Molecular Technology for Isolation and Characterization of Mitogen-Activated Protein Kinase Kinase 4 from Penaeus monodon, and the Response to Bacterial Infection and Low-Salinity Challenge
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and PmMKK4 cDNA Cloning
2.2. Bioinformatic Analysis
2.3. Sample Collection and cDNA Synthesis
2.4. Bacterial Infection Challenge
2.5. Low-Salinity Stress
2.6. qRT-PCR Analysis of PmMKK4 mRNA Expression
2.7. Low-Salinity Stress Testing in P. monodon following RNA Interference (RNAi)-Mediated Knockdown of PmMKK4
3. Results
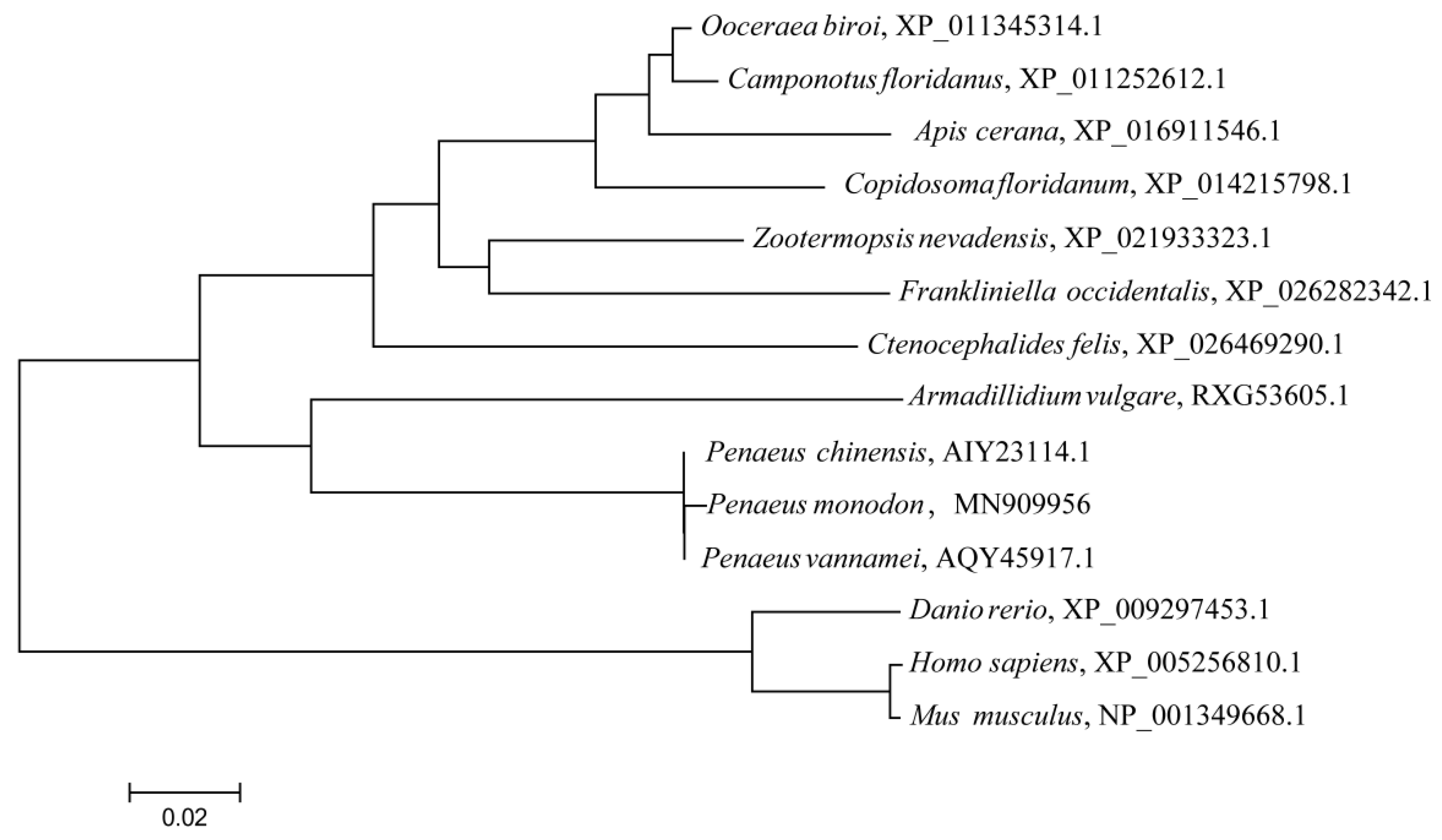
3.1. PmMKK4 Sequence Analysis
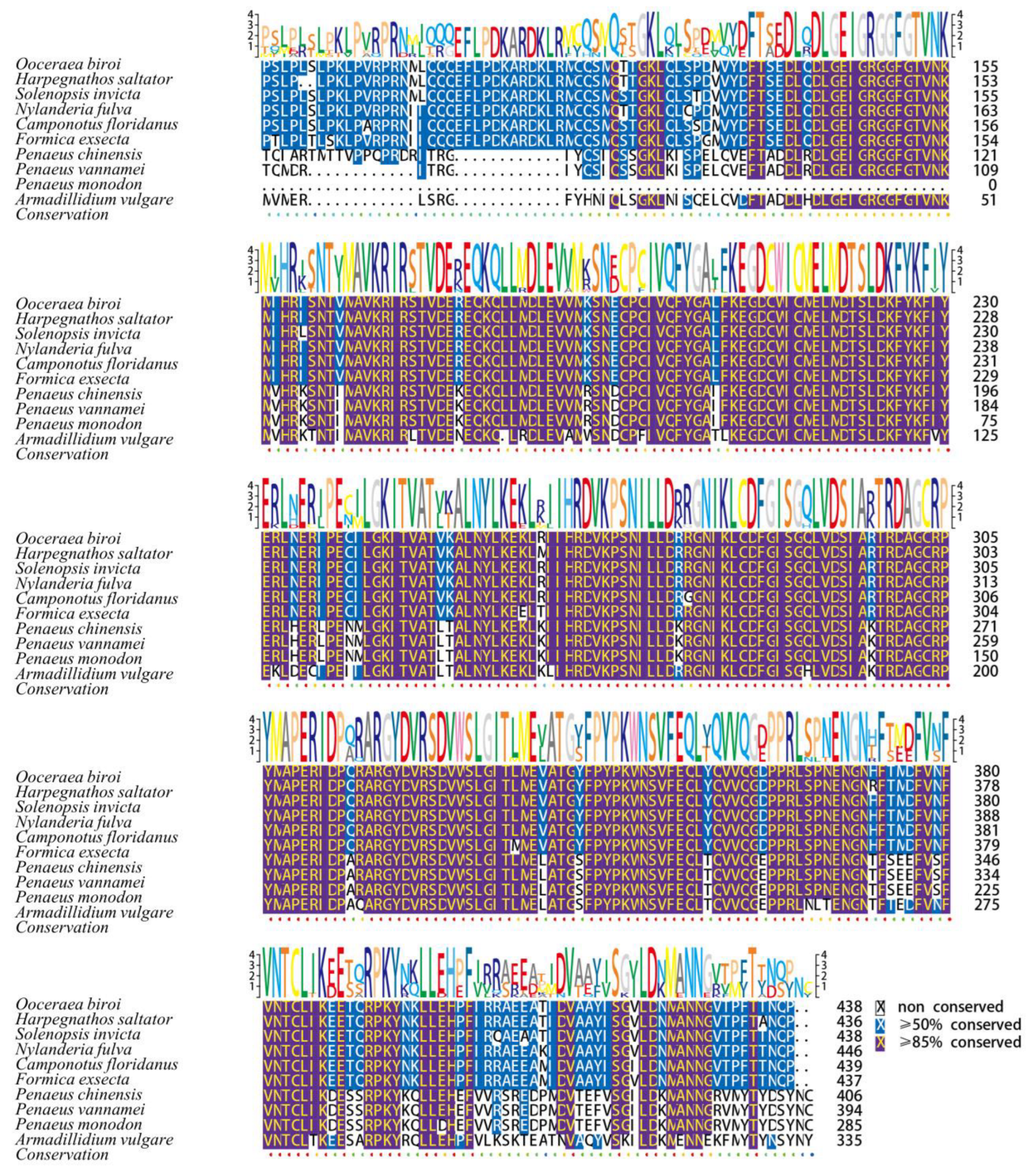
3.2. Sequence Alignment and Analysis
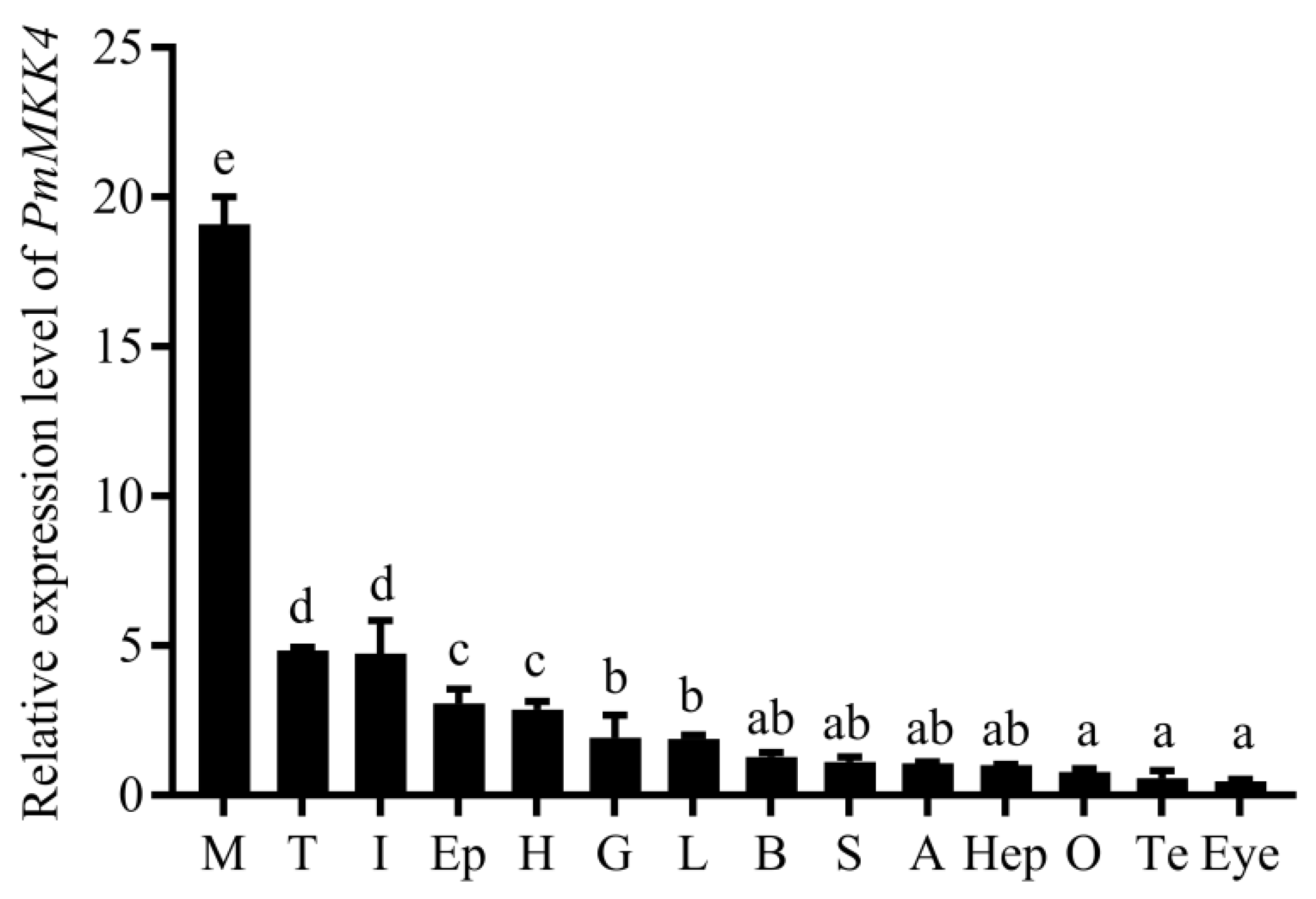
3.3. Tissue Expression Analysis of PmMKK4
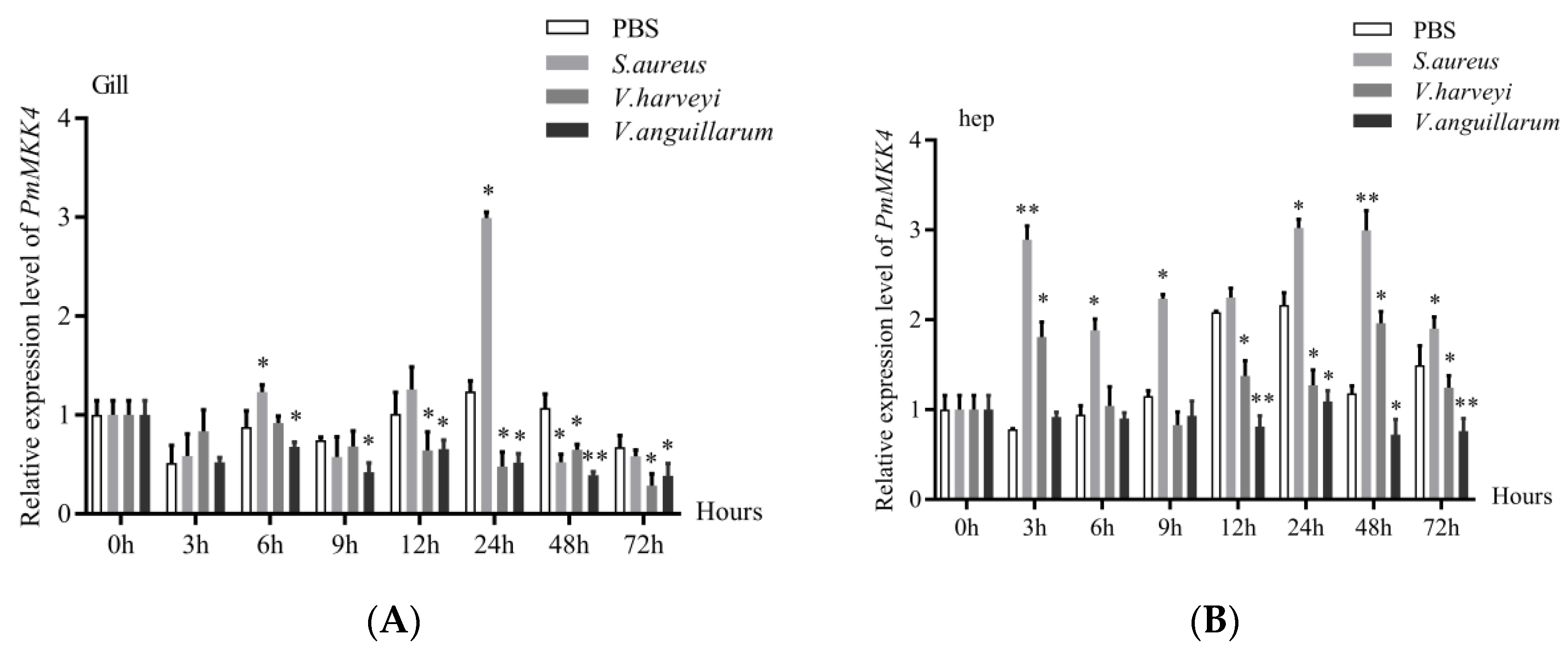
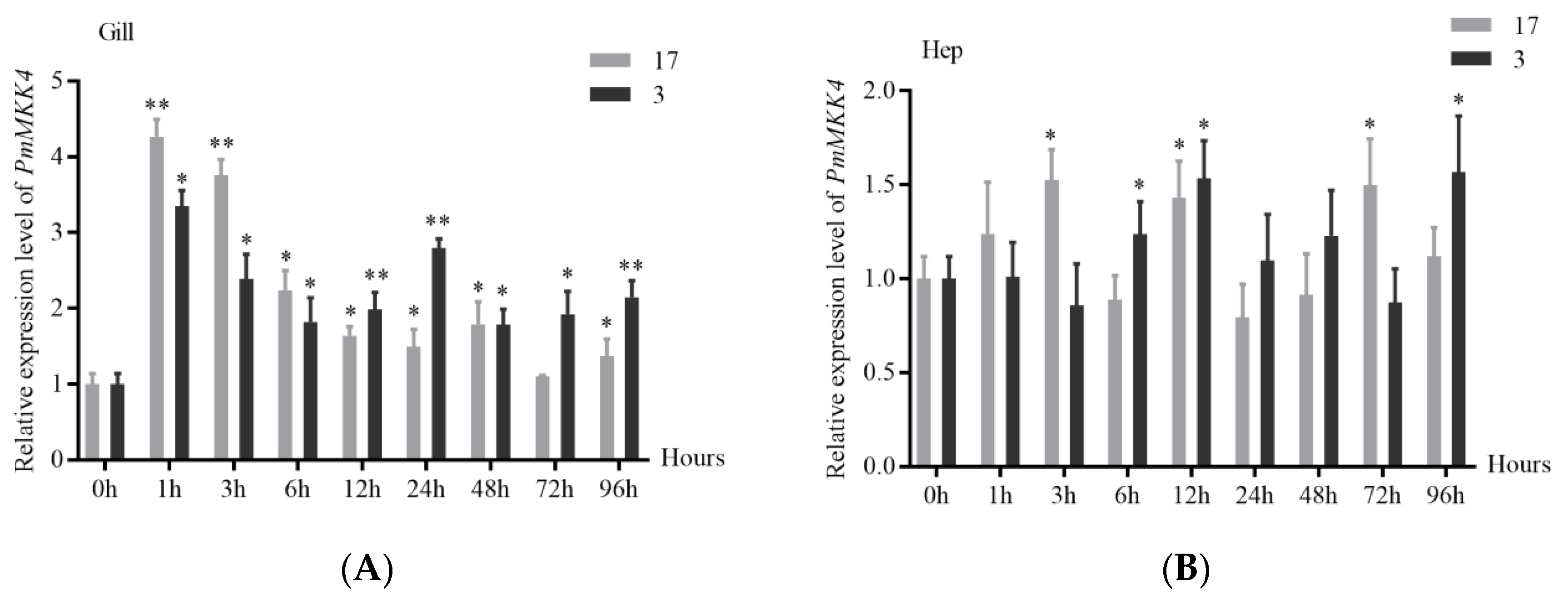
3.4. Expression Analysis of PmMKK4 under Bacterial Stimulation
3.5. Expression Analysis of PmMKK4 under Low Salt Stress
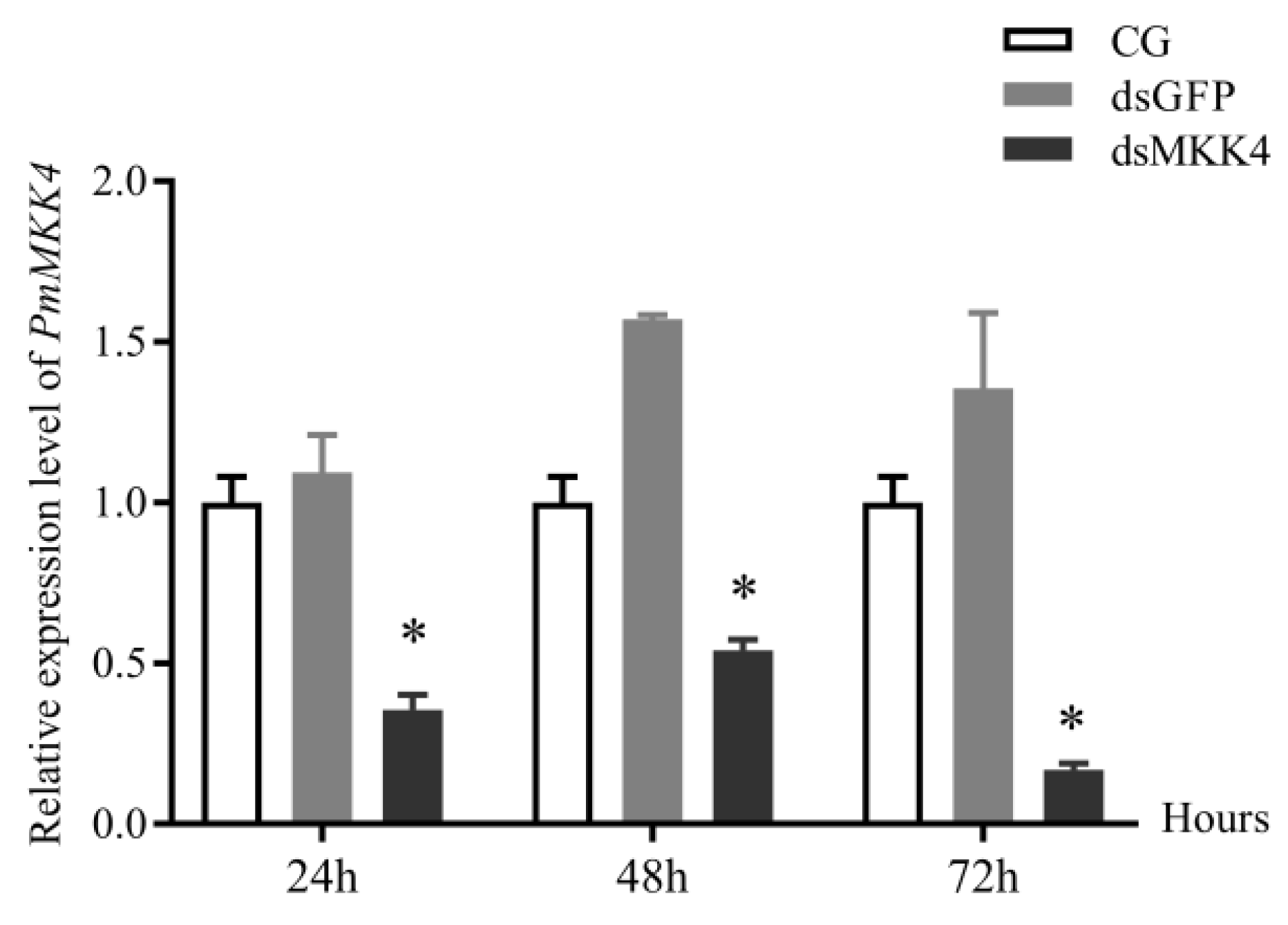
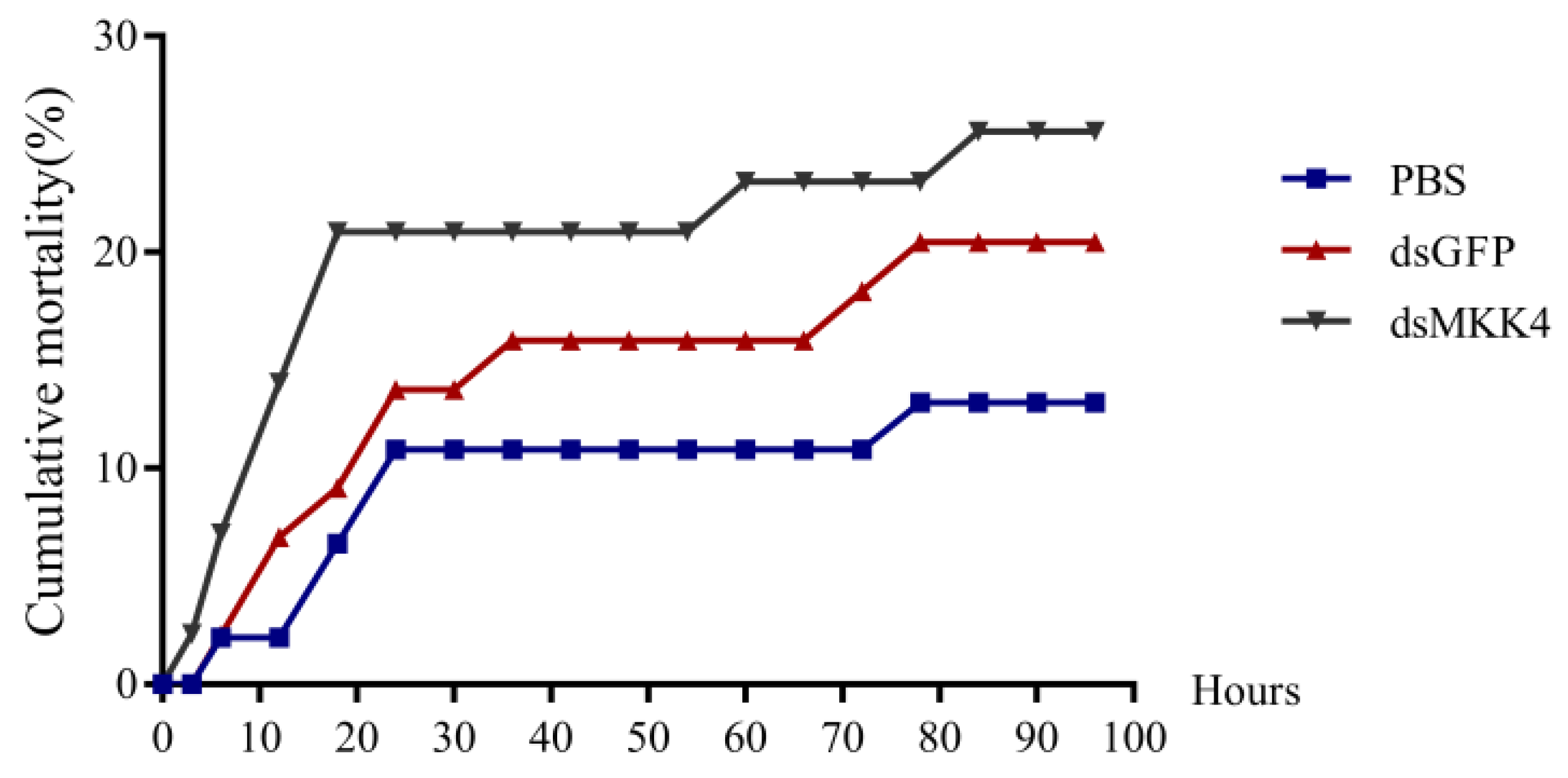
3.6. Expression Analysis after PmMKK4 Interference
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhang, Y.L.; Chen, D. Map kinases in immune responses. Cell. Mol. Immunol. 2005, 1, 22–29. Available online: https://pubmed.ncbi.nlm.nih.gov/16212907/ (accessed on 20 September 2022).
- Krzyzowska, M.; Swiatek, W.; Fijalkowska, B.; Niemialtowski, M.; Schollenberger, A. The role of map kinases in immune response. Adv. Cell Biol. 2010, 2, 125–138. [Google Scholar] [CrossRef][Green Version]
- Wang, S.; Yin, B.; Li, H.Y.; Xiao, B.; Lǚ, K.; Feng, C.G.; He, J.G.; Li, C.Y. Mkk4 from litopenaeus vannamei is a regulator of p38 mapk kinase and involved in anti-bacterial response. Dev. Comp. Immunol. 2017, 78, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Derijard, B.; Raingeaud, J.; Barrett, T.; Wu, I.H.; Han, J.H.; Ulevitch, R.J. Independent human map kinase signal transduction pathways defined by mek and mkk isoforms. Science 1995, 267, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, I.; Hughes, R.T.; Mayer, B.J.; Yee, K.; Woodgett, J.R.; Avruch, J.; Kyrlakls, J.M.; Zon, L.I. Role of sapk/erk kinase-1 in the stress-activated pathway regulating transcription factor c-jun. Nature 1994, 372, 794–798. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Pfrender, M.E.; Gilbert, D.; Thomas, W.K.; Tucker, A.; Oakley, T.H.; Tokishita, H.; Aerts, A.; Arnold, G.J.; Basu, M.K.; et al. The ecoresponsive genome of Daphnia pulex. Science 2011, 331, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.L.; He, Y.Y.; Liu, P.; Li, J.; Wang, Q.Y. cDNA cloning and expression analysis of MKK4 gene under ammonia-N stress in Fenneropenaeus Chinensis. J. Fish. China 2015, 39, 779–789. [Google Scholar] [CrossRef]
- Su, W.W.; Yang, L.S.; Su, T.F.; Yang, Q.B.; Zhu, C.Y.; Zhou, F.L. Expression analysis of MKK4 gene in Penaeus monodon. Guangdong Agric. Sci. 2012, 39, 155–157. [Google Scholar] [CrossRef]
- Wang, S.; Qian, Z.; Li, H.Y.; Lǚ, K.; Xu, X.P.; Weng, S.P.; He, J.G.; Li, C.Z. Identification and characterization of mkk7 as an upstream activator of jnk in litopenaeus vannamei. Fish Shellfish Immunol. 2016, 48, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.F.; Tang, J.Z.; Peng, X.Y.; Zhang, H.; Shi, L.P.; Huang, Z.Z.; Xu, W.Q.; Chen, H.Q.; Shen, Y.; Yan, J.P.; et al. Two novel mkks (mkk4 and mkk7) from ctenopharyngodon idella are involved in the intestinal immune response to bacterial muramyl dipeptide challenge. Dev. Comp. Immunol. 2019, 93, 103–114. [Google Scholar] [CrossRef]
- Qin, Y.K.; Jiang, S.G.; Huang, J.H.; Zhou, F.L.; Yang, Q.B.; Jiang, S.; Yang, L.S. C-type lectin response to bacterial infection and ammonia nitrogen stress in tiger shrimp (penaeus monodon). Fish Shellfish Immunol. 2019, 90, 188–198. [Google Scholar] [CrossRef]
- He, P.; Jiang, S.G.; Li, Y.D.; Yang, Q.B.; Jiang, S.; Yang, L.S.; Huang, J.H.; Zhou, F.L. Molecular cloning and expression pattern analysis of GLUT1 in black tiger shrimp (Penaeus monodon). South China Fish. Sci. 2019, 15, 72–82. Available online: https://www.cabdirect.org/cabdirect/welcome/?target=%2fcabdirect%2fabstract%2f20193290256 (accessed on 20 September 2022).
- Si, M.R.; Li, Y.D.; Jiang, S.G.; Yang, Q.B.; Jiang, S.; Yang, L.S.; Huang, J.H.; Chen, X.; Zhou, F.L. A CSDE1/Unr gene from Penaeus monodon: Molecular characterization, expression and association with tolerance to low salt stress. Aquaculture 2022, 561, 738660. [Google Scholar] [CrossRef]
- Grindheim, A.K.; Saraste, J.; Vedeler, A. Protein phosphorylation and its role in the regulation of annexin a2 function. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2017, 1861, 2515–2529. [Google Scholar] [CrossRef]
- Shi, G.F.; Zhao, C.; Fu, M.J.; Qiu, L.H. The immune response of the c-jun in the black tiger shrimp (penaeus monodon) after bacterial infection. Fish Shellfish Immunol. 2017, 61, 181–186. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.D.; Shi, Y.; Liu, W.G.; He, M.X. Identification and analysis of an mkk4 homologue in response to the nucleus grafting operation and antigens in the pearl oyster, pinctada fucata. Fish Shellfish Immunol. 2017, 73, 279–287. [Google Scholar] [CrossRef]
- Fan, H.; Li, Y.; Yang, Q.; Jiang, S.; Yang, L.; Huang, J.; Jiang, S.; Zhou, F. Isolation and characterization of a MAPKK gene from Penaeus monodon in response to bacterial infection and low-salinity challenge. Aquac. Rep. 2021, 20, 100671. [Google Scholar] [CrossRef]
- Tian, Y.; Wen, H.S.; Qi, X.; Zhang, X.Y.; Li, Y. Identification of mapk gene family in lateolabrax maculatus and their expression profiles in response to hypoxia and salinity challenges. Gene 2018, 684, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Li, Y.; Yang, Q.; Jiang, S.; Huang, J.; Yang, L.; Chen, X.; Zhou, F.; Jiang, S. Expression Analysis of a Novel Oxidoreductase Glutaredoxin 2 in Black Tiger Shrimp, Penaeus monodon. Antioxidants 2022, 11, 1857. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.-Y.; Huang, J.-H.; Zhou, F.-L.; Yang, Q.-B.; Li, Y.-D.; Jiang, S.; Jiang, S.-G.; Yang, L.-S. Identification and Expression Analysis of Dsx and Its Positive Transcriptional Regulation of IAG in Black Tiger Shrimp (Penaeus monodon). Int. J. Mol. Sci. 2022, 23, 12701. [Google Scholar] [CrossRef]


| Primer | Sequence (5′-3′) | Function |
|---|---|---|
| PmMKK4-F1 | AATAACGACCGTCCACAGAAC | ORF validation |
| PmMKK4-R1 | AATAACGACCGTCCACAGAAC | |
| PmMKK4-F2 | TGCGCTCTCCACAAGTGTCTC | |
| PmMKK4-R2 | AAACCTCCTCTTCCAATCTCCC | |
| PmMKK4-5G1 | AAGTCAAGGTTGTTCTGTGGACGGTCG | 5′ clone |
| PmMKK4-5G2 | CATGCTGAGAGTCCCTGGCCTGG | |
| PmMKK4-3G1 | TGGTGCCATATTCAAGGAGGGTGAC | 3′ clone |
| PmMKK4-3G2 | GCTCTGTGATTTCGGCATTTCTGGC | |
| UPM Long | CTAATACGACTCACTATAGGGCAAGCAGTG GTATCAACGCAGAGT | Universal primer |
| UPM Short | CTAATACGACTCACTATAGGGC | |
| NUP | AAGCAGTGGTATCAACGCAGAGT | |
| qMKK4-F | GCCACACTAACAGCACTA | qRT-PCR |
| qMKK4-R | GTCTACATCCAGCATCTCT | |
| qEF-1α-F | AAGCCAGGTATGGTTGTCAACTTT | Reference gene |
| qEF-1α-R | CGTGGTGCATCTCCACAGACT | |
| MKK4-f: | TCGCAAGAGCAACACAATC | RNAi |
| MKK4-r | GAACATCATATCCTCGGGC | |
| dsMKK4-f | TAATACGACTCACTATAGGGTCGCAAGAGCAACACAATC | RNAi |
| dsMKK4-r | TAATACGACTCACTATAGGGGAACATCATATCCTCGGGC | |
| dsGFP-F | TGGAGTGGTCCCAGTTCTTGTTGA | RNAi |
| dsGFP-R | GCCATTCTTTGGTTTGTCTCCCAT | |
| qMKK7-F | TAACCTAGAAGAGACCAAGC | qRT-PCR |
| qMKK7-R | CTGTGACGAAGCATCCTA | |
| qJNK-F | CCGTCCCCGCTACCCTGGCTATTC | qRT-PCR |
| qJNK-R | AGGTCTCGTGCTTGACTCGCTTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhou, F.; Fan, H.; Jiang, S.; Yang, Q.; Huang, J.; Yang, L.; Chen, X.; Zhang, W.; Jiang, S. Molecular Technology for Isolation and Characterization of Mitogen-Activated Protein Kinase Kinase 4 from Penaeus monodon, and the Response to Bacterial Infection and Low-Salinity Challenge. J. Mar. Sci. Eng. 2022, 10, 1642. https://doi.org/10.3390/jmse10111642
Li Y, Zhou F, Fan H, Jiang S, Yang Q, Huang J, Yang L, Chen X, Zhang W, Jiang S. Molecular Technology for Isolation and Characterization of Mitogen-Activated Protein Kinase Kinase 4 from Penaeus monodon, and the Response to Bacterial Infection and Low-Salinity Challenge. Journal of Marine Science and Engineering. 2022; 10(11):1642. https://doi.org/10.3390/jmse10111642
Chicago/Turabian StyleLi, Yundong, Falin Zhou, Hongdi Fan, Song Jiang, Qibin Yang, Jianhua Huang, Lishi Yang, Xu Chen, Wenwen Zhang, and Shigui Jiang. 2022. "Molecular Technology for Isolation and Characterization of Mitogen-Activated Protein Kinase Kinase 4 from Penaeus monodon, and the Response to Bacterial Infection and Low-Salinity Challenge" Journal of Marine Science and Engineering 10, no. 11: 1642. https://doi.org/10.3390/jmse10111642
APA StyleLi, Y., Zhou, F., Fan, H., Jiang, S., Yang, Q., Huang, J., Yang, L., Chen, X., Zhang, W., & Jiang, S. (2022). Molecular Technology for Isolation and Characterization of Mitogen-Activated Protein Kinase Kinase 4 from Penaeus monodon, and the Response to Bacterial Infection and Low-Salinity Challenge. Journal of Marine Science and Engineering, 10(11), 1642. https://doi.org/10.3390/jmse10111642






