Heterologous Display of Chlamydia trachomatis PmpD Passenger at the Surface of Salmonella OMVs
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Reagents and Sera
2.3. Plasmid Construction
2.4. Recombinant Protein Expression
2.5. Isolation of Cell Envelopes and Outer Membrane Fractions
2.6. OMVs Isolation
2.7. Proteinase K Accessibility Assay
3. Results
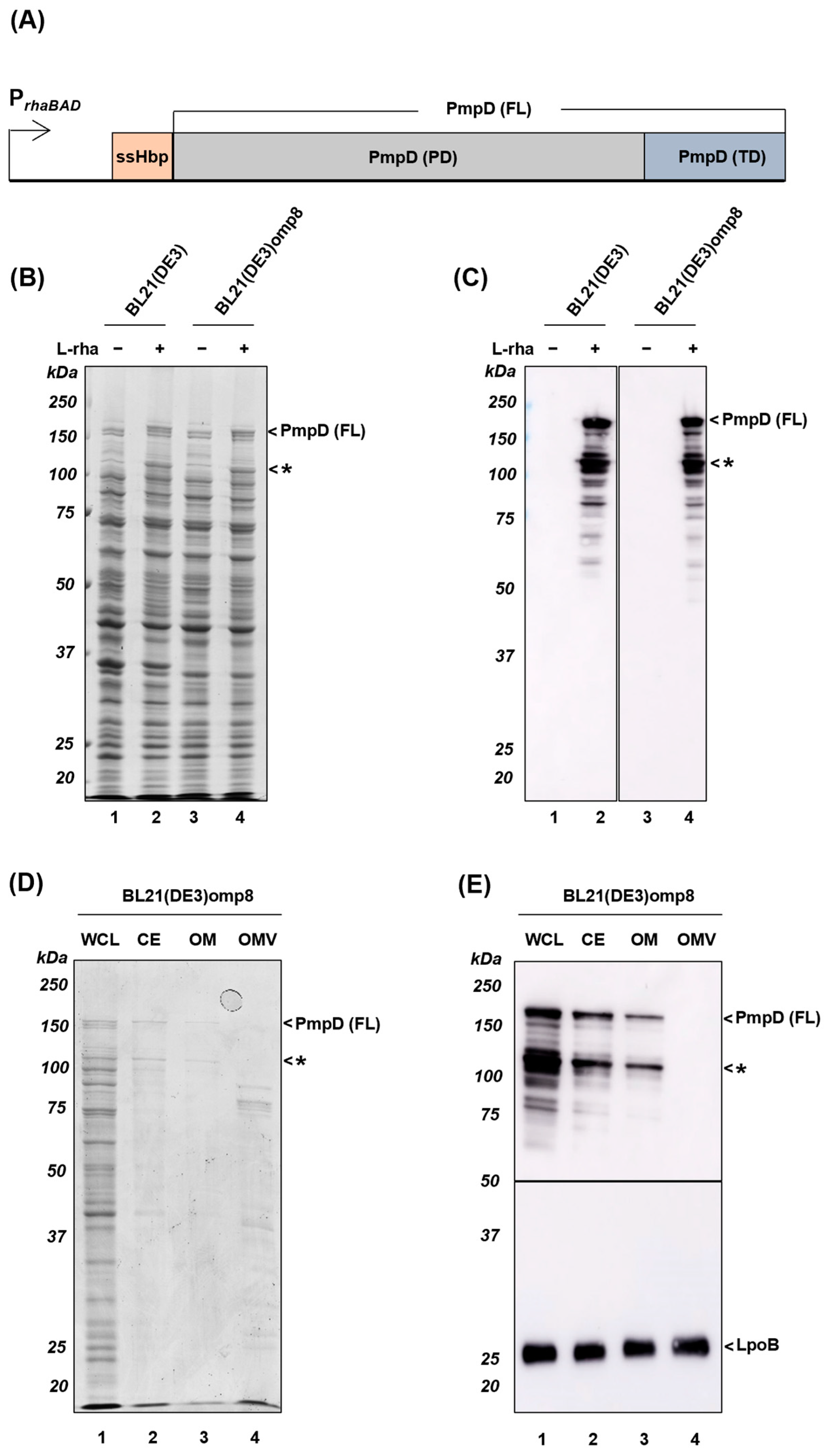
3.1. Expression and Localization of Full-Length PmpD (FL)
3.2. Display of Antigenic Passengers upon Fusion to Hbp in E. coli
3.3. Display of Truncated Passengers upon Fusion to Hbp in Salmonella
3.4. Display of Antigenic Passengers upon Seamless Fusion to the β-Helical Stem of Hbp
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Panzetta, M.E.; Valdivia, R.H.; Saka, H.A. Chlamydia Persistence: A Survival Strategy to Evade Antimicrobial Effects in-Vitro and in-Vivo. Front. Microbiol. 2018, 9, 3101. [Google Scholar] [CrossRef] [PubMed]
- Murray, S.M.; McKay, P.F. Chlamydia Trachomatis: Cell Biology, Immunology and Vaccination. Vaccine 2021, 39, 2965–2975. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.; Quigley, B.L.; Timms, P. Seventy Years of Chlamydia Vaccine Research–Limitations of the Past and Directions for the Future. Front. Microbiol. 2019, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; Juel, H.B.; Bang, P.; Cheeseman, H.M.; Dohn, R.B.; Cole, T.; Kristiansen, M.P.; Korsholm, K.S.; Lewis, D.; Olsen, A.W.; et al. Safety and Immunogenicity of the Chlamydia Vaccine Candidate CTH522 Adjuvanted with CAF01 Liposomes or Aluminium Hydroxide: A First-in-Human, Randomised, Double-Blind, Placebo-Controlled, Phase 1 Trial. Lancet Infect. Dis. 2019, 19, 1091–1100. [Google Scholar] [CrossRef]
- Feher, V.A.; Randall, A.; Baldi, P.; Bush, R.M.; de la Maza, L.M.; Amaro, R.E. A 3-Dimensional Trimeric β-Barrel Model for Chlamydia MOMP Contains Conserved and Novel Elements of Gram-Negative Bacterial Porins. PLoS ONE 2013, 8, e68934. [Google Scholar] [CrossRef]
- Hafner, L.M.; Wilson, D.P.; Timms, P. Development Status and Future Prospects for a Vaccine against Chlamydia Trachomatis Infection. Vaccine 2014, 32, 1563–1571. [Google Scholar] [CrossRef]
- Vasilevsky, S.; Stojanov, M.; Greub, G.; Baud, D. Chlamydial Polymorphic Membrane Proteins: Regulation, Function and Potential Vaccine Candidates. Virulence 2016, 7, 11–22. [Google Scholar] [CrossRef]
- Kiselev, A.O.; Stamm, W.E.; Yates, J.R.; Lampe, M.F. Expression, Processing, and Localization of PmpD of Chlamydia Trachomatis Serovar L2 during the Chlamydial Developmental Cycle. PLoS ONE 2007, 2, e568. [Google Scholar] [CrossRef]
- Van Ulsen, P.; Zinner, K.M.; Jong, W.S.P.; Luirink, J. On Display: Autotransporter Secretion and Application. FEMS Microbiol. Lett. 2018, 365, fny165. [Google Scholar] [CrossRef]
- Clarke, K.R.; Hor, L.; Pilapitiya, A.; Luirink, J.; Paxman, J.J.; Leo, J.C. Phylogenetic Classi Fi Cation and Functional Review of Autotransporters. Front. Immunol. 2022, 13, 921272. [Google Scholar] [CrossRef]
- Crane, D.D.; Carlson, J.H.; Fischer, E.R.; Bavoil, P.; Hsia, R.C.; Tan, C.; Kuo, C.C.; Caldwell, H.D. Chlamydia Tracomatis Polymorphic Membrane Protein D Is a Species-Common Pan-Neutralizing Antigen. Proc. Natl. Acad. Sci. USA 2006, 103, 1894–1899. [Google Scholar] [CrossRef]
- Swanson, K.A.; Taylor, L.D.; Frank, S.D.; Sturdevant, G.L.; Fischer, E.R.; Carlson, J.H.; Whitmire, W.M.; Caldwell, H.D. Chlamydia Trachomatis Polymorphic Membrane Protein D Is an Oligomeric Autotransporter with a Higher-Order Structure. Infect. Immun. 2009, 77, 508–516. [Google Scholar] [CrossRef]
- Paes, W.; Brown, N.; Brzozowski, A.M.; Coler, R.; Reed, S.; Carter, D.; Bland, M.; Kaye, P.M.; Lacey, C.J.N. Recombinant Polymorphic Membrane Protein D in Combination with a Novel, Second-Generation Lipid Adjuvant Protects against Intra-Vaginal Chlamydia Trachomatis Infection in Mice. Vaccine 2016, 34, 4123–4131. [Google Scholar] [CrossRef]
- Pais, R.; Omosun, Y.; Igietseme, J.U.; Fujihashi, K.; Eko, F.O. Route of Vaccine Administration Influences the Impact of Fms-like Tyrosine Kinase 3 Ligand (Flt3L) on Chlamydial-Specific Protective Immune Responses. Front. Immunol. 2019, 10, 1577. [Google Scholar] [CrossRef]
- Russi, R.C.; Del Balzo, D.; Luján, A.; Reidel, I.G.; García, M.I.; Veaute, C.; Damiani, M.T. Heterologous Prime-Boost Vaccination Based on Polymorphic Protein D Protects against Intravaginal Chlamydia Trachomatis Infection in Mice. Sci. Rep. 2022, 12, 6664. [Google Scholar] [CrossRef]
- Waltz, E. How Nasal-Spray Vaccines Could Change the Pandemic. Nature 2022, 609, 240–242. [Google Scholar] [CrossRef]
- van der Ley, P.A.; Zariri, A.; van Riet, E.; Oosterhoff, D.; Kruiswijk, C.P. An Intranasal OMV-Based Vaccine Induces High Mucosal and Systemic Protecting Immunity against a SARS-CoV-2 Infection. Front. Immunol. 2021, 12, 781280. [Google Scholar] [CrossRef]
- Portilho, A.I.; Correa, V.A.; Cirqueira, S. Intranasal and Intramuscular Immunization with Outer Membrane Vesicles from Serogroup C Meningococci Induced Functional Antibodies and Immunologic Memory Intranasal and Intramuscular Immunization with Outer Membrane Vesicles from Serogroup C Meningococci. Immunol. Investig. 2022, 51, 2066–2085. [Google Scholar] [CrossRef]
- Krishnan, N.; Kubiatowicz, L.J.; Holay, M.; Zhou, J.; Fang, R.H.; Zhang, L. Bacterial Membrane Vesicles for Vaccine Applications. Adv. Drug Deliv. Rev. 2022, 185, 114294. [Google Scholar] [CrossRef]
- Bai, X.; Findlow, J.; Borrow, R. Recombinant Protein Meningococcal Serogroup B Vaccine Combined with Outer Membrane Vesicles. Expert Opin. Biol. Ther. 2011, 11, 969–985. [Google Scholar] [CrossRef]
- Kuipers, K.; Jong, W.S.P.; van der Gaast-de Jongh, C.E.; Houben, D.; van Opzeeland, F.; Simonetti, E.; van Selm, S.; de Groot, R.; Koenders, M.I.; Azarian, T.; et al. Th17-Mediated Cross Protection against Pneumococcal Carriage by Vaccination with a Variable Antigen. Infect. Immun. 2017, 85, e00281-17. [Google Scholar] [CrossRef] [PubMed]
- Daleke-Schermerhorn, M.H.; Felix, T.; Soprova, Z.; ten Hagen-Jongman, C.M.; Vikström, D.; Majlessi, L.; Beskers, J.; Follmann, F.; de Punder, K.; van der Wel, N.N.; et al. Decoration of Outer Membrane Vesicles with Multiple Antigens by Using an Autotransporter Approach. Appl. Environ. Microbiol. 2014, 80, 5854–5865. [Google Scholar] [CrossRef] [PubMed]
- Jong, W.S.P.; Schillemans, M.; ten Hagen-Jongman, C.M.; Luirink, J.; van Ulsen, P. Comparing Autotransporter β-Domain Configurations for Their Capacity to Secrete Heterologous Proteins to the Cell Surface. PLoS ONE 2018, 13, e0191622. [Google Scholar] [CrossRef] [PubMed]
- van den Berg van Saparoea, H.B.; Houben, D.; de Jonge, M.I.; Jong, W.S.P.; Luirink, J. Display of Recombinant Proteins on Bacterial Outer Membrane Vesicles by Using Protein Ligation. Appl. Environ. Microbiol. 2018, 84, 1–17. [Google Scholar] [CrossRef]
- Kuipers, K.; Daleke-Schermerhorn, M.H.; Jong, W.S.P.; ten Hagen-Jongman, C.M.; van Opzeeland, F.; Simonetti, E.; Luirink, J.; de Jonge, M.I. Salmonella Outer Membrane Vesicles Displaying High Densities of Pneumococcal Antigen at the Surface Offer Protection against Colonization. Vaccine 2015, 33, 2022–2029. [Google Scholar] [CrossRef]
- Hays, M.P.; Houben, D.; Yang, Y.; Luirink, J.; Hardwidge, P.R. Immunization with Skp Delivered on Outer Membrane Vesicles Protects Mice against Enterotoxigenic Escherichia Coli CHALLENGE. Front. Cell. Infect. Microbiol. 2018, 8, 132. [Google Scholar] [CrossRef]
- Jiang, L.; Driedonks, T.A.P.; Jong, W.S.P.; Dhakal, S.; Bart van den Berg van Saparoea, H.; Sitaras, I.; Zhou, R.; Caputo, C.; Littlefield, K.; Lowman, M.; et al. A Bacterial Extracellular Vesicle-based Intranasal Vaccine against SARS-CoV-2 Protects against Disease and Elicits Neutralizing Antibodies to Wild-type and Delta Variants. J. Extracell. Vesicles 2022, 11, e12192. [Google Scholar] [CrossRef]
- Prilipov, A.; Phale, P.S.; Van Gelder, P.; Rosenbusch, J.P.; Koebnik, R. Coupling Site-Directed Mutagenesis with High-Level Expression: Large Scale Production of Mutant Porins from E. Coli. FEMS Microbiol. Lett. 1998, 163, 65–72. [Google Scholar] [CrossRef]
- Strauch, K.L.; Johnson, K.I.T.; Beckwith, J.O.N. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 1989, 171, 2689–2696. [Google Scholar] [CrossRef]
- Otto, B.R.; Van Dooren, S.J.M.; Dozois, C.M.; Luirink, J.; Oudega, B. Escherichia Coli Hemoglobin Protease Autotransporter Contributes to Synergistic Abscess Formation and Heme-Dependent Growth of Bacteroides Fragilis. Infect. Immun. 2002, 70, 5–10. [Google Scholar] [CrossRef]
- Van Dooren, S.J.M.; Tame, J.R.H.; Luirink, J.; Oudega, B.; Otto, B.R. Purification of the Autotransporter Protein Hbp of Escherichia Coli. FEMS Microbiol. Lett. 2001, 205, 147–150. [Google Scholar] [CrossRef]
- Consoli, E.; Collet, J.F.; Blaauwen, T. Den the Escherichia Coli Outer Membrane Β-barrel Assembly Machinery (Bam) Anchors the Peptidoglycan Layer by Spanning It with All Subunits. Int. J. Mol. Sci. 2021, 22, 1853. [Google Scholar] [CrossRef]
- Egan, A.J.F.; Jean, N.L.; Koumoutsi, A.; Bougault, C.M.; Biboy, J.; Sassine, J.; Solovyova, A.S.; Breukink, E.; Typas, A.; Vollmer, W.; et al. Outer-Membrane Lipoprotein LpoB Spans the Periplasm to Stimulate the Peptidoglycan Synthase PBP1B. Proc. Natl. Acad. Sci. USA 2014, 111, 8197–8202. [Google Scholar] [CrossRef]
- Robert, V.; Volokhina, E.B.; Senf, F.; Bos, M.P.; Van Gelder, P.; Tommassen, J. Assembly Factor Omp85 Recognizes Its Outer Membrane Protein Substrates by a Species-Specific C-Terminal Motif. PLoS Biol. 2006, 4, 1984–1995. [Google Scholar] [CrossRef]
- Wagner, S.; Klepsch, M.M.; Schlegel, S.; Appel, A.; Draheim, R.; Tarry, M.; Högbom, M.; Van Wijk, K.J.; Slotboom, D.J.; Persson, J.O.; et al. Tuning Escherichia Coli for Membrane Protein Overexpression. Proc. Natl. Acad. Sci. USA 2008, 105, 14371–14376. [Google Scholar] [CrossRef]
- Schlegel, S.; Löfblom, J.; Lee, C.; Hjelm, A.; Klepsch, M.; Strous, M.; Drew, D.; Slotboom, D.J.; De Gier, J.W. Optimizing Membrane Protein Overexpression in the Escherichia Coli Strain Lemo21(DE3). J. Mol. Biol. 2012, 423, 648–659. [Google Scholar] [CrossRef]
- Hashemzadeh-Bonehi, L.; Mehraein-Ghomi, F.; Mitsopoulos, C.; Jacob, J.P.; Hennessey, E.S.; Broome-Smith, J.K. Importance of Using Lac Rather than Ara Promoter Vectors for Modulating the Levels of Toxic Gene Products in Escherichia Coli. Mol. Microbiol. 1998, 30, 676–678. [Google Scholar] [CrossRef]
- Mooi, F.R.; Van Oirschot, H.; Heuvelman, K.; Van der Heide, H.G.J.; Gaastra, W.; Willems, R.J.L. Polymorphism in the Bordetella Pertussis Virulence Factors P.69/Pertactin and Pertussis Toxin in The Netherlands: Temporal Trends and Evidence for Vaccine-Driven Evolution. Infect. Immun. 1998, 66, 670–675. [Google Scholar] [CrossRef]
- Zarzecka, U.; Modrak-Wójcik, A.; Figaj, D.; Apanowicz, M.; Lesner, A.; Bzowska, A.; Lipinska, B.; Zawilak-Pawlik, A.; Backert, S.; Skorko-Glonek, J. Properties of the HtrA Protease from Bacterium Helicobacter Pylori Whose Activity Is Indispensable for Growth under Stress Conditions. Front. Microbiol. 2019, 10, 961. [Google Scholar] [CrossRef]
- Huynh, D.T.; Jong, W.S.P.; Koningstein, G.M.; van Ulsen, P.; Luirink, J. Overexpression of the Bam Complex Improves the Production of Chlamydia Trachomatis MOMP in the E. coli Outer Membrane. Int. J. Mol. Sci. 2022, 23, 7393. [Google Scholar] [CrossRef]
- König, E.; Gagliardi, A.; Riedmiller, I.; Andretta, C.; Tomasi, M.; Irene, C.; Frattini, L.; Zanella, I.; Berti, F.; Grandi, A.; et al. Multi-Antigen Outer Membrane Vesicle Engineering to Develop Polyvalent Vaccines: The Staphylococcus Aureus Case. Front. Immunol. 2021, 12, 752168. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, N.; Chng, S.S.; Hinikera, A.; Kahne, D.; Silhavy, T.J. Nonconsecutive Disulfide Bond Formation in an Essential Integral Outer Membrane Protein. Proc. Natl. Acad. Sci. USA 2010, 107, 12245–12250. [Google Scholar] [CrossRef]
- Thoma, J.; Manioglu, S.; Kalbermatter, D.; Bosshart, P.D.; Fotiadis, D.; Müller, D.J. Protein-Enriched Outer Membrane Vesicles as a Native Platform for Outer Membrane Protein Studies. Commun. Biol. 2018, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Jong, W.S.P.; Ten Hagen-Jongman, C.M.; Den Blaauwen, T.; Jan Slotboom, D.; Tame, J.R.H.; Wickström, D.; De Gier, J.W.; Otto, B.R.; Luirink, J. Limited Tolerance towards Folded Elements during Secretion of the Autotransporter Hbp. Mol. Microbiol. 2007, 63, 1524–1536. [Google Scholar] [CrossRef]
- Paramasivam, N.; Habeck, M.; Linke, D. Is the C-Terminal Insertional Signal in Gram-Negative Bacterial Outer Membrane Proteins Species-Specific or Not? BMC Genom. 2012, 13, 510. [Google Scholar] [CrossRef]
- Tomasek, D.; Kahne, D. The Assembly of β-Barrel Outer Membrane Proteins. Curr. Opin. Microbiol. 2021, 60, 16–23. [Google Scholar] [CrossRef]
- Roussel-Jazédé, V.; Van Gelder, P.; Sijbrandi, R.; Rutten, L.; Otto, B.R.; Luirink, J.; Gros, P.; Tommassen, J.; Van Ulsen, P. Channel Properties of the Translocator Domain of the Autotransporter Hbp of Escherichia Coli. Mol. Membr. Biol. 2011, 28, 157–169. [Google Scholar] [CrossRef]
- Kiselev, A.O.; Skinner, M.C.; Lampe, M.F. Analysis of PmpD Expression and PmpD Post-Translational Processing during the Life Cycle of Chlamydia Trachomatis Serovars A, D, and L2. PLoS ONE 2009, 4, e5191. [Google Scholar] [CrossRef]
- Fantappie, L.; Irene, C.; De Santis, M.; Armini, A.; Gagliardi, A.; Tomasi, M.; Parri, M.; Cafardi, V.; Bonomi, S.; Ganfini, L.; et al. Some Gram-Negative Lipoproteins Keep Their Surface Topology When Transplanted from One Species to Another and Deliver Foreign Polypeptides to the Bacterial Surface. Mol. Cell. Proteom. 2017, 16, 1348–1364. [Google Scholar] [CrossRef]
- Doyle, M.T.; Bernstein, H.D. BamA Forms a Translocation Channel for Polypeptide Export across the Bacterial Outer Membrane. Mol. Cell 2021, 81, 2000–2012.e3. [Google Scholar] [CrossRef]
- Hart, E.M.; Silhavy, T.J. Functions of the BamBCDE Lipoproteins Revealed by Bypass Mutations in BamA. J. Bacteriol. 2020, 202, 1–16. [Google Scholar] [CrossRef]
- Wang, X.; Bernstein, H.D. The E. Coli Outer Membrane Protein OmpA Acquires Secondary Structure Prior to Its Integration into the Membrane. J. Biol. Chem. 2022, 298, 101802. [Google Scholar] [CrossRef]
- Harkness, R.W.; Toyama, Y.; Ripstein, Z.A.; Zhao, H.; Sever, A.I.M.; Luan, Q.; Brady, J.P.; Clark, P.L.; Schuck, P.; Kay, L.E. Competing Stress-Dependent Oligomerization Pathways Regulate Self-Assembly of the Periplasmic Protease-Chaperone DegP. Proc. Natl. Acad. Sci. USA 2021, 118, e2109732118. [Google Scholar] [CrossRef]
- Baclayon, M.; Van Ulsen, P.; Mouhib, H.; Shabestari, M.H.; Verzijden, T.; Abeln, S.; Roos, W.H.; Wuite, G.J.L. Mechanical Unfolding of an Autotransporter Passenger Protein Reveals the Secretion Starting Point and Processive Transport Intermediates. ACS Nano 2016, 10, 5710–5719. [Google Scholar] [CrossRef]
- Tozakidis, I.E.P.; Sichwart, S.; Jose, J. Going beyond E. Coli: Autotransporter Based Surface Display on Alternative Host Organisms. N. Biotechnol. 2015, 32, 644–650. [Google Scholar] [CrossRef]
- Nicolay, T.; Vanderleyden, J.; Spaepen, S. Autotransporter-Based Cell Surface Display in Gram-Negative Bacteria. Crit. Rev. Microbiol. 2015, 41, 109–123. [Google Scholar] [CrossRef]
- Sauri, A.; Soprova, Z.; Wickström, D.; de Gier, J.W.; Van der Schors, R.C.; Smit, A.B.; Jong, W.S.P.; Luirink, J. The Bam (Omp85) Complex Is Involved in Secretion of the Autotransporter Haemoglobin Protease. Microbiology 2009, 155, 3982–3991. [Google Scholar] [CrossRef]
- Phan, T.H.; Kuijl, C.; Huyn, D.; Jong, W.S.P.; Luirink, J.; Van Ulsen, P. Overproducing the BAM Complex Improves Secretion of Difficult-to-Secrete Recombinant Autotransporter Chimeras. Microb. Cell Fact. 2021, 20, 176. [Google Scholar] [CrossRef]
- Ma, H.; Cummins, D.D.; Edelstein, N.B.; Gomez, J.; Khan, A.; Llewellyn, M.D.; Picudella, T.; Willsey, S.R.; Nangia, S. Modeling Diversity in Structures of Bacterial Outer Membrane Lipids. J. Chem. Theory Comput. 2017, 13, 811–824. [Google Scholar] [CrossRef]
- Saurí, A.; Oreshkova, N.; Soprova, Z.; Jong, W.S.P.; Sani, M.; Peters, P.J.; Luirink, J.; Van Ulsen, P. Autotransporter β-Domains Have a Specific Function in Protein Secretion beyond Outer-Membrane Targeting. J. Mol. Biol. 2011, 412, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Juodeikis, R.; Carding, S.R. Outer Membrane Vesicles: Biogenesis, Functions, and Issues. Microbiol. Mol. Biol. Rev. 2022, 86, 1092–2172. [Google Scholar] [CrossRef] [PubMed]
- Soprova, Z.; Sauri, A.; Van Ulsen, P.; Tame, J.R.H.; Den Blaauwen, T.; Jong, W.S.P.; Luirink, J. A Conserved Aromatic Residue in the Autochaperone Domain of the Autotransporter Hbp Is Critical for Initiation of Outer Membrane Translocation. J. Biol. Chem. 2010, 285, 38224–38233. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, D.T.; Jong, W.S.P.; Oudejans, M.A.H.; Berg van Saparoea, H.B.v.d.; Luirink, J.; Ulsen, P.v. Heterologous Display of Chlamydia trachomatis PmpD Passenger at the Surface of Salmonella OMVs. Membranes 2023, 13, 366. https://doi.org/10.3390/membranes13040366
Huynh DT, Jong WSP, Oudejans MAH, Berg van Saparoea HBvd, Luirink J, Ulsen Pv. Heterologous Display of Chlamydia trachomatis PmpD Passenger at the Surface of Salmonella OMVs. Membranes. 2023; 13(4):366. https://doi.org/10.3390/membranes13040366
Chicago/Turabian StyleHuynh, Dung T., Wouter S. P. Jong, Manon A. H. Oudejans, H. Bart van den Berg van Saparoea, Joen Luirink, and Peter van Ulsen. 2023. "Heterologous Display of Chlamydia trachomatis PmpD Passenger at the Surface of Salmonella OMVs" Membranes 13, no. 4: 366. https://doi.org/10.3390/membranes13040366
APA StyleHuynh, D. T., Jong, W. S. P., Oudejans, M. A. H., Berg van Saparoea, H. B. v. d., Luirink, J., & Ulsen, P. v. (2023). Heterologous Display of Chlamydia trachomatis PmpD Passenger at the Surface of Salmonella OMVs. Membranes, 13(4), 366. https://doi.org/10.3390/membranes13040366





