-Bento, M.; Bonetti, E.J.; Paster, B.J.; Bolivar, I.; Baratti-Mayer, D.; Pittet, D.; Schrenzel, J. Novel Microarray design strategy to study complex bacterial communities. Appl. Environ. Microbiol. 2008, 74, 1876–1885. [Google Scholar] [CrossRef]
- Xia, S.; Duan, L.; Song, Y.; Li, J.; Piceno, Y.M.; Andersen, G.L.; Alvarez-Cohen, L.; Moreno-Andrade, I.; Huang, C.-L.; Hermanowicz, S.W. Bacterial Community Structure in Geographically Distributed Biological Wastewater Treatment Reactors. Environ. Sci. Technol. 2010, 44, 7391–7396. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Song, Y.; Xia, S.; Hermanowicz, S.W. Characterization of nitrifying microbial community in a submerged membrane bioreactor at short solids retention times. Bioresour. Technol. 2013, 149, 200–207. [Google Scholar] [CrossRef]
- Salman, M.Y.; Taşkan, E.; Hasar, H. Comparative potentials of H2- and O2-MBfRs in removing multiple tetracycline antibiotics. Process Saf. Environ. Prot. 2022, 167, 184–191. [Google Scholar] [CrossRef]
- Spain, A.M.; Krumholz, L.R.; Elshahed, M.S. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME J. 2009, 3, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Loy, A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002, 13, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; Van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Luo, L.; Yao, J.; Liu, W.; Yang, L.; Li, H.; Liang, M.; Ma, H.; Liu, Z.; Chen, Y. Comparison of bacterial communities and antibiotic resistance genes in oxidation ditches and membrane bioreactors. Sci. Rep. 2021, 11, 8955. [Google Scholar] [CrossRef]
- Johan, L.; Sara, M.L. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol. 2020, 110, 63–98. [Google Scholar]
- Justice, N.B.; Norman, A.; Brown, C.T.; Singh, A.; Thomas, B.C.; Banfield, J.F. Comparison of environmental and isolate Sulfobacillus genomes reveals diverse carbon, sulfur, nitrogen, and hydrogen metabolisms. BMC Genom. 2014, 15, 1107. [Google Scholar] [CrossRef]
- Wang, Q.; Han, Y.; Lan, S.; Hu, C. Metagenomic Insight into Patterns and Mechanism of Nitrogen Cycle During Biocrust Succession. Front. Microbiol. 2021, 12, 633428. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Wen, L.-L.; Shi, L.-D.; Zhao, K.-K.; Wang, Y.-Q.; Yang, X.; Rittmann, B.E.; Zhou, C.; Tang, Y.; Zheng, P.; et al. Selenate and Nitrate Bioreductions Using Methane as the Electron Donor in a Membrane Biofilm Reactor. Environ. Sci. Technol. 2016, 50, 10179–10186. [Google Scholar] [CrossRef]
- Van Ginkel, S.W.; Lamendella, R.; Kovacik Jr, W.P.; Santo Domingo, J.W.; Rittmann, B.E. Microbial community structure during nitrate and perchlorate reduction in ion-exchange brine using the hydrogen-based membrane biofilm reactor (MBfR). Bioresour. Technol. 2010, 101, 3747–3750. [Google Scholar] [CrossRef]
- Li, S.; Duan, L.; Song, Y.; Hermanowicz, S.W. High-Density Microarray Analysis of Microbial Community Structures in Membrane Bioreactor at Short Sludge Retention Time. Membranes 2023, 13, 146. [Google Scholar] [CrossRef]
- Afzaal, M.; Saeed, F.; Ahmed, A.; Saeed, M.; Ateeq, H. Chapter 21—Hydrogels as Carrier for the Delivery of Probiotics; Singh, J., Vyas, A., Eds.; Advances in Dairy Microbial Products; Woodhead Publishing: Sawston, UK, 2022; pp. 303–315. [Google Scholar]
- Celmer, D.; Oleszkiewicz, J.A.; Cicek, N. Impact of shear force on the biofilm structure and performance of a membrane biofilm reactor for tertiary hydrogen-driven denitrification of municipal wastewater. Water Res. 2008, 42, 3057–3065. [Google Scholar] [CrossRef]
- Yoon, D.-N.; Park, S.-J.; Kim, S.-J.; Jeon, C.O.; Chae, J.-C.; Rhee, S.-K. Isolation, characterization, and abundance of filamentous members of Caldilineae in activated sludge. J. Microbiol. 2010, 48, 275–283. [Google Scholar] [CrossRef]
- Green, S.J.; Prakash, O.; Gihring, T.M.; Akob, D.M.; Jasrotia, P.; Jardine, P.M.; Watson, D.B.; Brown, S.D.; Palumbo, A.V.; Kostka, J.E. Denitrifying Bacteria Isolated from Terrestrial Subsurface Sediments Exposed to Mixed-Waste Contamination. Appl. Environ. Microbiol. 2010, 76, 3244–3254. [Google Scholar] [CrossRef]
- Sun, D.D.; Khor, S.L.; Hay, C.T.; Leckie, J.O. Impact of prolonged sludge retention time on the performance of a submerged membrane bioreactor. Desalination 2006, 208, 101–112. [Google Scholar]
- Martinage, V.; Paul, E. Effect of environmental parameters on autotrophic decay rate (b(A)). Environ. Technol. 2000, 21, 31–41. [Google Scholar] [CrossRef]
- Park, J.-H.; Choi, O.; Lee, T.-H.; Kim, H.; Sang, B.-I. Pyrosequencing analysis of microbial communities in hollow fiber-membrane biofilm reactors system for treating high-strength nitrogen wastewater. Chemosphere 2016, 163, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xia, S.; Zhong, F.; Yang, N. A polyvinyl chloride hollow membrane biofilm reactor for autohydrogenotrophic denitrification of drinking water. In Proceedings of the ICEET: 2009 International Conference on Energy and Environment Technology, Guilin, China, 16–18 October 2009; Volume 2, p. 385. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, Y.; Yuan, Y.; Chen, Y.; Lin, H.; Zheng, J.; Li, H.; Zhang, X. Nitrate Removal and Dynamics of Microbial Community of a Hydrogen-Based Membrane Biofilm Reactor at Diverse Nitrate Loadings and Distances from Hydrogen Supply End. Water 2020, 12, 3196. [Google Scholar] [CrossRef]
- Runzhu, M.; Jianfang, W.; Jiaqi, C.; Junjie, Z. Denitrification efficiency and impact factors of a hydrogen-based membrane biofilm reactor. Chin. J. Environ. Eng. 2022, 16, 2425–2435. [Google Scholar]
- Kämpfer, P.; Schulze, R.; Jäckel, U.; Malik, K.A.; Amann, R.; Spring, S. Hydrogenophaga defluvii sp nov and Hydrogenophaga atypica sp nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2005, 55, 341–344. [Google Scholar] [CrossRef]

| Genus | Species | Function | Relative Fluorescence Intensity | Sample | Reference |
|---|---|---|---|---|---|
| Marinobacter | Marinobacter hydrocarbonoclasticus | Nitrate-reducing bacteria | 2112–2098 | 30d–60d | [27] |
| Thiobacillus | Thiobacillus denitrificans | Hydrogenotrophic denitrifying bacteria | 1165 | 30d | [35] |
| Hydrogenophaga | Hydrogenophaga pseudoflava | Hydrogenotrophic denitrifying bacteria | 4766–3036 | 30d–60d | [35,36] |
| Hydrogenophaga electricum | 4552–2532 | ||||
| Hydrogenophaga taeniospiralis | 5552–2864 | ||||
| Rhodobacter | Paracoccus denitrificans | Autotrophic denitrifying bacteria | 1593–2643 | 30d–60d | [35,37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Duan, L.; Zhao, Y.; Gao, F.; Hermanowicz, S.W. Analysis of Microbial Communities in Membrane Biofilm Reactors Using a High-Density Microarray. Membranes 2023, 13, 324. https://doi.org/10.3390/membranes13030324
Li S, Duan L, Zhao Y, Gao F, Hermanowicz SW. Analysis of Microbial Communities in Membrane Biofilm Reactors Using a High-Density Microarray. Membranes. 2023; 13(3):324. https://doi.org/10.3390/membranes13030324
Chicago/Turabian StyleLi, Shilong, Liang Duan, Yang Zhao, Fu Gao, and Slawomir W. Hermanowicz. 2023. "Analysis of Microbial Communities in Membrane Biofilm Reactors Using a High-Density Microarray" Membranes 13, no. 3: 324. https://doi.org/10.3390/membranes13030324
APA StyleLi, S., Duan, L., Zhao, Y., Gao, F., & Hermanowicz, S. W. (2023). Analysis of Microbial Communities in Membrane Biofilm Reactors Using a High-Density Microarray. Membranes, 13(3), 324. https://doi.org/10.3390/membranes13030324










