Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism
Abstract
:1. Introduction
2. Methods
2.1. Continuum Membrane Model
2.2. Quantitative Description of Membrane Deformation
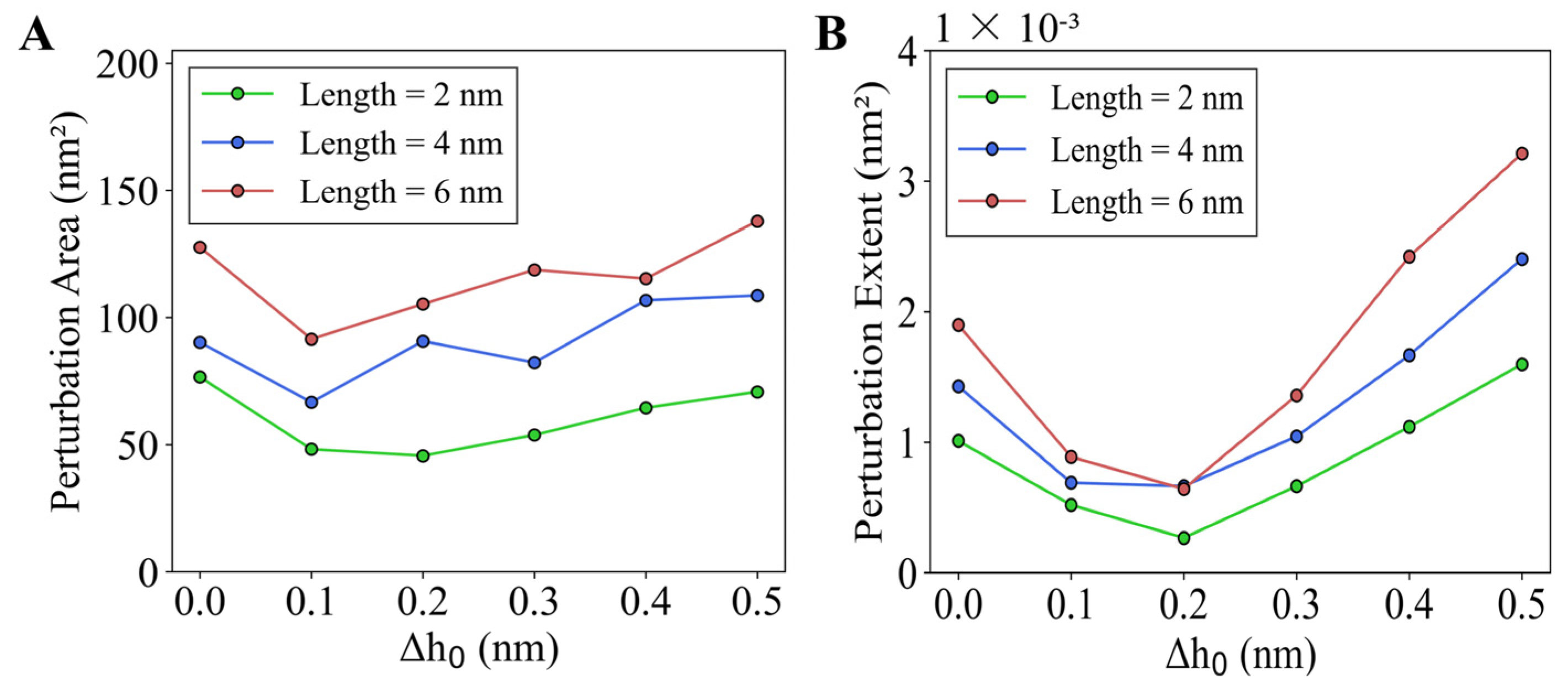
2.2.1. Perturbation Area (PA)
2.2.2. Perturbation Extent (PE)
3. Results
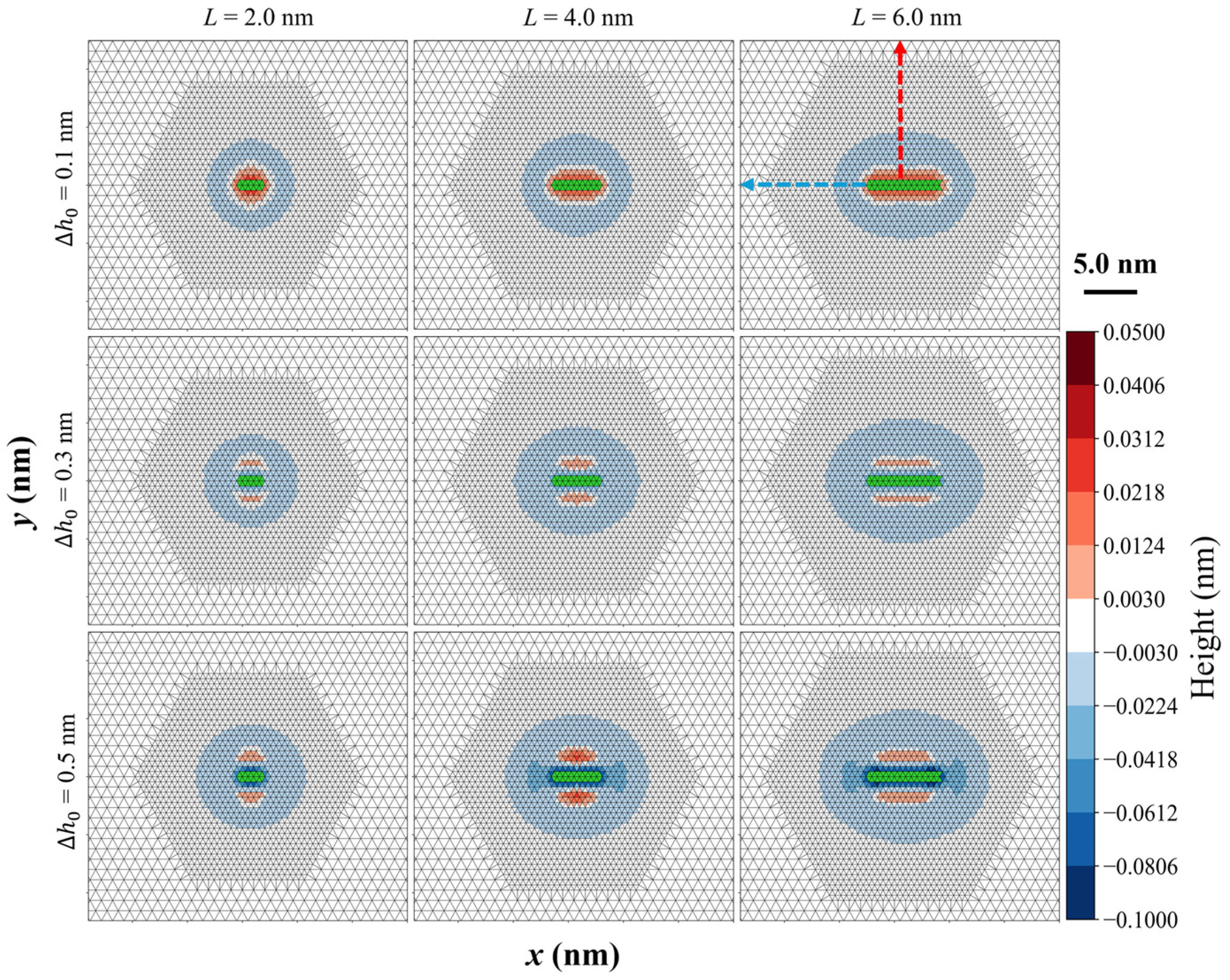
3.1. Membrane Deformation Induced by a Single Helix
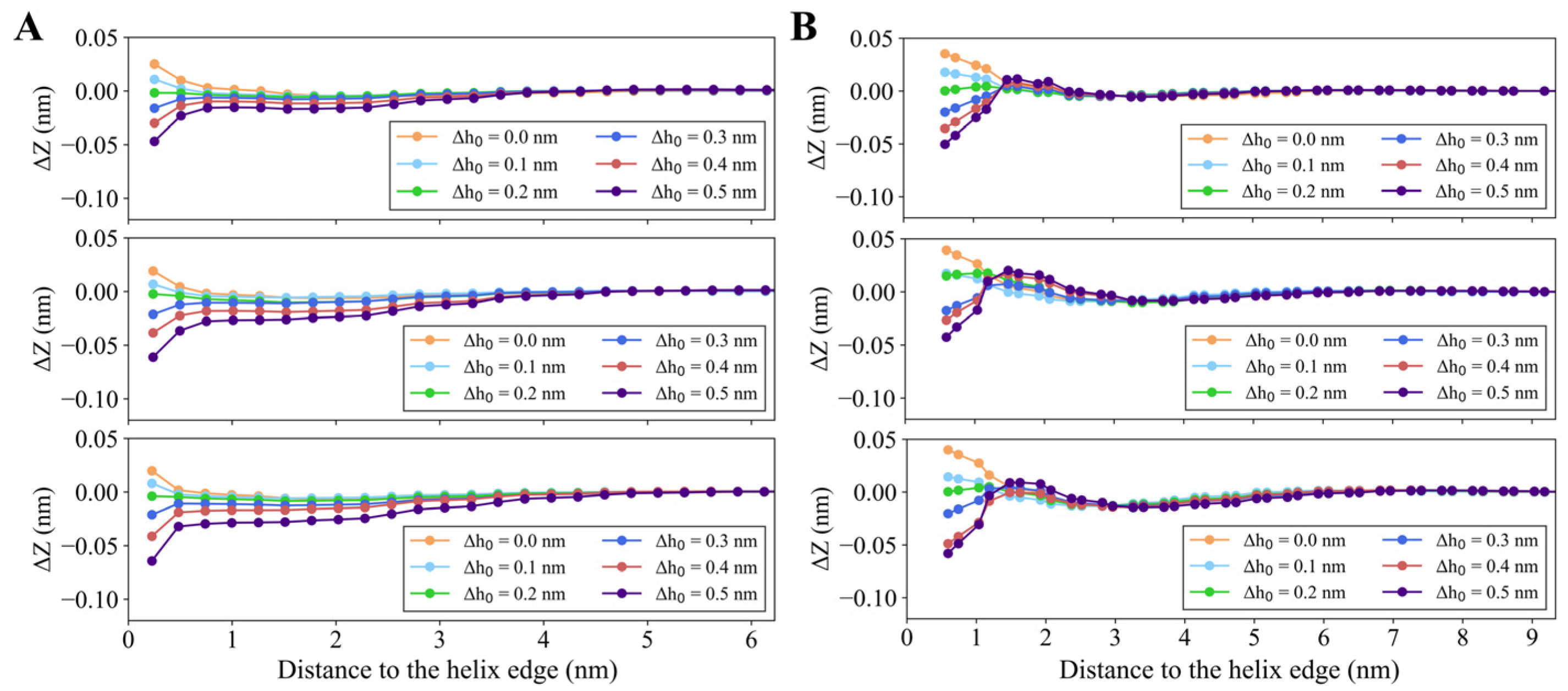
3.1.1. Radial Perturbation Induced by Helix Insertion
3.1.2. Anisotropic Perturbation Induced by Helix Insertion
3.1.3. Quantitative Analysis of Membrane Deformation
3.2. Membrane Deformation Induced by Two Helices
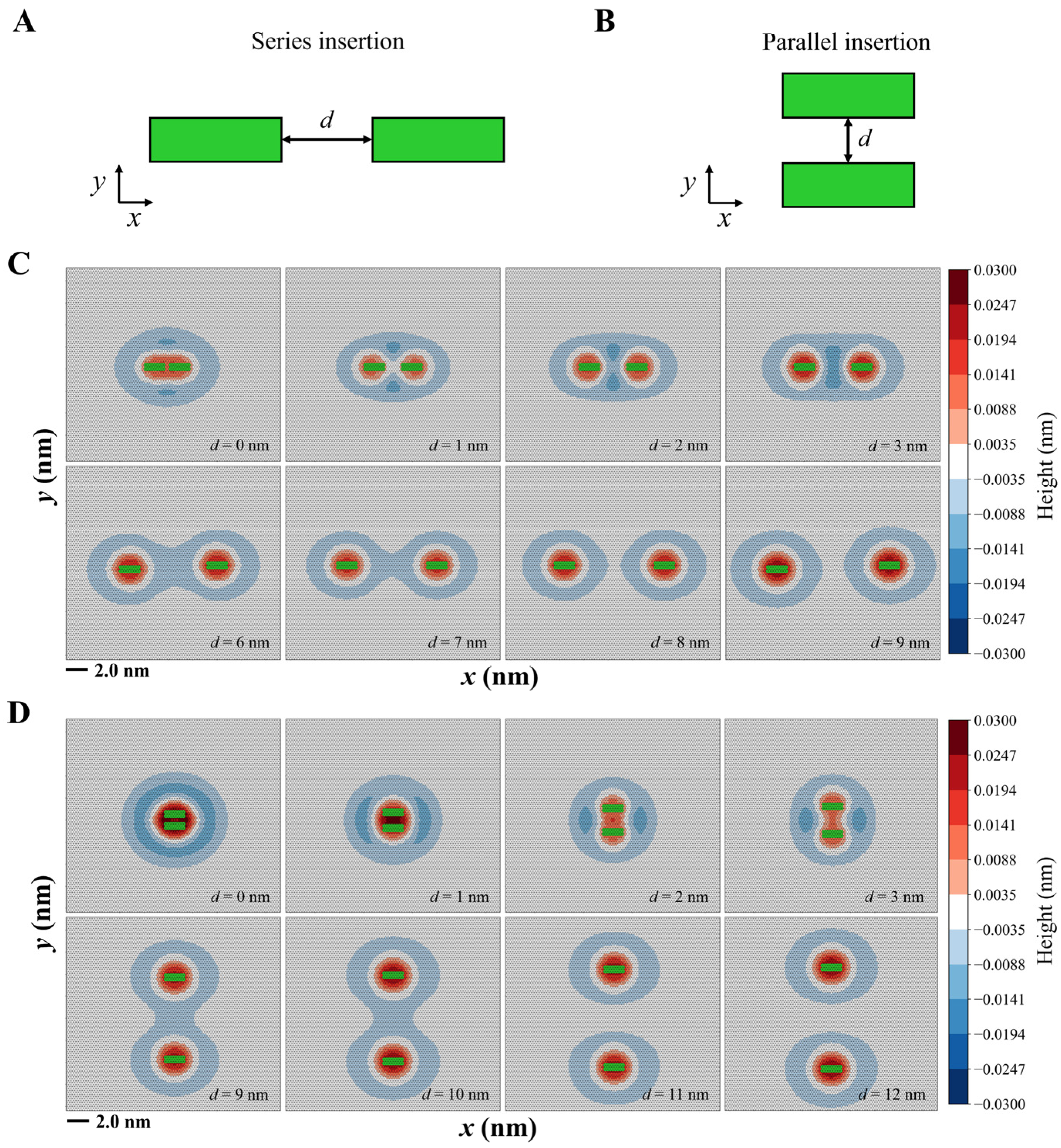
3.2.1. Two Helices Induce Mixed Membrane Deformation
3.2.2. Mixed Membrane Deformation Exhibits a Stepwise Process
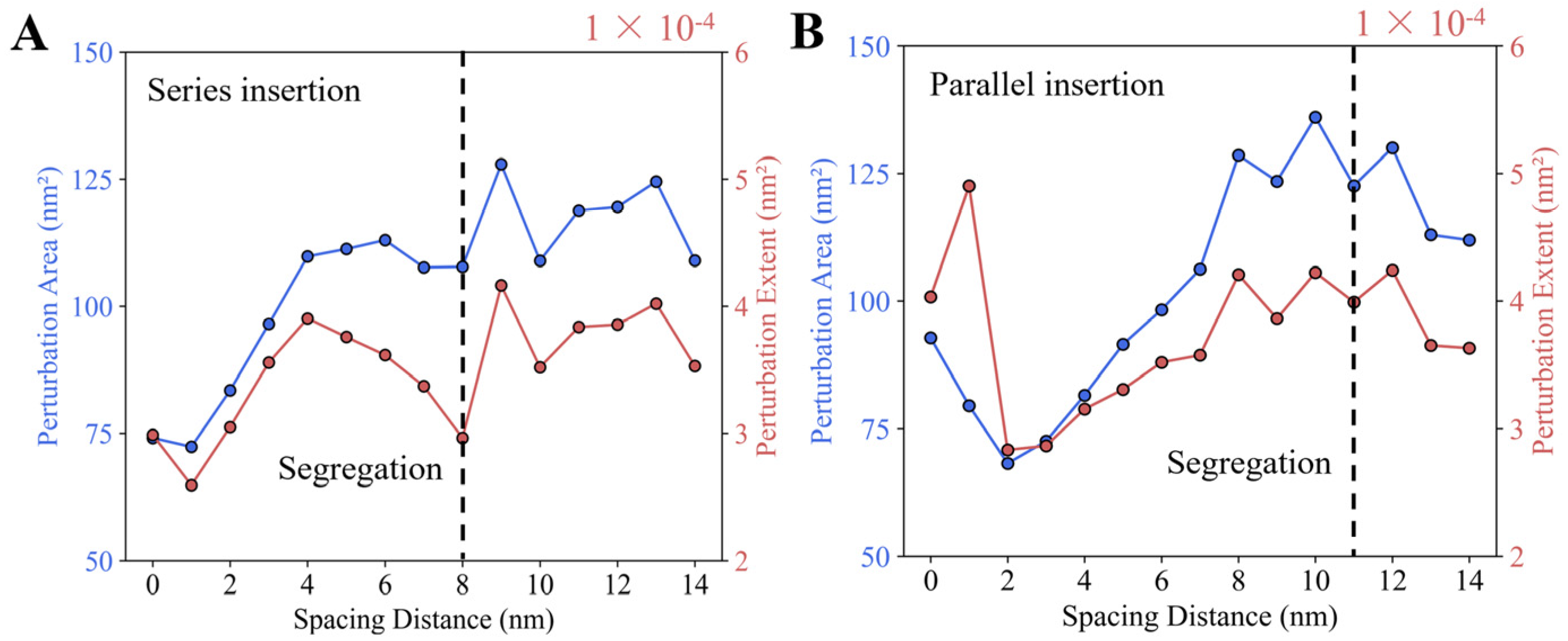
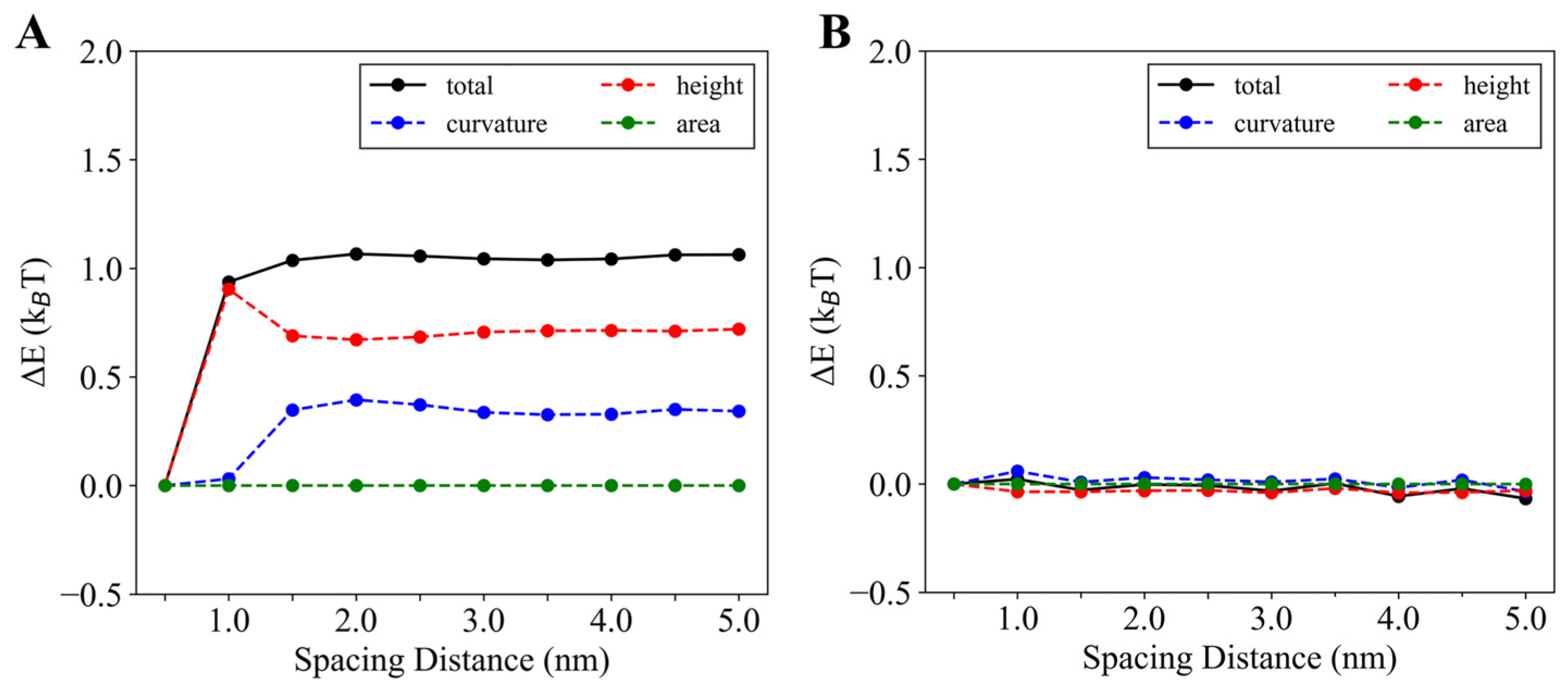
3.2.3. Parallel Insertions Exhibit Potential Cooperativity in Membrane Binding
3.2.4. Longer Helices and Deeper Insertions Enhance Cooperativity
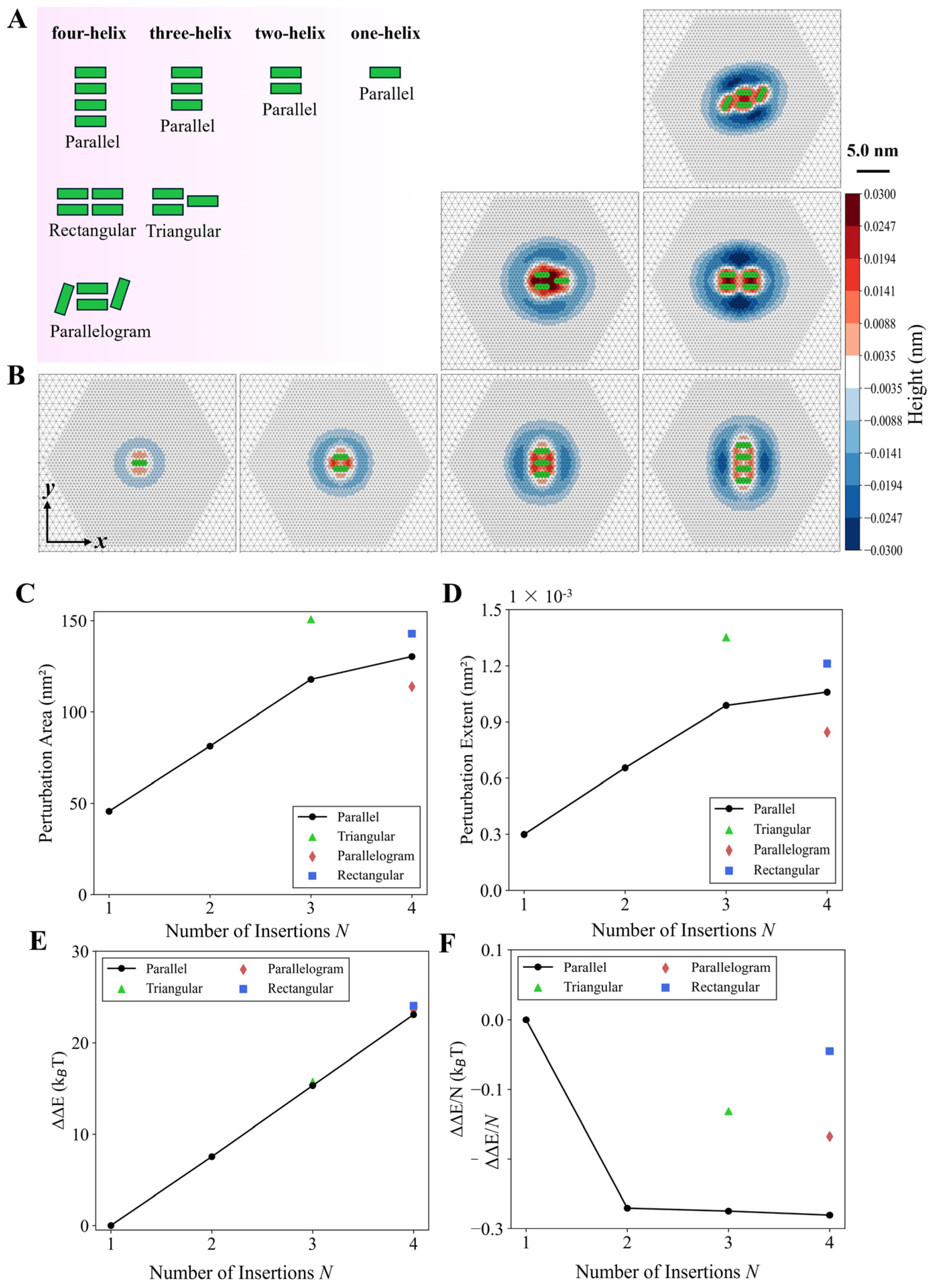
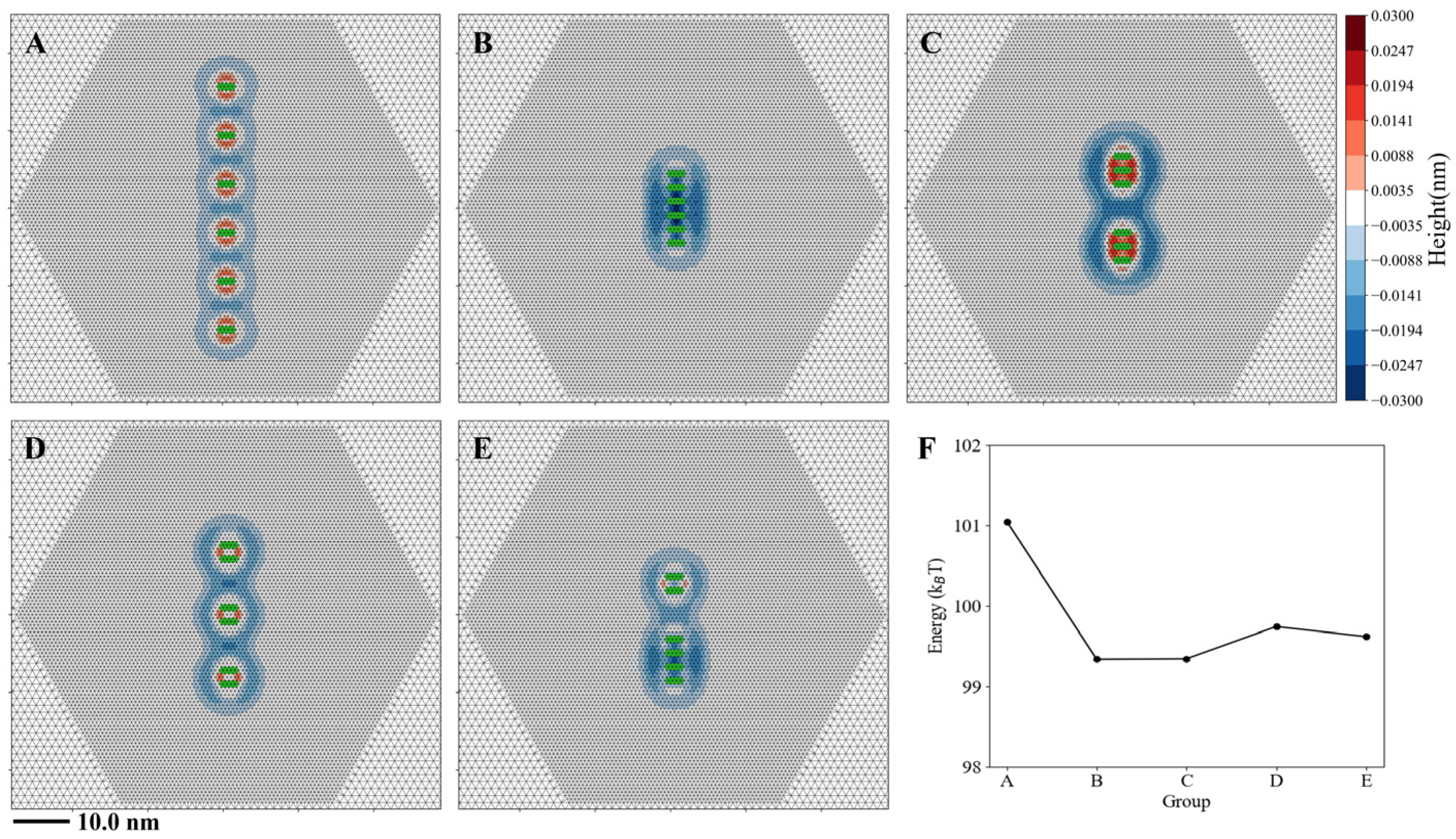
3.3. Multi-Helix Insertions Cooperate to Form Clusters on the Membrane
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Lin, Z.; Mao, Z.; Ma, R. Inferring biophysical properties of membranes during endocytosis using machine learning. Soft Matter 2024, 20, 651–660. [Google Scholar] [CrossRef]
- Groves, J.T.; Kuriyan, J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010, 17, 659–665. [Google Scholar] [CrossRef]
- Carlton, J.G.; Jones, H.; Eggert, U.S. Membrane and organelle dynamics during cell division. Nat. Rev. Mol. Cell Biol. 2020, 21, 151–166. [Google Scholar] [CrossRef]
- Stachowiak, J.C.; Schmid, E.M.; Ryan, C.J.; Ann, H.S.; Sasaki, D.Y.; Sherman, M.B.; Geissler, P.L.; Fletcher, D.A.; Hayden, C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012, 14, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Gallop, J.L.; Jao, C.C.; Kent, H.M.; Butler, P.J.; Evans, P.R.; Langen, R.; McMahon, H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006, 25, 2898–2910. [Google Scholar] [CrossRef]
- Woods, B.L.; Gladfelter, A.S. The state of the septin cytoskeleton from assembly to function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Baumgart, T.; Capraro, B.R.; Zhu, C.; Das, S.L. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu. Rev. Phys. Chem. 2011, 62, 483–506. [Google Scholar] [CrossRef]
- Antonny, B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011, 80, 101–123. [Google Scholar] [CrossRef]
- Xiao, K.; Wu, C.-X.; Ma, R. Vesiculation mechanisms mediated by anisotropic proteins. Phys. Rev. Res. 2023, 5, 023176. [Google Scholar] [CrossRef]
- Fu, Y.; Zeno, W.F.; Stachowiak, J.C.; Johnson, M.E. A continuum membrane model can predict curvature sensing by helix insertion. Soft Matter 2021, 17, 10649–10663. [Google Scholar] [CrossRef]
- Liu, M.; Huang, L.; Zhang, W.; Wang, X.; Geng, Y.; Zhang, Y.; Wang, L.; Zhang, W.; Zhang, Y.-J.; Xiao, S.; et al. A transistor-like pH-sensitive nanodetergent for selective cancer therapy. Nat. Nanotechnol. 2022, 17, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, Y.; Huang, Y.; Xu, C.; Chen, J.; Zhang, X.; Huang, B.; Gan, Z.; Dong, X.; Huang, S.; et al. Helicity-directed recognition of bacterial phospholipid via radially amphiphilic antimicrobial peptides. Sci. Adv. 2024, 10, eadn9435. [Google Scholar] [CrossRef]
- Ma, D.-F.; Zhang, S.; Xu, S.-Y.; Huang, Z.; Tao, Y.; Chen, F.; Zhang, S.; Li, D.; Chen, T.; Liu, C.; et al. Self-limiting multimerization of α-synuclein on membrane and its implication in Parkinson’s diseases. Sci. Adv. 2024, 10, eado4893. [Google Scholar] [CrossRef] [PubMed]
- Capraro, B.R.; Yoon, Y.; Cho, W.; Baumgart, T. Curvature sensing by the epsin N-terminal homology domain measured on cylindrical lipid membrane tethers. J. Am. Chem. Soc. 2010, 132, 1200–1201. [Google Scholar] [CrossRef] [PubMed]
- Hatzakis, N.S.; Bhatia, V.K.; Larsen, J.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.H.; Castillo, J.; Gether, U.; Hedegård, P.; Stamou, D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat. Chem. Biol. 2009, 5, 835–841. [Google Scholar] [CrossRef]
- Zeno, W.F.; Baul, U.; Snead, W.T.; DeGroot, A.C.M.; Wang, L.; Lafer, E.M.; Thirumalai, D.; Stachowiak, J.C. Synergy between intrinsically disordered domains and structured proteins amplifies membrane curvature sensing. Nat. Commun. 2018, 9, 4152. [Google Scholar] [CrossRef]
- Mim, C.; Unger, V.M. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012, 37, 526–533. [Google Scholar] [CrossRef]
- Sodt, A.J.; Pastor, R.W. Molecular Modeling of Lipid Membrane Curvature Induction by a Peptide: More than Simply Shape. Biophys. J. 2014, 106, 1958–1969. [Google Scholar] [CrossRef]
- Cui, H.S.; Lyman, E.; Voth, G.A. Mechanism of Membrane Curvature Sensing by Amphipathic Helix Containing Proteins. Biophys. J. 2011, 100, 1271–1279. [Google Scholar] [CrossRef]
- Gonzalez-Rubio, P.; Gautier, R.; Etchebest, C.; Fuchs, P.F.J. Amphipathic-Lipid-Packing-Sensor interactions with lipids assessed by atomistic molecular dynamics. Biochim. Biophys. Acta BBA—Biomembr. 2011, 1808, 2119–2127. [Google Scholar] [CrossRef]
- Vanni, S.; Vamparys, L.; Gautier, R.; Drin, G.; Etchebest, C.; Fuchs, P.F.J.; Antonny, B. Amphipathic Lipid Packing Sensor Motifs: Probing Bilayer Defects with Hydrophobic Residues. Biophys. J. 2013, 104, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Campelo, F.; McMahon, H.T.; Kozlov, M.M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008, 95, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Campelo, F.; Kozlov, M.M. Sensing Membrane Stresses by Protein Insertions. PLoS Comput. Biol. 2014, 10, e1003556. [Google Scholar]
- Kahraman, O.; Koch, P.D.; Klug, W.S.; Haselwandter, C.A. Bilayer-thickness-mediated interactions between integral membrane proteins. Phys. Rev. E 2016, 93, 042410. [Google Scholar] [CrossRef]
- Wu, Z.; Yi, X. Membrane-mediated interaction of intercellular cylindrical nanoparticles. Phys. Rev. E 2021, 104, 034403. [Google Scholar] [CrossRef]
- Kondrashov, O.V.; Galimzyanov, T.R.; Jiménez-Munguía, I.; Batishchev, O.V.; Akimov, S.A. Membrane-mediated interaction of amphipathic peptides can be described by a one-dimensional approach. Phys. Rev. E 2019, 99, 022401. [Google Scholar]
- Pinigin, K.V.; Kuzmin, P.I.; Akimov, S.A.; Galimzyanov, T.R. Additional contributions to elastic energy of lipid membranes: Tilt-curvature coupling and curvature gradient. Phys. Rev. E 2020, 102, 042406. [Google Scholar] [PubMed]
- Kahraman, O.; Langen, R.; Haselwandter, C.A. Directed Supramolecular Organization of N-BAR Proteins through Regulation of H0 Membrane Immersion Depth. Sci. Rep. 2018, 8, 16383. [Google Scholar] [CrossRef]
- Fu, Y.; Johnson, D.H.; Beaven, A.H.; Sodt, A.J.; Zeno, W.F.; Johnson, M.E. Predicting protein curvature sensing across membrane compositions with a bilayer continuum model. bioRxiv 2024. [Google Scholar] [CrossRef]
- Foley, S.L.; Deserno, M. Asymmetric membrane “sticky tape” enables simultaneous relaxation of area and curvature in simulation. J. Chem. Phys. 2024, 160, 064111. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Deserno, M. Determining the pivotal plane of fluid lipid membranes in simulations. J. Chem. Phys. 2015, 143, 164109. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, W. Elastic properties of lipid bilayers: Theory and possible experiments. Z. Naturforsch. C 1973, 28, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Canham, P.B. The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J. Theor. Biol. 1970, 26, 61–81. [Google Scholar] [CrossRef]
- Zhong-can, O.-Y.; Helfrich, W. Bending energy of vesicle membranes: General expressions for the first, second, and third variation of the shape energy and applications to spheres and cylinders. Phys. Rev. A 1989, 39, 5280–5288. [Google Scholar] [CrossRef]
- Feng, F.; Klug, W.S. Finite element modeling of lipid bilayer membranes. J. Comput. Phys. 2006, 220, 394–408. [Google Scholar] [CrossRef]
- Lipowsky, R. Remodeling of Membrane Shape and Topology by Curvature Elasticity and Membrane Tension. Adv. Biol. 2022, 6, 2101020. [Google Scholar] [CrossRef]
- Hamm, M.; Kozlov, M.M. Elastic energy of tilt and bending of fluid membranes. Eur. Phys. J. E 2000, 3, 323–335. [Google Scholar] [CrossRef]
- Wu, H.; Shiba, H.; Noguchi, H. Mechanical properties and microdomain separation of fluid membranes with anchored polymers. Soft Matter 2013, 9, 9907–9917. [Google Scholar] [CrossRef]
- Nagle, J.F.; Tristram-Nagle, S. Structure of lipid bilayers. Biochim. Biophys. Acta BBA-Rev. Biomembr. 2000, 1469, 159–195. [Google Scholar] [CrossRef]
- Kucerka, N.; Liu, Y.F.; Chu, N.J.; Petrache, H.I.; Tristram-Nagle, S.T.; Nagle, J.F. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005, 88, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- Kaltenegger, M.; Kremser, J.; Frewein, M.P.K.; Ziherl, P.; Bonthuis, D.J.; Pabst, G. Intrinsic lipid curvatures of mammalian plasma membrane outer leaflet lipids and ceramides. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183709. [Google Scholar] [CrossRef] [PubMed]
- Nagle, J.F. Experimentally determined tilt and bending moduli of single-component lipid bilayers. Chem. Phys. Lipids 2017, 205, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Rawicz, W.; Olbrich, K.C.; McIntosh, T.; Needham, D.; Evans, E. Effect of Chain Length and Unsaturation on Elasticity of Lipid Bilayers. Biophys. J. 2000, 79, 328–339. [Google Scholar] [CrossRef]
- Doktorova, M.; LeVine, M.V.; Khelashvili, G.; Weinstein, H. A New Computational Method for Membrane Compressibility: Bilayer Mechanical Thickness Revisited. Biophys. J. 2019, 116, 487–502. [Google Scholar] [CrossRef]

| Parameter | DOPC | DLPC |
|---|---|---|
| Bilayer hydrocarbon height | 2.71 nm [40] | 2.09 nm [41] |
| Monolayer spontaneous curvature | −0.04 nm−1 [42] | +0.11 nm−1 [42] |
| Bilayer bending modulus | 19.4 kBT [43] | 20.4 kBT [43] |
| Bilayer area modulus | 265 pN/nm [44] | 234 pN/nm [45] |
| Monolayer height modulus | 267 pN/nm [30] | 165 pN/nm [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.; Fu, Y. Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism. Membranes 2025, 15, 101. https://doi.org/10.3390/membranes15040101
Hu J, Fu Y. Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism. Membranes. 2025; 15(4):101. https://doi.org/10.3390/membranes15040101
Chicago/Turabian StyleHu, Jie, and Yiben Fu. 2025. "Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism" Membranes 15, no. 4: 101. https://doi.org/10.3390/membranes15040101
APA StyleHu, J., & Fu, Y. (2025). Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism. Membranes, 15(4), 101. https://doi.org/10.3390/membranes15040101





