The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties
Abstract
1. Introduction
2. Bioactive Peptides (BAPs)
2.1. Physicochemical Properties of BAPs
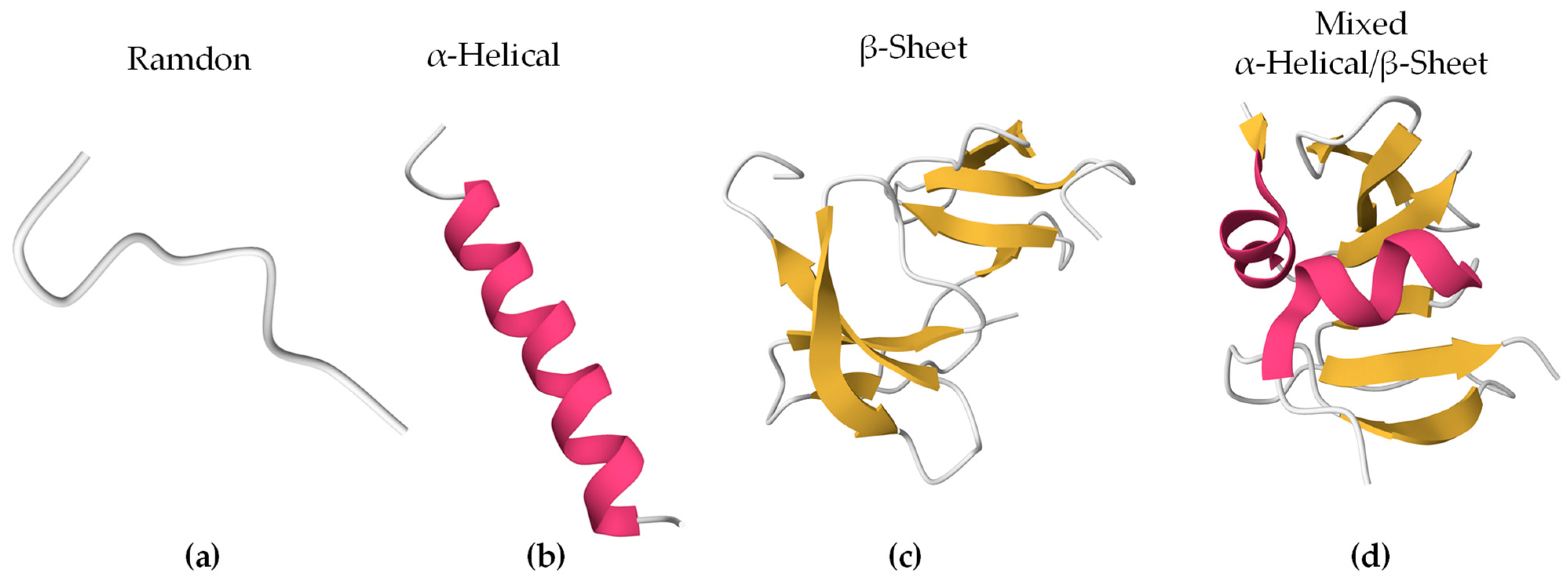
2.2. Structural Characteristics
3. Eukaryotic Membranes
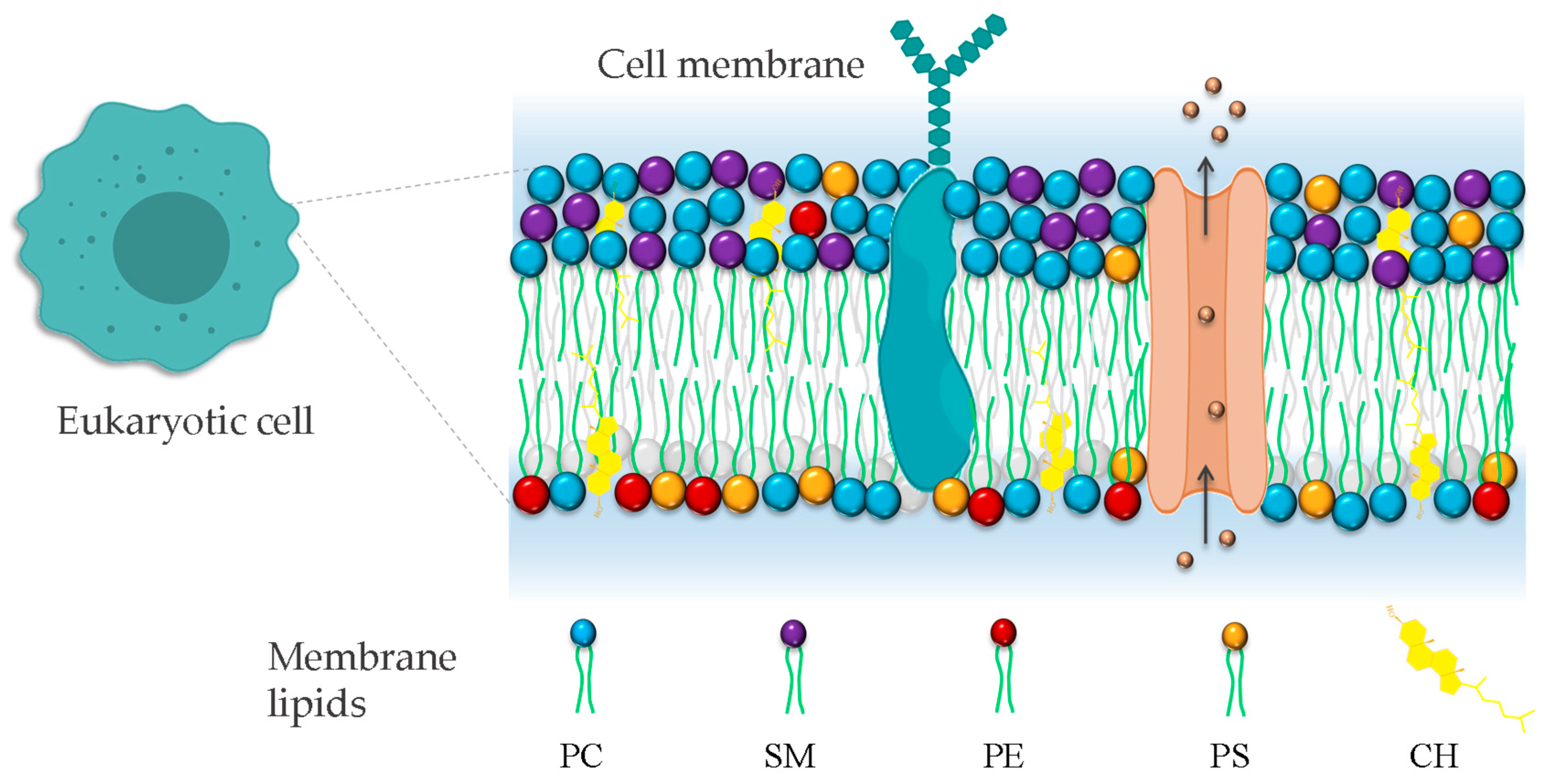
3.1. Structure of Membranes
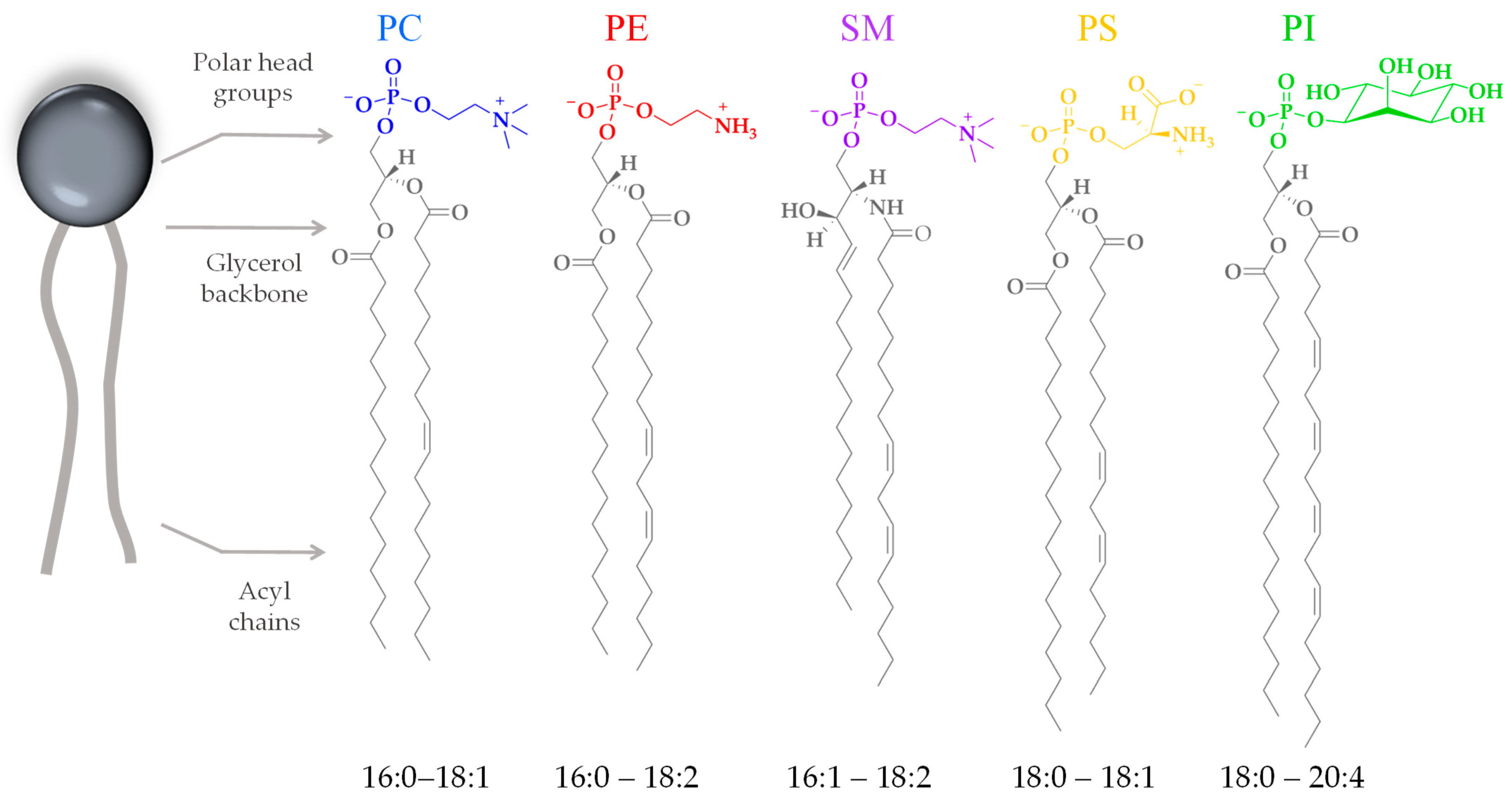
3.2. Lipid Composition
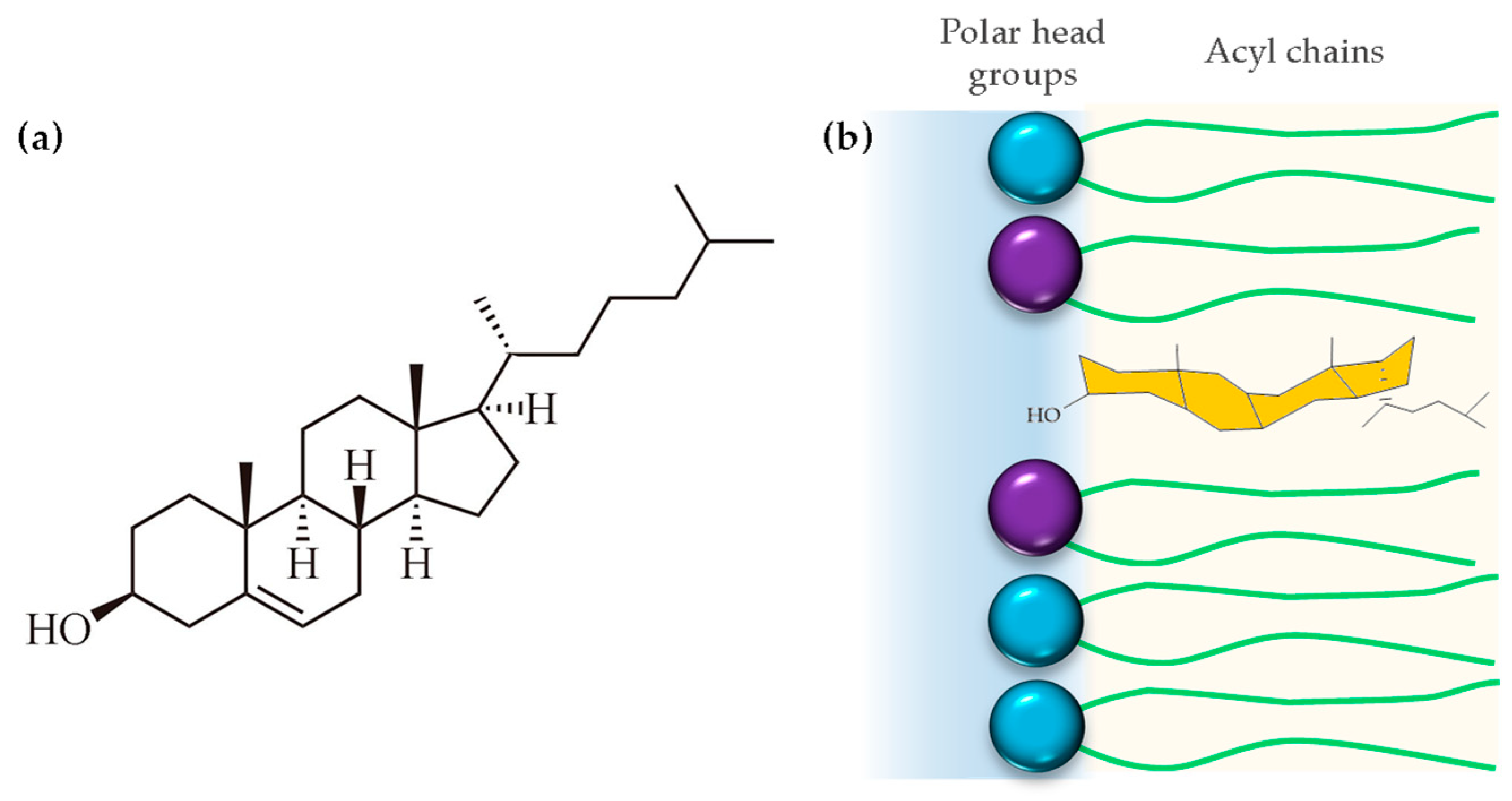
4. Cholesterol
4.1. Cholesterol as a Physiological and Structural Component
4.2. Cholesterol as a Membrane Fluidity Regulator
4.3. Cholesterol Asymmetry
5. Interaction of Cholesterol and BAPs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- van Meer, G. Membrane lipids, where they are and how they behave: Sphingolipids on the move. FASEB J. 2010, 24, 312.1. [Google Scholar] [CrossRef]
- Casares, D.; Escribá, P.V.; Rosselló, C.A. Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 2019, 20, 2167. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High cholesterol/low cholesterol: Effects in biological membranes: A review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef]
- Doktorova, M.; Symons, J.L.; Levental, I. Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 2020, 16, 1321–1330. [Google Scholar] [CrossRef]
- Boughter, C.T.; Monje-Galvan, V.; Im, W.; Klauda, J.B. Influence of Cholesterol on Phospholipid Bilayer Structure and Dynamics. J. Phys. Chem. B 2016, 120, 11761–11772. [Google Scholar] [CrossRef]
- Sugahara, M.; Uragami, M.; Yan, X.; Regen, S.L. The structural role of cholesterol in biological membranes. J. Am. Chem. Soc. 2001, 123, 7939–7940. [Google Scholar] [CrossRef]
- Baron, S.F.; Hylemon, P.B. Biotransformation of bile acids, cholesterol, and steroid hormones. In Gastrointestinal Microbiology: Volume 1 Gastrointestinal Ecosystems and Fermentations; Springer: Heidelberg, Germany, 1997; pp. 470–510. [Google Scholar]
- King, R.J.; Singh, P.K.; Mehla, K. The cholesterol pathway: Impact on immunity and cancer. Trends Immunol. 2022, 43, 78–92. [Google Scholar] [CrossRef]
- Akbarian, M.; Khani, A.; Eghbalpour, S.; Uversky, V.N. Bioactive peptides: Synthesis, sources, applications, and proposed mechanisms of action. Int. J. Mol. Sci. 2022, 23, 1445. [Google Scholar] [CrossRef]
- Nourmohammadi, E.; Mahoonak, A.S. Health implications of bioactive peptides: A review. Int. J. Vitam. Nutr. Res. 2019, 88, 223–343. [Google Scholar] [CrossRef]
- Garcia-Mora, P.; Peñas, E.; Frías, J.; Gómez, R.; Martinez-Villaluenga, C. High-pressure improves enzymatic proteolysis and the release of peptides with angiotensin I converting enzyme inhibitory and antioxidant activities from lentil proteins. Food Chem. 2015, 171, 224–232. [Google Scholar] [CrossRef]
- Mojica, L.; De Mejía, E.G. Optimization of enzymatic production of anti-diabetic peptides from black bean (Phaseolus vulgaris L.) proteins, their characterization and biological potential. Food Funct. 2016, 7, 713–727. [Google Scholar] [CrossRef]
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.-S.; Yang, H.; Sun, L. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef]
- Rademacher, F.; Gläser, R.; Harder, J. Antimicrobial peptides and proteins: Interaction with the skin microbiota. Exp. Dermatol. 2021, 30, 1496–1508. [Google Scholar] [CrossRef]
- Toosi, S.; Behravan, J. Osteogenesis and bone remodeling: A focus on growth factors and bioactive peptides. Biofactors 2020, 46, 326–340. [Google Scholar] [CrossRef]
- Gomes, B.; Augusto, M.T.; Felício, M.R.; Hollmann, A.; Franco, O.L.; Gonçalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef]
- Mammari, N.; Krier, Y.; Albert, Q.; Devocelle, M.; Varbanov, M.; OEMONOM. Plant-derived antimicrobial peptides as potential antiviral agents in systemic viral infections. Pharmaceuticals 2021, 14, 774. [Google Scholar] [CrossRef]
- Dadar, M.; Shahali, Y.; Chakraborty, S.; Prasad, M.; Tahoori, F.; Tiwari, R.; Dhama, K. Antiinflammatory peptides: Current knowledge and promising prospects. Inflamm. Res. 2019, 68, 125–145. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial peptides as anticancer agents: Functional properties and biological activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Shin, M.K.; Jang, B.-Y.; Bu, K.-B.; Lee, S.-H.; Han, D.-H.; Oh, J.W.; Sung, J.S. De novo design of AC-P19M, a novel anticancer peptide with apoptotic effects on lung cancer cells and anti-angiogenic activity. Int. J. Mol. Sci. 2022, 23, 15594. [Google Scholar] [CrossRef]
- Prabha, N.; Sannasimuthu, A.; Kumaresan, V.; Elumalai, P.; Arockiaraj, J. Intensifying the Anticancer Potential of Cationic Peptide Derived from Serine Threonine Protein Kinase of Teleost by Tagging with Oligo Tryptophan. Int. J. Pept. Res. Ther. 2020, 26, 75–83. [Google Scholar] [CrossRef]
- Gach-Janczak, K.; Biernat, M.; Kuczer, M.; Adamska-Bartłomiejczyk, A.; Kluczyk, A. Analgesic Peptides: From Natural Diversity to Rational Design. Molecules 2024, 29, 1544. [Google Scholar] [CrossRef]
- Pavlicevic, M.; Marmiroli, N.; Maestri, E. Immunomodulatory peptides—A promising source for novel functional food production and drug discovery. Peptides 2022, 148, 170696. [Google Scholar] [CrossRef]
- Pountos, I.; Panteli, M.; Lampropoulos, A.; Jones, E.; Calori, G.M.; Giannoudis, P.V. The role of peptides in bone healing and regeneration: A systematic review. BMC Med. 2016, 14, 103. [Google Scholar] [CrossRef]
- Shoari, A.; Khodabakhsh, F.; Cohan, R.A.; Salimian, M.; Karami, E. Anti-angiogenic peptides application in cancer therapy; A review. Res. Pharm. Sci. 2021, 16, 559–574. [Google Scholar] [CrossRef]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Aisenbrey, C.; Marquette, A.; Bechinger, B. The mechanisms of action of cationic antimicrobial peptides refined by novel concepts from biophysical investigations. In Antimicrobial Peptides: Basics for Clinical Application; Springer: Berlin/Heidelberg, Germany, 2019; pp. 33–64. [Google Scholar] [CrossRef]
- Hemshekhar, M.; Anaparti, V.; Mookherjee, N. Functions of cationic host defense peptides in immunity. Pharmaceuticals 2016, 9, 40. [Google Scholar] [CrossRef]
- Abarca-Cabrera, L.; Fraga-García, P.; Berensmeier, S. Bio-nano interactions: Binding proteins, polysaccharides, lipids and nucleic acids onto magnetic nanoparticles. Biomater. Res. 2021, 25, 12. [Google Scholar] [CrossRef]
- Savini, F.; Loffredo, M.R.; Troiano, C.; Bobone, S.; Malanovic, N.; Eichmann, T.O.; Caprio, L.; Canale, V.C.; Park, Y.; Mangoni, M.L.; et al. Binding of an antimicrobial peptide to bacterial cells: Interaction with different species, strains and cellular components. Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183291. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-action of antimicrobial peptides: Membrane disruption vs. intracellular mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Wenzel, M.; Chiriac, A.I.; Otto, A.; Zweytick, D.; May, C.; Schumacher, C.; Gust, R.; Albada, H.B.; Penkova, M.; Krämer, U.; et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc. Natl. Acad. Sci. USA 2014, 111, E1409–E1418. [Google Scholar] [CrossRef]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial peptides (AMPs): Roles, functions and mechanism of action. Int. J. Pept. Res. Ther. 2020, 26, 1451–1463. [Google Scholar] [CrossRef]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Goki, N.H.; Tehranizadeh, Z.A.; Saberi, M.R.; Khameneh, B.; Bazzaz, B.S. Structure, Function, and Physicochemical Properties of Pore-forming Antimicrobial Peptides. Curr. Pharm. Biotechnol. 2024, 25, 1041–1057. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Z.; Peng, H.; Lv, Y.; Feng, Y.; Kang, J.; Lu, N.; Ma, R.; Hou, S.; Sun, W.; et al. Bomidin: An optimized antimicrobial peptide with broad antiviral activity against enveloped viruses. Front. Immunol. 2022, 13, 851642. [Google Scholar] [CrossRef]
- El-Dirany, R.; Shahrour, H.; Dirany, Z.; Abdel-Sater, F.; Gonzalez-Gaitano, G.; Brandenburg, K.; Martinez de Tejada, G.; Nguewa, P.A. Activity of anti-microbial peptides (AMPs) against Leishmania and other parasites: An overview. Biomolecules 2021, 11, 984. [Google Scholar] [CrossRef]
- Patiño, A.; Hernández, L.; Patiño, E.; Ortíz, B.; Manrique-Moreno, M. Isolation and Evaluation of Galleria mellonella Peptides with Anti-leishmanial Activity. Anal. Biochem. 2016, 546, 35–42. [Google Scholar] [CrossRef]
- Patiño-Márquez, I.A.; Manrique-Moreno, M.; Patiño González, E.; Jemioła-Rzemińska, M.; Strzałka, K. Effect of antimicrobial peptides from Galleria mellonella on molecular models of Leishmania membrane. Thermotropic and fluorescence anisotropy study. J. Antibiot. 2018, 71, 642–652. [Google Scholar] [CrossRef]
- Manrique-Moreno, M.; Santa-González, G.A.; Gallego, V. Bioactive cationic peptides as potential agents for breast cancer treatment. Biosci. Rep. 2021, 41, BSR20211218C. [Google Scholar] [CrossRef]
- Koehbach, J.; Craik, D.J. The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci. 2019, 40, 517–528. [Google Scholar] [CrossRef]
- Xu, R.; Tang, J.; Hadianamrei, R.; Liu, S.; Lv, S.; You, R.; Pan, F.; Zhang, P.; Wang, N.; Cai, Z.; et al. Antifungal activity of designed α-helical antimicrobial peptides. Biomater. Sci. 2023, 11, 2845–2859. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef]
- Fillion, M.; Valois-Paillard, G.; Lorin, A.; Noël, M.; Voyer, N.; Auger, M. proteins, a. Membrane interactions of synthetic peptides with antimicrobial potential: Effect of electrostatic interactions and amphiphilicity. Probiotics Antimicrob. Proteins 2015, 7, 66–74. [Google Scholar] [CrossRef]
- Duque, H.M.; Rodrigues, G.; Santos, L.S.; Franco, O.L. The biological role of charge distribution in linear antimicrobial peptides. Expert Opin. Drug Discov. 2023, 18, 287–302. [Google Scholar] [CrossRef]
- Huang, Y.-B.; Wang, X.-F.; Wang, H.-Y.; Liu, Y.; Chen, Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol. Cancer Ther. 2011, 10, 416–426. [Google Scholar] [CrossRef]
- Wieprecht, T.; Dathe, M.; Beyermann, M.; Krause, E.; Maloy, W.L.; MacDonald, D.L.; Bienert, M. Peptide hydrophobicity controls the activity and selectivity of magainin 2 amide in interaction with membranes. Biochemistry 1997, 36, 6124–6132. [Google Scholar] [CrossRef]
- Datta, G.; Chaddha, M.; Hama, S.; Navab, M.; Fogelman, A.M.; Garber, D.W.; Mishra, V.K.; Epand, R.M.; Epand, R.F.; Lund-Katz, S.L.; et al. Effects of increasing hydrophobicity on the physical-chemical and biological properties of a class A amphipathic helical peptide. J. Lipid Res. 2001, 42, 1096–1104. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef]
- Bechinger, B.; Juhl, D.W.; Glattard, E.; Aisenbrey, C. Revealing the mechanisms of synergistic action of two magainin antimicrobial peptides. Front. Med. Technol. 2020, 2, 615494. [Google Scholar] [CrossRef]
- Lee, H.-T.; Lee, C.-C.; Yang, J.-R.; Lai, J.Z.; Chang, K.Y. A large-scale structural classification of antimicrobial peptides. BioMed Res. Int. 2015, 2015, 475062. [Google Scholar] [CrossRef]
- Singh, V. The origin of eukaryotic cells. Resonance 2021, 26, 479–489. [Google Scholar] [CrossRef]
- Singer, S.J.; Nicolson, G.L. The fluid mosaic model of the structure of cell membranes. Science 1972, 175, 720–731. [Google Scholar] [CrossRef]
- Nicolson, G.L. The Fluid—Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 1451–1466. [Google Scholar] [CrossRef]
- Cho, W.; Stahelin, R.V. Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 119–151. [Google Scholar] [CrossRef]
- Jury, E.C.; Flores-Borja, F.; Kabouridis, P.S. Lipid rafts in T cell signalling and disease. Semin. Cell Dev. Biol. 2007, 18, 608–615. [Google Scholar] [CrossRef]
- Ahmadpour, S.T.; Mahéo, K.; Servais, S.; Brisson, L.; Dumas, J.-F. Cardiolipin, the Mitochondrial Signature Lipid: Implication in Cancer. Int. J. Mol. Sci. 2020, 21, 8031. [Google Scholar] [CrossRef]
- Vance, J.E. Phospholipid synthesis and transport in mammalian cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Marquardt, D.; Geier, B.; Pabst, G. Asymmetric lipid membranes: Towards more realistic model systems. Membranes 2015, 5, 180–196. [Google Scholar] [CrossRef]
- St. Clair, J.R.; Wang, Q.; Li, G.; London, E. Preparation and physical properties of asymmetric model membrane vesicles. In The Biophysics of Cell Membranes: Biological Consequences; Springer: Heidelberg, Germany, 2017; pp. 1–27. [Google Scholar]
- Nguyen, M.H.; Rickeard, B.W.; DiPasquale, M.; Marquardt, D. Asymmetric Model Membranes: Frontiers and Challenges. In Biomimetic Lipid Membranes: Fundamentals, Applications, and Commercialization; Springer: Heidelberg, Germany, 2019; pp. 47–71. [Google Scholar]
- Doktorova, M.; Symons, J.L.; Zhang, X.; Wang, H.-Y.; Schlegel, J.; Lorent, J.H.; Heberle, F.A.; Sezgin, E.; Lyman, E.; Levental, K.R.; et al. Cell Membranes Sustain Phospholipid Imbalance Via Cholesterol Asymmetry. bioRxiv 2023. [Google Scholar] [CrossRef]
- Vahedi, A.; Bigdelou, P.; Farnoud, A.M. Quantitative analysis of red blood cell membrane phospholipids and modulation of cell-macrophage interactions using cyclodextrins. Sci. Rep. 2020, 10, 15111. [Google Scholar] [CrossRef]
- Lorent, J.H.; Levental, K.R.; Ganesan, L.; Rivera-Longsworth, G.; Sezgin, E.; Doktorova, M.; Lyman, E.; Levental, I. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 2020, 16, 644–652. [Google Scholar] [CrossRef]
- Kobayashi, T.; Menon, A.K. Transbilayer lipid asymmetry. Curr. Biol. 2018, 28, R386–R391. [Google Scholar] [CrossRef]
- Murate, M.; Abe, M.; Kasahara, K.; Iwabuchi, K.; Umeda, M.; Kobayashi, T. Transbilayer distribution of lipids at nano scale. J. Cell Sci. 2015, 128, 1627–1638. [Google Scholar] [CrossRef]
- Nelson, D.; Cox, M. Lehninger Principles of Biochemistry, 8th ed.; WH Freeman: New York City, NY, USA, 2021. [Google Scholar]
- Daleke, D.L. Regulation of phospholipid asymmetry in the erythrocyte membrane. Curr. Opin. Hematol. 2008, 15, 191–195. [Google Scholar] [CrossRef]
- Verkleij, A.J.; Post, J.A. Membrane phospholipid asymmetry and signal transduction. J. Membr. Biol. 2000, 178, 1–10. [Google Scholar] [CrossRef]
- Zwaal, R.F.A.; Roelofsen, B.; Comfurius, P.; Van Deenen, L.L.M. Organization of phospholipids in human red cell membranes as detected by the action of various purified phospholipases. Biochim. Biophys. Acta BBA Biomembr. 1975, 406, 83–96. [Google Scholar] [CrossRef]
- Verkleij, A.J.; Zwaal, R.F.; Roelofsen, B.; Comfurius, P.; Kastelijn, D.; van Deenen, L.L.M. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta BBA Biomembr. 1973, 323, 178–193. [Google Scholar] [CrossRef]
- Gordesky, S.E.; Marinetti, G.V. The asymetric arrangement of phospholipids in the human erythrocyte membrane. Biochem. Biophys. Res. Commun. 1973, 50, 1027–1031. [Google Scholar] [CrossRef]
- Zwaal, R.F.; Roelofsen, B.; Colley, C.M. Localization of red cell membrane constituents. Biochim. Biophys. Acta BBA Rev. Biomembr. 1973, 300, 159–182. [Google Scholar] [CrossRef]
- Zwaal, R.F.; Bevers, E.M.; Comfurius, P.C. Platelets and coagulation. In Blood Coagulation; Elsevier Science: Amsterdam, The Netherlands, 1986; Chapter 6; pp. 141–169. [Google Scholar]
- Wang, C.T.; Shiao, Y.J.; Chen, J.C.; Tsai, W.J.; Yang, C.C. Estimation of the phospholipid distribution in the human platelet plasma membrane based on the effect of phospholipase A2 from Naja nigricollis. Biochim. Biophys. Acta BBA Biomembr. 1986, 856, 244–258. [Google Scholar] [CrossRef]
- Bevers, E.M.; Comfurius, P.; Zwaal, R.F. Changes in membrane phospholipid distribution during platelet activation. Biochim. Biophys. Acta BBA Biomembr. 1983, 736, 57–66. [Google Scholar] [CrossRef]
- Chap, H.J.; Zwaal, R.F.A.; van Deenen, L.L.M. Action of highly purified phospholipases on blood platelets. Evidence for an asymmetric distribution of phospholipids in the surface membrane. Biochim. Biophys. Acta BBA Biomembr. 1977, 467, 146–164. [Google Scholar] [CrossRef]
- Otnaess, A.B.; Holm, T. The effect of phospholipase C on human blood platelets. J. Clin. Investig. 1976, 57, 1419–1425. [Google Scholar] [CrossRef]
- Schick, P.K.; Kurica, K.B.; Chacko, G.K. Location of phosphatidylethanolamine and phosphatidylserine in the human platelet plasma membrane. J. Clin. Investig. 1976, 57, 1221–1226. [Google Scholar] [CrossRef]
- Li, G.; Kim, J.; Huang, Z.; St Clair, J.R.; Brown, D.A.; London, E. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc. Natl. Acad. Sci. USA 2016, 113, 14025–14030. [Google Scholar] [CrossRef]
- Léonard, A.; Escrive, C.; Laguerre, M.; Pebay-Peyroula, E.; Néri, W.; Pott, T.; Katsaras, J.; Dufourc, E.J. Location of Cholesterol in DMPC Membranes. A Comparative Study by Neutron Diffraction and Molecular Mechanics Simulation. Langmuir 2001, 17, 2019–2030. [Google Scholar] [CrossRef]
- Harroun, T.A.; Katsaras, J.; Wassall, S.R. Cholesterol Is Found To Reside in the Center of a Polyunsaturated Lipid Membrane. Biochemistry 2008, 47, 7090–7096. [Google Scholar] [CrossRef]
- Carson, J.A.S.; Lichtenstein, A.H.; Anderson, C.A.; Appel, L.J.; Kris-Etherton, P.M.; Meyer, K.A.; Petersen, K.; Polonsky, T.; Van Horn, L. Dietary cholesterol and cardiovascular risk: A science advisory from the American Heart Association. Circulation 2020, 141, e39–e53. [Google Scholar] [CrossRef]
- Cerqueira, N.M.; Oliveira, E.F.; Gesto, D.S.; Santos-Martins, D.; Moreira, C.; Moorthy, H.N.; Ramos, M.J.; Fernandes, P. Cholesterol biosynthesis: A mechanistic overview. Biochemistry 2016, 55, 5483–5506. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Ishimatu, T. Effects of cholesterol on membrane stability of human erythrocytes. Biol. Pharm. Bull. 2020, 43, 1604–1608. [Google Scholar] [CrossRef]
- Chakraborty, S.; Doktorova, M.; Molugu, T.R.; Heberle, F.A.; Scott, H.L.; Dzikovski, B.; Nagao, M.; Stingaciu, L.-R.; Standaert, R.F.; Barrera, F.N.; et al. How cholesterol stiffens unsaturated lipid membranes. Proc. Natl. Acad. Sci. USA 2020, 117, 21896–21905. [Google Scholar] [CrossRef]
- Kumar, G.A.; Chattopadhyay, A. Membrane cholesterol regulates endocytosis and trafficking of the serotonin1A receptor: Insights from acute cholesterol depletion. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2021, 1866, 158882. [Google Scholar] [CrossRef]
- Islam, M.M.; Hlushchenko, I.; Pfisterer, S.G. Low-density lipoprotein internalization, degradation and receptor recycling along membrane contact sites. Front. Cell Dev. Biol. 2022, 10, 826379. [Google Scholar] [CrossRef]
- Rituper, B.; Guček, A.; Lisjak, M.; Gorska, U.; Šakanović, A.; Bobnar, S.T.; Lasič, E.; Božić, M.; Abbineni, P.S.; Jorgačevski, J.; et al. Vesicle cholesterol controls exocytotic fusion pore. Cell Calcium 2022, 101, 102503. [Google Scholar] [CrossRef]
- Olkkonen, V.M.; Ikonen, E. Cholesterol transport in the late endocytic pathway: Roles of ORP family proteins. J. Steroid Biochem. Mol. Biol. 2022, 216, 106040. [Google Scholar] [CrossRef]
- Ogunmowo, T.H.; Jing, H.; Raychaudhuri, S.; Kusick, G.F.; Imoto, Y.; Li, S.; Itoh, K.; Ma, Y.; Jafri, H.; Dalva, M.B.; et al. Membrane compression by synaptic vesicle exocytosis triggers ultrafast endocytosis. Nat. Commun. 2023, 14, 2888. [Google Scholar] [CrossRef]
- García-Arribas, A.B.; Alonso, A.; Goñi, F.M. Cholesterol interactions with ceramide and sphingomyelin. Chem. Phys. Lipids 2016, 199, 26–34. [Google Scholar] [CrossRef]
- Ayuyan, A.G.; Cohen, F.S. Raft Composition at Physiological Temperature and pH in the Absence of Detergents. Biophys. J. 2008, 94, 2654–2666. [Google Scholar] [CrossRef]
- McIntosh, T.J.; Simon, S.A.; Needham, D.; Huang, C.H. Structure and cohesive properties of sphingomyelin/cholesterol bilayers. Biochemistry 1992, 31, 2012–2020. [Google Scholar] [CrossRef]
- Olkkonen, V.M.; Ikonen, E. Inter-and intra-membrane lipid transport. In Biochemistry of Lipids, Lipoproteins and Membranes; Elsevier: Amsterdam, The Netherlands, 2021; pp. 457–486. [Google Scholar]
- Doole, F.T.; Kumarage, T.; Ashkar, R.; Brown, M.F. Cholesterol stiffening of lipid membranes. J. Membr. Biol. 2022, 255, 385–405. [Google Scholar] [CrossRef]
- McMullen, T.P.; Lewis, R.N.; McElhaney, R.N. Cholesterol–phospholipid interactions, the liquid-ordered phase and lipid rafts in model and biological membranes. Curr. Opin. Colloid Interface Sci. 2004, 8, 459–468. [Google Scholar] [CrossRef]
- Subczynski, W.K.; Hyde, J.S.; Kusumi, A. Effect of alkyl chain unsaturation and cholesterol intercalation on oxygen transport in membranes: A pulse ESR spin labeling study. Biochemistry 1991, 30, 8578–8590. [Google Scholar] [CrossRef]
- Khan, N.; Shen, J.; Chang, T.Y.; Chang, C.C.; Fung, P.C.; Grinberg, O.; Demidenko, E.; Swartz, H. Plasma membrane cholesterol: A possible barrier to intracellular oxygen in normal and mutant CHO cells defective in cholesterol metabolism. Biochemistry 2003, 42, 23–29. [Google Scholar] [CrossRef]
- Menchaca, H.J.; Michalek, V.N.; Rohde, T.D.; Hirsch, A.T.; Tuna, N.; Buchwald, H. Improvement of blood oxygen diffusion capacity and anginal symptoms by cholesterol lowering with simvastatin. J. Appl. Res. Clin. Exp. Ther. 2004, 4, 410–418. [Google Scholar]
- Widomska, J.; Raguz, M.; Subczynski, W.K. Oxygen permeability of the lipid bilayer membrane made of calf lens lipids. Biochim. Biophys. Acta BBA Biomembr. 2007, 1768, 2635–2645. [Google Scholar] [CrossRef]
- Róg, T.; Pasenkiewicz-Gierula, M. Cholesterol effects on the phospholipid condensation and packing in the bilayer: A molecular simulation study. FEBS Lett. 2001, 502, 68–71. [Google Scholar] [CrossRef]
- Scott, H.L.; Kennison, K.B.; Enoki, T.A.; Doktorova, M.; Kinnun, J.J.; Heberle, F.A.; Katsaras, J. Model membrane systems used to study plasma membrane lipid asymmetry. Symmetry 2021, 13, 1356. [Google Scholar] [CrossRef]
- Clarke, R.J.; Hossain, K.R.; Cao, K. Physiological roles of transverse lipid asymmetry of animal membranes. Biochim. Et Biochim. Biophys. Acta BBA Biomembr. 2020, 1862, 183382. [Google Scholar] [CrossRef]
- Blumer, M.; Harris, S.; Li, M.; Martinez, L.; Untereiner, M.; Saeta, P.N.; Carpenter, T.S.; Ingólfsson, H.I.; Bennett, W.D. Simulations of asymmetric membranes illustrate cooperative leaflet coupling and lipid adaptability. Front. Cell Dev. Biol. 2020, 8, 575. [Google Scholar] [CrossRef]
- Barrantes, F.J. Cholesterol and nicotinic acetylcholine receptor: An intimate nanometer-scale spatial relationship spanning the billion year time-scale. Biomed. Spectrosc. Imaging 2016, 5, S67–S86. [Google Scholar] [CrossRef]
- Budelier, M.M.; Cheng, W.W.; Chen, Z.-W.; Bracamontes, J.R.; Sugasawa, Y.; Krishnan, K.; Mydock-McGrane, L.; Covey, D.F.; Evers, A.S. Common binding sites for cholesterol and neurosteroids on a pentameric ligand-gated ion channel. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2019, 1864, 128–136. [Google Scholar] [CrossRef]
- Thiriet, M. Ion carriers and receptors. In Biology Mechanics of Blood Flows: Part I: Biology; Springer: Heidelberg, Germany, 2008; pp. 93–153. [Google Scholar]
- Sakuragi, T.; Nagata, S. Regulation of phospholipid distribution in the lipid bilayer by flippases and scramblases. Nat. Rev. Mol. Cell Biol. 2023, 24, 576–596. [Google Scholar] [CrossRef]
- Arashiki, N.; Saito, M.; Koshino, I.; Kamata, K.; Hale, J.; Mohandas, N.; Manno, S.; Takakuwa, Y. An unrecognized function of cholesterol: Regulating the mechanism controlling membrane phospholipid asymmetry. Biochemistry 2016, 55, 3504–3513. [Google Scholar] [CrossRef]
- Thallmair, S.; Ingólfsson, H.I.; Marrink, S.J. Cholesterol flip-flop impacts domain registration in plasma membrane models. J. Phys. Chem. Lett. 2018, 9, 5527–5533. [Google Scholar] [CrossRef]
- Li, J.; Lu, X.; Ma, W.; Chen, Z.; Sun, S.; Wang, Q.; Yuan, B.; Yang, K. Cholesterols Work as a Molecular Regulator of the Antimicrobial Peptide-Membrane Interactions. Front. Mol. Biosci. 2021, 8, 638988. [Google Scholar] [CrossRef]
- Henderson, J.M.; Iyengar, N.S.; Lam, K.L.H.; Maldonado, E.; Suwatthee, T.; Roy, I.; Waring, A.J.; Lee, K.Y.C. Beyond electrostatics: Antimicrobial peptide selectivity and the influence of cholesterol-mediated fluidity and lipid chain length on protegrin-1 activity. Biochim. Biophys. Acta Biomembr. 2019, 1861, 182977. [Google Scholar] [CrossRef]
- Fernandez-Perez, E.J.; Sepulveda, F.J.; Peters, C.; Bascunan, D.; Riffo-Lepe, N.O.; Gonzalez-Sanmiguel, J.; Sanchez, S.A.; Peoples, R.W.; Vicente, B.; Aguayo, L.G. Effect of Cholesterol on Membrane Fluidity and Association of Abeta Oligomers and Subsequent Neuronal Damage: A Double-Edged Sword. Front. Aging Neurosci. 2018, 10, 226. [Google Scholar] [CrossRef]
- Qian, S.; Heller, W.T. Melittin-induced cholesterol reorganization in lipid bilayer membranes. Biochim. Biophys. Acta BBA Biomembr. 2015, 1848, 2253–2260. [Google Scholar] [CrossRef]
- Sharma, V.K.; Mamontov, E.; Anunciado, D.B.; O’Neill, H.; Urban, V.S. Effect of antimicrobial peptide on the dynamics of phosphocholine membrane: Role of cholesterol and physical state of bilayer. Soft Matter 2015, 11, 6755–6767. [Google Scholar] [CrossRef]
- McHenry, A.J.; Sciacca, M.F.; Brender, J.R.; Ramamoorthy, A. Does cholesterol suppress the antimicrobial peptide induced disruption of lipid raft containing membranes? Biochim. Biophys. Acta BBA Biomembr. 2012, 1818, 3019–3024. [Google Scholar] [CrossRef]
- Arsov, Z.; Nemec, M.; Schara, M.; Johansson, H.; Langel, U.; Zorko, M. Cholesterol prevents interaction of the cell-penetrating peptide transportan with model lipid membranes. J. Pept. Sci. 2008, 14, 1303–1308. [Google Scholar] [CrossRef]
- Verly, R.M.; Rodrigues, M.A.; Daghastanli, K.R.; Denadai, A.M.; Cuccovia, I.M.; Bloch, C., Jr.; Frezard, F.; Santoro, M.M.; Pilo-Veloso, D.; Bemquerer, M.P. Effect of cholesterol on the interaction of the amphibian antimicrobial peptide DD K with liposomes. Peptides 2008, 29, 15–24. [Google Scholar] [CrossRef]
- Zhao, H.; Mattila, J.P.; Holopainen, J.M.; Kinnunen, P.K. Comparison of the membrane association of two antimicrobial peptides, magainin 2 and indolicidin. Biophys. J. 2001, 81, 2979–2991. [Google Scholar] [CrossRef]
- Prenner, E.J.; Lewis, R.N.; Jelokhani-Niaraki, M.; Hodges, R.S.; McElhaney, R.N. Cholesterol attenuates the interaction of the antimicrobial peptide gramicidin S with phospholipid bilayer membranes. Biochim. Biophys. Acta BBA Biomembr. 2001, 1510, 83–92. [Google Scholar] [CrossRef]
- Leeb, F.; Maibaum, L. Spatially resolving the condensing effect of cholesterol in lipid bilayers. Biophys. J. 2018, 115, 2179–2188. [Google Scholar] [CrossRef]
- Heinrich, F.; Salyapongse, A.; Kumagai, A.; Dupuy, F.G.; Shukla, K.; Penk, A.; Huster, D.; Ernst, R.K.; Pavlova, A.; Gumbart, J.C.; et al. Synergistic biophysical techniques reveal structural mechanisms of engineered cationic antimicrobial peptides in lipid model membranes. Chem. Eur. J. 2020, 26, 6247–6256. [Google Scholar] [CrossRef]
- Allsopp, R.; Pavlova, A.; Cline, T.; Salyapongse, A.M.; Gillilan, R.E.; Di, Y.P.; Deslouches, B.; Klauda, J.B.; Gumbart, J.C.; Tristram-Nagle, S. Antimicrobial peptide mechanism studied by scattering-guided molecular dynamics simulation. J. Phys. Chem. B 2022, 126, 6922–6935. [Google Scholar] [CrossRef]
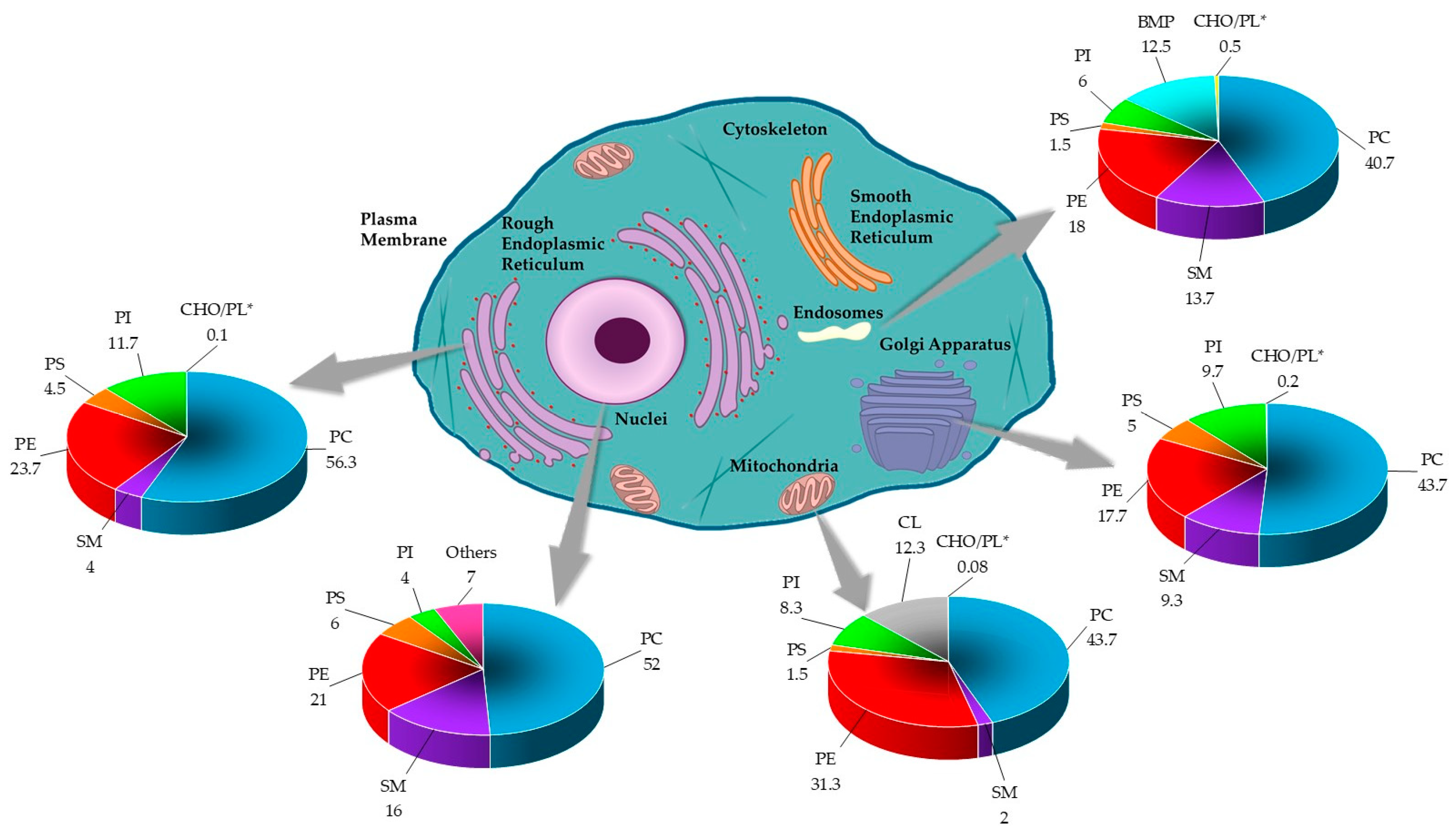
 ), SM (
), SM ( ), PE (
), PE ( ), and PS (
), and PS ( ). Data are presented in ratios based on PC abundance.
). Data are presented in ratios based on PC abundance.
 ), SM (
), SM ( ), PE (
), PE ( ), and PS (
), and PS ( ). Data are presented in ratios based on PC abundance.
). Data are presented in ratios based on PC abundance.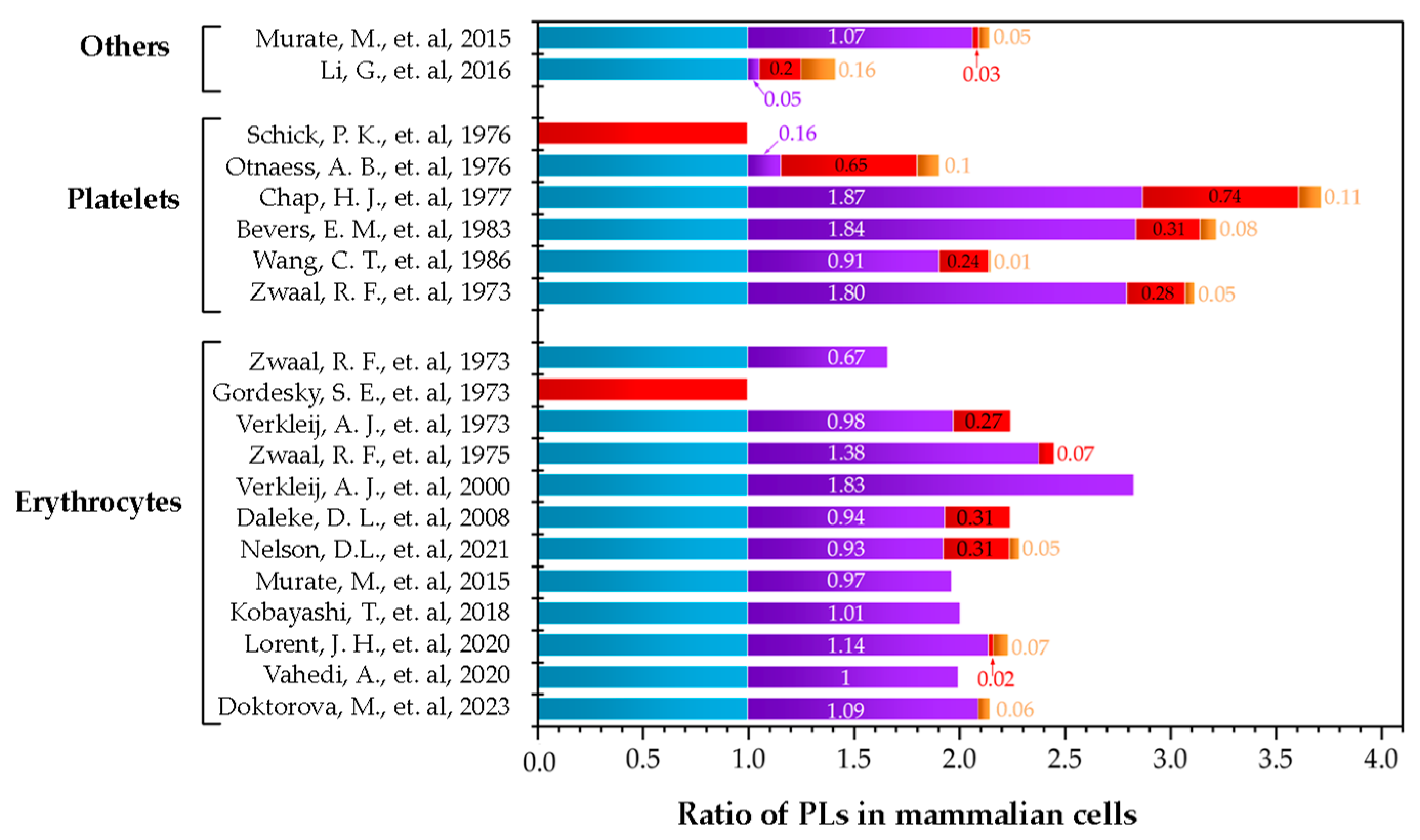
| Cell Type | Results OL | Unit | Technique Used for Quantification | Year | Ref. |
|---|---|---|---|---|---|
| Erythrocytes | PC: 8.5 SM: 9.3 PS: 0.48 | Mol % | Enzymatic digestion/Mass spectrometry | 2023 | [65] |
| PC: 1 SM: 1 | Ratio | Cyclodextrin chemistry/Thin-layer chromatography/Mass spectrometry | 2020 | [66] | |
| PC: 42 SM: 48 PE: 1 PS: 3 | Mol % | Enzymatic digestion/Mass spectrometry | 2020 | [67] | |
| PC: 98 SM:99 | Total PL (%) | Sodium dodecyl sulphate-digested freeze–fracture/Specifics monoclonal antibodies | 2018 | [68] | |
| PC: 98 SM: 95 | Total PL (%) | Sodium dodecyl sulphate-digested freeze-fracture/Specifics monoclonal antibodies | 2015 | [69] | |
| PC: 19.2 SM: 20 PE: 7 PS: 1.3 | Total PL (%) | Not reported | 2012 | [70] | |
| PC: 22.5 SM: 21.1 PE: 7 | Total PL (%) | Not reported | 2008 | [71] | |
| PC: 15.1 SM: 27.7 | Total PL (%) | Not reported | 2000 | [72] | |
| PC: 61.5 SM: 85 PE: 4.5 PS: 0 | Total PL (%) | Enzymatic degradation/Thin-layer chromatography and determined as phosphorus | 1975 | [73] | |
| PC: 21 SM: 20.5 PE: 5.7 | Total PL (%) | Enzymatic degradation and freeze-etching electron microscopy/Two-dimensional thin-layer chromatography and determined as phosphorus | 1973 | [74] | |
| PE: 33 PS: 0 | Total PL (%) | Titrobenzenesulfonate-diazosulfanilic acid/UV–Vis spectrometry | 1973 | [75] | |
| PC: 3 SM: 2 | Ratio | Enzymatic degradation/Two-dimensional thin-layer chromatography and UV–Vis spectrometry | 1973 | [76] | |
| Platelets | PC: 6.5 SM: 11.7 PE: 1.8 PS: 0.3 | Total PL (%) | Enzymatic degradation | 1987 | [77] |
| PC: 45.9 SM: 41.8 PE: 10.8 PS: 0.5 | Total PL (%) | Enzymatic degradation/Thin-layer chromatography and determined as phosphorus | 1986 | [78] | |
| PC: 31 SM: 57.1 PE: 9.5 PS: 2.4 | Total PL (%) | Enzymatic degradation/Thin-layer chromatography and determined as phosphorus | 1983 | [79] | |
| PC: 12.7 SM: 23.8 PE: 9.4 PS: 1.4 | Total PL (%) | Enzymatic degradation/Thin-layer chromatography and determined as phosphorus | 1977 | [80] | |
| PC: 26.7 SM: 4.2 PE: 17.3 PS: 2.8 | Total PL (%) | Enzymatic degradation/Thin-layer chromatography and determined as phosphorus | 1976 | [81] | |
| PE: 17.9 PS: 0 | Total PL (%) | Titrobenzenesulfonate/Thin-layer chromatography and determined as phosphorus | [82] | ||
| Others | PC: 60.1 SM: 3.3 PE: 11.9 PS: 9.9 | Total PL (%) | Cyclodextrin chemistry/Thin-layer chromatography/Mass spectrometry | 2016 | [83] |
| PC: 82.3 SM: 87.8 PE: 2.5 PS: 4.2 | Total PL (%) | Sodium dodecyl sulphate-digested freeze-fracture/Specific monoclonal antibodies | 2015 | [69] |
| Peptide | Sequence | Net Charge | Technique | Result | Ref. |
|---|---|---|---|---|---|
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | +6 | Dynamic giant uni-lamellar vesicle leakage assay and coarse-grained (CG)/Fluorescence spectroscopy/Molecular dynamics (MD) | A higher peptide concentration is necessary to induce calcein release in the DOPC/CHO system as opposed to the membrane model with DOPC alone. CHO hinders peptide-induced pore formation in the membrane model | [115] |
| Protegrin-1 (PG-1) | RGGRLCYCRRRFCVCVGR-NH2 | +7 | Isothermal titration calorimetry (ITC) and atomic force microscopy (AFM) | Increasing CHO content decreases the favorability of the peptide–lipid interaction | [116] |
| Amyloid-beta (Aβ) | DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA | −3 | MβCD-cholesterol complex/Cell viability assay (MTT)/Patch-perforated/Generalized polarization (GP) | Effect on Aβ depends on the amount of CHO in the membranes. Low CHO induced a facilitation of the membrane perforation andhigh CHO inhibited membrane disruption | [117] |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | +6 | Small-angle neutron scattering (SANS)/Circular dichroism (CD) | Increasing CHO content decreases the amount of transmembrane peptide. CHO reduces the penetration depth of melittin, which would decrease the amount by which it thins the bilayer | [118] |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ | +6 | Circular dichroism (CD)/Elastic incoherent neutron scattering (EINS)/Quasi-elastic neutron scattering (QENS) | Adding CHO to DMPC vesicles mitigates peptide interaction and prevents melittin embedding in the membrane depth | [119] |
| MSI-78 MSI-594 MSI-367 MSI-843 | GIGKFLKKAKKFGKAFVKILKK-NH2 GIGKFLKKAKKGIGAVLKVLTTGL-NH2 KFAKKFAKFAKKFAKFAKKFA-NH2 Oct-OOLLOOLOOL-NH2 | +10 +7 +10 +7 | Fluorescence spectroscopy | A strong reduction in membrane alteration is only observed for all peptides tested above 20% CHO content. CHO’s protective, membrane stabilizing effect does not occur to an appreciable level in lipid systems containing raft domains | [120] |
| Cys-TP | GWTLNSAGYLLGCINLKALAALAKISIL-NH2 | +3 | Spin-label electron paramagnetic resonance (EPR) | An increase in CHO in the DMPC lipid system resists the incorporation of Cys-TP into the lipid bilayer | [121] |
| DD K | H-GLWSKIKAAGKEAAKAAGKAALNAVSEAV-NH2 | +5 | Surface plasmon resonance (SPR)/Atomic force microscopy (AFM)/Isothermal titration microcalorimetry (ITC)/Fluorescence spectroscopy/Dynamic light scattering (DLS) | Peptide interaction with mimetic PC membranes is reduced by CHO presence in concentrations typically found in mammalian cells | [122] |
| Magainin 2 Indolicidin | GIGKFLHSAKKFGKAFVGEIMNS ILPWKWPWWPWRR | +4 +3 | Magnetically stirred circular Teflon wells/Fluorescence spectroscopy | The CHO presence prevents the membrane-perturbing action of magainin 2 and, in the same system with CHO, indolicidin exerts a disturbing effect on the membrane model | [123] |
| Gramicidin S | Cyclo-(VO*LF*P)2 | - | Fourier transform infrared spectroscopic (FT-IR)/Circular dichroism (CD)/Nuclear magnetic resonance (31 P-NMR)/Fluorescence spectroscopy | The presence of CHO attenuates the interaction of GS with PC bilayers. CHO-containing POPC vesicles are more resistant to peptide-induced permeabilization than CHO-free vesicles | [124] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giraldo-Lorza, J.M.; Leidy, C.; Manrique-Moreno, M. The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties. Membranes 2024, 14, 220. https://doi.org/10.3390/membranes14100220
Giraldo-Lorza JM, Leidy C, Manrique-Moreno M. The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties. Membranes. 2024; 14(10):220. https://doi.org/10.3390/membranes14100220
Chicago/Turabian StyleGiraldo-Lorza, Juan M., Chad Leidy, and Marcela Manrique-Moreno. 2024. "The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties" Membranes 14, no. 10: 220. https://doi.org/10.3390/membranes14100220
APA StyleGiraldo-Lorza, J. M., Leidy, C., & Manrique-Moreno, M. (2024). The Influence of Cholesterol on Membrane Targeted Bioactive Peptides: Modulating Peptide Activity Through Changes in Bilayer Biophysical Properties. Membranes, 14(10), 220. https://doi.org/10.3390/membranes14100220







