Establishment and Application of Microsatellite Multiplex PCR System for Cheilinus undulatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Experimental Methods
DNA Extraction and Sequencing
2.3. Primer Design
2.4. Primer Selection
2.5. Establishment of a Microsatellite Multiplex PCR Amplification System
2.6. Polymorphism Analysis
3. Results
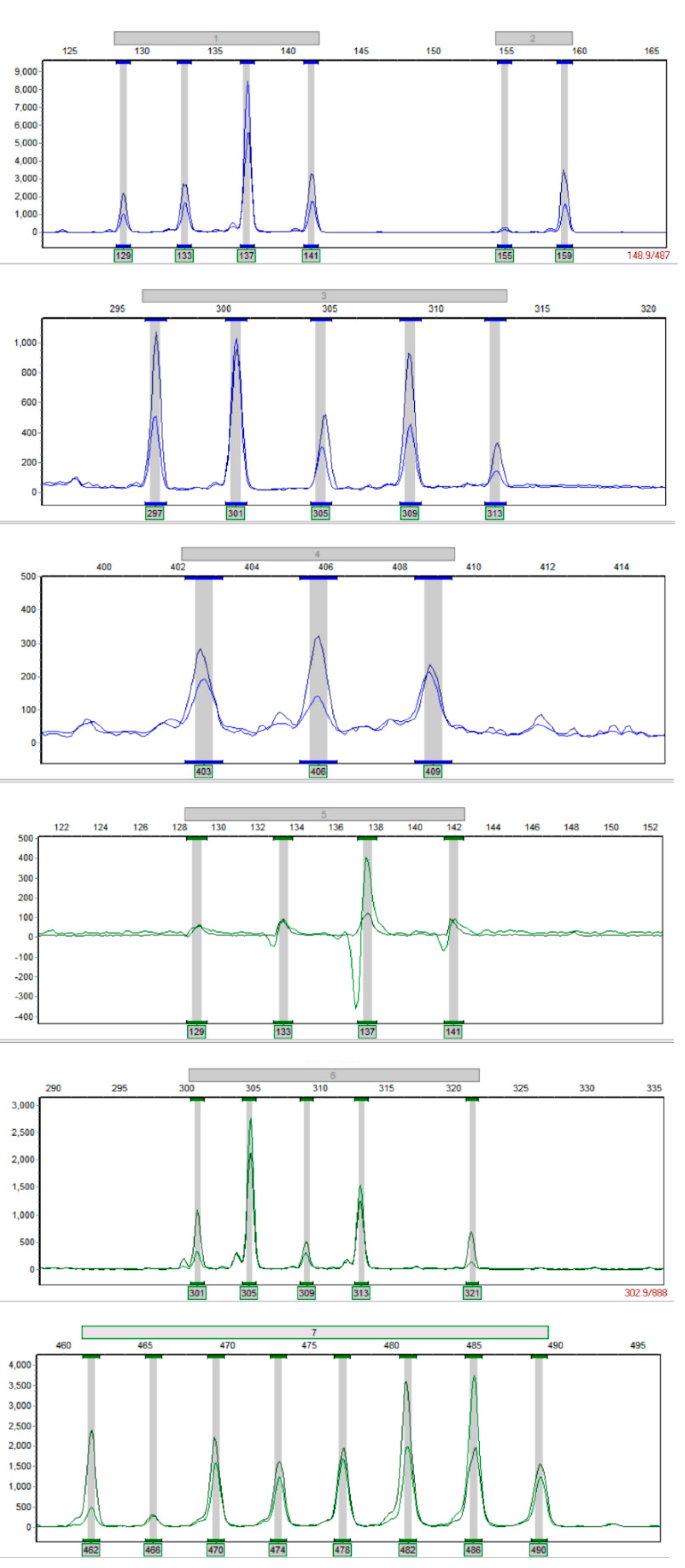
3.1. Multiplex PCR System Establishment
3.2. Polymorphism Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Donaldson, T.J.; Sadovy, Y. Threatened Fishes of the World: Cheilinus Undulatus Rüppell, 1835 (Labridae). Environ. Biol. Fishes 2001, 62, 428. [Google Scholar] [CrossRef]
- Hu, J.; Hou, X.; Yin, S.; Zhu, F.; Jia, Y.; Hu, Y. Genetic diversity of different geographical populations of Chelilinus undulatus revealed by microsatellite analysis. Adv. Mar. Sci. 2013, 31, 538–545. [Google Scholar]
- Dorenbosch, M.; Grol, M.G.G.; Nagelkerken, I.; van der Velde, G. Seagrass Beds and Mangroves as Potential Nurseries for the Threatened Indo-Pacific Humphead Wrasse, Cheilinus Undulatus and Caribbean Rainbow Parrotfish, Scarus Guacamaia. Biol. Conserv. 2006, 129, 277–282. [Google Scholar] [CrossRef]
- Ou, Y.; Li, J. Analysis and evaluation of nutrition composition of double-headed parrotfish Cheilinus undulates. J. Trop. Oceanogr. 2010, 29, 97–102. [Google Scholar]
- Liu, D.; Wang, X.; Guo, H.; Zhang, X.; Zhang, M.; Tang, W. Chromosome-level genome assembly of the endangered humphead wrasse Cheilinus undulatus: Insight into the expansion of opsin genes in fishes. Mol. Ecol. Resour. 2021, 21, 2388–2406. [Google Scholar] [CrossRef]
- Peng, Y.; Luo, J.; Yin, S.; Zhu, X.; Hu, J.; Liu, Z.; Zhu, F.; Qi, X.; Hu, Y. Screening and suitability analysis of microsatellite markers in Cheilinus undulatus. Open J. Mar. Sci. 2011, 36, 109–116. [Google Scholar]
- Indriatmoko; Syam, A.R.; Syahputra, K. Control Region-Mitochondrial Partial DNA Analysis of Humphead Wrasse [Cheilinus Undulates (Ruppel, 1835)] from Anambas Islands, Indonesia. Aquat. Procedia 2016, 7, 125–131. [Google Scholar] [CrossRef]
- Shen, Y.; Ma, K.; Yue, G.H. Status, Challenges and Trends of Aquaculture in Singapore. Aquaculture 2021, 533, 736210. [Google Scholar] [CrossRef]
- Nakamura, H.; Teshima, K.; Tachida, H. Effects of Cyclic Changes in Population Size on Neutral Genetic Diversity. Ecol. Evol. 2018, 8, 9362–9371. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Kamsongkram, P.; Subpayakom, N.; Sillapaprayoon, S.; Muangmai, N.; Kongphoemph, A.; Wongsodchuen, A.; Intapan, S.; Chamchumroon, W.; Safoowong, M.; et al. Predictive Genetic Plan for a Captive Population of the Chinese Goral (Naemorhedus Griseus) and Prescriptive Action for Ex Situ and in Situ Conservation Management in Thailand. PLoS ONE 2020, 15, e0234064. [Google Scholar] [CrossRef]
- Othman, O.E.M.; Payet-Duprat, N.; Harkat, S.; Laoun, A.; Maftah, A.; Lafri, M.; Silva, A.D. Sheep diversity of five Egyptian breeds: Genetic proximity revealed between desert breeds. Small Rumin. Res. 2016, 144, 346–352. [Google Scholar] [CrossRef]
- Sill, S.R.; Dawson, T.P. Climate Change Impacts on the Ecological Dynamics of Two Coral Reef Species, the Humphead Wrasse (Cheilinus Undulatus) and Crown-of-Thorns Starfish (Ancanthaster Planci). Ecol. Inf. 2021, 65, 101399. [Google Scholar] [CrossRef]
- Faria, J.; Pita, A.; Martins, G.M.; Ribeiro, P.A.; Hawkins, S.J.; Presa, P.; Neto, A.I. Inbreeding in the Exploited Limpet Patella Aspera across the Macaronesia Archipelagos (NE Atlantic): Implications for Conservation. Fish. Res. 2018, 198, 180–188. [Google Scholar] [CrossRef]
- Wang, L.; Wu, Z.; Zou, C.; Lu, Y.; Yue, X.; Song, Z.; Yang, R.; You, F. Genetic Diversity and Signatures of Selection in the Mito-Gynogenetic Olive Flounder Paralichthys Olivaceus Revealed by Genome-Wide SNP Markers. Aquaculture 2022, 553, 738062. [Google Scholar] [CrossRef]
- Machmoum, M.; Boujenane, I.; Azelhak, R.; Badaoui, B.; Petit, D.; Piro, M. Genetic Diversity and Population Structure of Arabian Horse Populations Using Microsatellite Markers. J. Equine Vet. Sci. 2020, 93, 103200. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, Q.; Qiu, G. Large-Scale Isolation of Microsatellites from Chinese Mitten Crab Eriocheir sinensis via a Solexa Genomic Survey. Int. J. Mol. Sci. 2012, 13, 16333–16345. [Google Scholar] [CrossRef]
- De Deus Vidal, J., Jr.; Cortez, M.B.; Alves, F.M.; Koehler, S.; de Souza, A.P.; Koch, I. Development and Cross-Validation of Microsatellite Markers for Rauvolfia Weddeliana Müll.Arg. (Apocynaceae) Species Complex. Braz. J. Bot. 2018, 41, 681–686. [Google Scholar] [CrossRef]
- Papetti, C.; Harms, L.; Jürgens, J.; Sandersfeld, T.; Koschnick, N.; Windisch, H.S.; Knust, R.; Pörtner, H.-O.; Lucassen, M. Microsatellite Markers for the Notothenioid Fish Lepidonotothen Nudifrons and Two Congeneric Species. BMC Res. Notes 2016, 9, 238. [Google Scholar] [CrossRef]
- Li, J.Z.; Sjakste, T.G.; Röder, M.S.; Ganal, M.W. Development and Genetic Mapping of 127 New Microsatellite Markers in Barley. Theor. Appl. Genet. 2003, 107, 1021–1027. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Hellemans, B.; Volckaert, F.A.M. Microsatellites and their genomic distribution, evolution, function and applications: A review with special reference to fish genetics. Aquaculture 2006, 255, 1–29. [Google Scholar] [CrossRef]
- Gupta, P.; Varshney, R. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 2000, 113, 163–185. [Google Scholar] [CrossRef]
- Gu, Z.X.; Gou, T.J.; Xi, X.B. Applications of microsatellite markers in studies of genetics and breeding of fish. Chin. J. Agric. Biotechnol. 2006, 3, 83–87. [Google Scholar]
- Chamberlain, J.S.; Gibbs, R.A.; Rainer, J.E.; Caskey, C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic. Acids. Res. 1988, 16, 11141–11156. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Liu, S.; Liu, Y.; Jia, Y.; Li, F.; Chi, M.; Zheng, J.; Cheng, S.; Gu, Z. Development of a Multiplex PCR Assay for Parentage Assignment of the Redclaw Crayfish (Cherax Quadricarinatus). Aquaculture 2022, 550, 737813. [Google Scholar] [CrossRef]
- Lata, C.; Kumar, A.; Gangwar, O.P.; Prasad, P.; Adhikari, S.; Kumar, S.; Kulshreshtha, N.; Bhardwaj, S.C. Multiplex PCR Assay for the Detection of Lr24 and Lr68 in Salt Tolerant Wheat Genotypes. Cereal Res. Commun. 2022, 50, 1019–1027. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Q.; Yang, J.; Zhang, N.; Liu, B.; Zhu, K.; Guo, H.; Jiang, S.; Zhang, D. MultiplexSSR: A Pipeline for Developing Multiplex SSR-PCR Assays from Resequencing Data. Ecol. Evol. 2020, 10, 3055–3067. [Google Scholar] [CrossRef]
- Sudo, R.; Miyao, M.; Uchino, T.; Yamada, Y.; Tsukamoto, K.; Sakamoto, T. Parentage Assignment of a Hormonally Induced Mass Spawning in Japanese Eel (Anguillla Japonica). Aquaculture 2018, 484, 317–321. [Google Scholar] [CrossRef]
- Ge, C.; Cui, Y.; Jing, P.; Hong, X. An alternative suite of universal primers for genotyping in multiplex PCR. PLoS ONE 2014, 9, e92826. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A Reliable Software Tool for the Analysis of Forensic STR Data: GENEMARKER® HID. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Jones, A.; Small, C.; Paczolt, K.; Ratterman, N. A practical guide to methods of parentage analysis. Mol. Ecol. Resour. 2010, 10, 6–30. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, J.; Li, W.; Zhang, X.; Hong, X.; Zhu, X. Development of a Multiplex Microsatellite PCR Assay Based on Microsatellite Markers for the Mud Carp, Cirrhinus molitorella. J. World Aquacult. Soc. 2016, 47, 277–286. [Google Scholar] [CrossRef]
- Domingos, J.; Carolyn, S.K.; Dean, R. Fate of genetic diversity within and between generations and implications for DNA parentage analysis in selective breeding of mass spawners: A case study of commercially farmed barramundi, Lates calcarifer. Aquaculture 2014, 424, 174–182. [Google Scholar] [CrossRef]
- Vallecillos, A.; María-Dolores, E.; Villa, J.; Rueda, F.M.; Carrillo, J.; Ramis, G.; Soula, M.; Afonso, J.M.; Armero, E. Development of the First Microsatellite Multiplex PCR Panel for Meagre (Argyrosomus Regius), a Commercial Aquaculture Species. Fishes 2022, 7, 117. [Google Scholar] [CrossRef]
- Fu, J.; Shen, Y.; Xu, X.; Chen, Y.; Li, D.; Li, J. Multiplex microsatellite PCR sets for parentage assignment of grass carp (Ctenopharyngodon idella). Aquacult. Int. 2013, 21, 1195–1207. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, D.H.; Yoon, S.; Kim, D.H.; Kim, S.; Hyun, Y.S.; Min, G.; Chung, K. Characterization of 20 microsatellite loci by multiplex PCR in swimming crab, Portunus trituberculatus. Genes Genom. 2013, 35, 77–85. [Google Scholar] [CrossRef]
- Hussain, I.A.; Jeyasekaran, G.; Shakila, R.J.; Raj, K.T.; Jeevithan, E. Detection of Hemolytic Strains of Aeromonas Hydrophila and A. Sobria along with Other Aeromonas Spp. from Fish and Fishery Products by Multiplex PCR. J. Food Sci. Technol. 2014, 51, 401–407. [Google Scholar] [CrossRef]
- An, H.S.; Lee, J.W.; Kim, H.Y.; Kim, J.B.; Chang, D.S.; Park, J.Y.; Myeong, J.I.; An, C.M. Population Genetic Structure of the Sea Bass (Lateolabrax Japonicus) in Korea Based on Multiplex PCR Assays with 12 Polymorphic Microsatellite Markers. Genes Genom. 2014, 36, 247–259. [Google Scholar] [CrossRef]

| Name | Number | Primer Pairs | Repetitive Units | Fragment Length Range (bp) | Primer Sequences | Primer Number |
|---|---|---|---|---|---|---|
| Multiplex PCR1 | 1 | Cun463 | CTAT | 129–141 | tgtaaaacgacggccagtcatgaaacaacccgtataccct | Cun463.F |
| aatagccctgctccatacttca | Cun463.R | |||||
| 2 | Cun378 | AGAT | 155–159 | ttgagaggatcgcatccatgtattgatcatgctttctgcc | Cun378.F | |
| taagtctgagccaatcgcatta | Cun378.R | |||||
| 3 | Cun500 | GATA | 297–313 | tgtaaaacgacggccagtaacacaacacgcagcttagaga | Cun500.F | |
| aatggatcctttgagagcgata | Cun500.R | |||||
| 4 | Cun626 | ATA | 403–409 | tgtaaaacgacggccagtctatttcctgcatgtctctccc | Cun626.F | |
| atggcccgtttatagacacaat | Cun626.R | |||||
| 5 | Cun672 | TAGA | 129–141 | ttgagaggatcgcatccacacttcatctgtcccaccatta | Cun672.F | |
| ctccctcctgcttcactgtact | Cun672.R | |||||
| 6 | Cun586 | AGAT | 301–321 | tgtaaaacgacggccagtcaagaattgacaaggtttccct | Cun586.F | |
| taactgcagtgatgaaccctgt | Cun586.R | |||||
| 7 | Cun148 | ATCT | 462–490 | tgtaaaacgacggccagttgcaagagcattggtgatattc | Cun148.F | |
| aagggacaacaaggacactgtta | Cun148.R | |||||
| 8 | Generic primers | FAM-5′-tgtaaaacgacggccagt | M13 | |||
| 9 | HEX-5′-ttgagaggatcgcatcca | PQE-F | ||||
| Multiplex PCR2 | 1 | Cun752 | ACAAA | 157–167 | tgtaaaacgacggccagttctggaagcacctcatgataga | Cun752.F |
| ctttgactgcaaggtttcctct | Cun752.R | |||||
| 2 | Cun230 | ATAG | 252–272 | tgtaaaacgacggccagtattaaacgcgctggttgttatt | Cun230.F | |
| accaccaactgtgtgaatgttt | Cun230.R | |||||
| 3 | Cun27 | GATA | 367–375 | tgtaaaacgacggccagtctctgtgctctcttgtcattgg | Cun27.F | |
| gtcatgtcattgcatctcacct | Cun27.R | |||||
| 4 | Cun485 | TGGA | 404–412 | ttgagaggatcgcatccacaggctaggaaggaagaaatca | Cun485.F | |
| tttatgcctctgtgacagcatt | Cun485.R | |||||
| 5 | Cun484 | GATA | 470–486 | ttgagaggatcgcatccacatgtatactctgccaccctcca | Cun484.F | |
| agaagttgccaggaaattggta | Cun484.R | |||||
| 6 | Generic primers | FAM-5′-tgtaaaacgacggccagt | M13 | |||
| 7 | HEX-5′-ttgagaggatcgcatcca | PQE-F | ||||
| G1 Group PCR System Reactants | Content (µL) | G2 Group PCR System Reactants | Content (µL) |
|---|---|---|---|
| Chun463.F (20 µM) | 0.06 | Cun752.F (10 µM) | 0.06 |
| Chun463.R (20 µM) | 0.24 | Cun752.R (10 µM) | 0.24 |
| Chun378.F (20 µM) | 0.06 | Cun230.F (20 µM) | 0.06 |
| Chun378.R (20 µM) | 0.24 | Cun230.R (20 µM) | 0.24 |
| Chun500.F (10 µM) | 0.06 | Cun27.F (10 µM) | 0.06 |
| Chun500.R (10 µM) | 0.24 | Cun27.R (10 µM) | 0.24 |
| Chun626.F (20 µM) | 0.06 | Cun485.F (10 µM) | 0.06 |
| Chun626.R (20 µM) | 0.24 | Cun485.R (10 µM) | 0.24 |
| Chun672.F (10 µM) | 0.06 | Cun484.F (10 µM) | 0.06 |
| Chun672.R (10 µM) | 0.24 | Cun484.R (10 µM) | 0.24 |
| Chun586.F (10 µM) | 0.06 | M13 (10 µM) | 0.36 |
| Chun586.R (10 µM) | 0.24 | PQE-F (10 µM) | 0.36 |
| Chun148.F (20 µM) | 0.06 | BSA (2 mg/mL) | 0.45 |
| Chun148.R (20 µM) | 0.24 | DNA (50 ng/µL) | 2.0 |
| M13 (10 µM) | 0.36 | Taq HS (Takara) | 12.5 |
| PQE-F (10 µM) | 0.36 | ddH2O | 7.83 |
| BSA (2 mg/mL) | 0.45 | Total | 25.0 |
| DNA (50 ng/µL) | 2.0 | ||
| Taq HS (Takara) | 12.5 | ||
| ddH2O | 7.23 | ||
| Total | 25.0 |
| Locus | N | Na | Ne | Ho | He | uHe | F | F (Null) | PIC | HW |
|---|---|---|---|---|---|---|---|---|---|---|
| Cun463 | 30 | 4.000 | 1.835 | 0.367 | 0.455 | 0.463 | 0.194 | 0.125 | 0.422 | ns |
| Cun378 | 30 | 2.000 | 1.724 | 0.267 | 0.420 | 0.427 | 0.365 | 0.189 | 0.460 | ns |
| Cun500 | 30 | 5.000 | 3.455 | 0.667 | 0.711 | 0.723 | 0.062 | 0.039 | 0.671 | ** |
| Cun626 | 30 | 3.000 | 2.817 | 0.433 | 0.645 | 0.656 | 0.328 | 0.192 | 0.572 | ns |
| Cun672 | 30 | 4.000 | 1.850 | 0.433 | 0.459 | 0.467 | 0.057 | 0.039 | 0.431 | ns |
| Cun586 | 30 | 5.000 | 2.521 | 0.500 | 0.603 | 0.614 | 0.171 | 0.103 | 0.542 | ns |
| Cun148 | 30 | 8.000 | 4.380 | 0.900 | 0.772 | 0.785 | −0.166 | −0.104 | 0.746 | ** |
| Cun752 | 30 | 3.000 | 2.203 | 0.300 | 0.546 | 0.555 | 0.451 | 0.256 | 0.486 | ** |
| Cun230 | 30 | 6.000 | 4.592 | 0.667 | 0.782 | 0.795 | 0.148 | 0.112 | 0.745 | ns |
| Cun27 | 30 | 3.000 | 2.436 | 0.100 | 0.589 | 0.599 | 0.830 | 0.716 | 0.542 | ** |
| Cun485 | 30 | 3.000 | 2.332 | 0.500 | 0.571 | 0.581 | 0.125 | 0.071 | 0.536 | ns |
| Cun484 | 30 | 5.000 | 2.320 | 0.567 | 0.569 | 0.579 | 0.004 | −0.102 | 0.534 | * |
| Mean | 30 | 4.250 | 2.705 | 0.475 | 0.594 | 0.604 | 0.214 | 0.136 | 0.557 | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Guo, L.; Zhang, N.; Zhu, K.; Yang, J.; Liu, B.; Guo, H.; Zhang, D. Establishment and Application of Microsatellite Multiplex PCR System for Cheilinus undulatus. J. Mar. Sci. Eng. 2022, 10, 2000. https://doi.org/10.3390/jmse10122000
Zhao F, Guo L, Zhang N, Zhu K, Yang J, Liu B, Guo H, Zhang D. Establishment and Application of Microsatellite Multiplex PCR System for Cheilinus undulatus. Journal of Marine Science and Engineering. 2022; 10(12):2000. https://doi.org/10.3390/jmse10122000
Chicago/Turabian StyleZhao, Fangcao, Liang Guo, Nan Zhang, Kecheng Zhu, Jingwen Yang, Baosuo Liu, Huayang Guo, and Dianchang Zhang. 2022. "Establishment and Application of Microsatellite Multiplex PCR System for Cheilinus undulatus" Journal of Marine Science and Engineering 10, no. 12: 2000. https://doi.org/10.3390/jmse10122000
APA StyleZhao, F., Guo, L., Zhang, N., Zhu, K., Yang, J., Liu, B., Guo, H., & Zhang, D. (2022). Establishment and Application of Microsatellite Multiplex PCR System for Cheilinus undulatus. Journal of Marine Science and Engineering, 10(12), 2000. https://doi.org/10.3390/jmse10122000








