Uncertainty in Marine Species Distribution Modelling: Trying to Locate Invasion Hotspots for Pterois miles in the Eastern Mediterranean Sea
Abstract
:1. Introduction
2. Materials and Methods
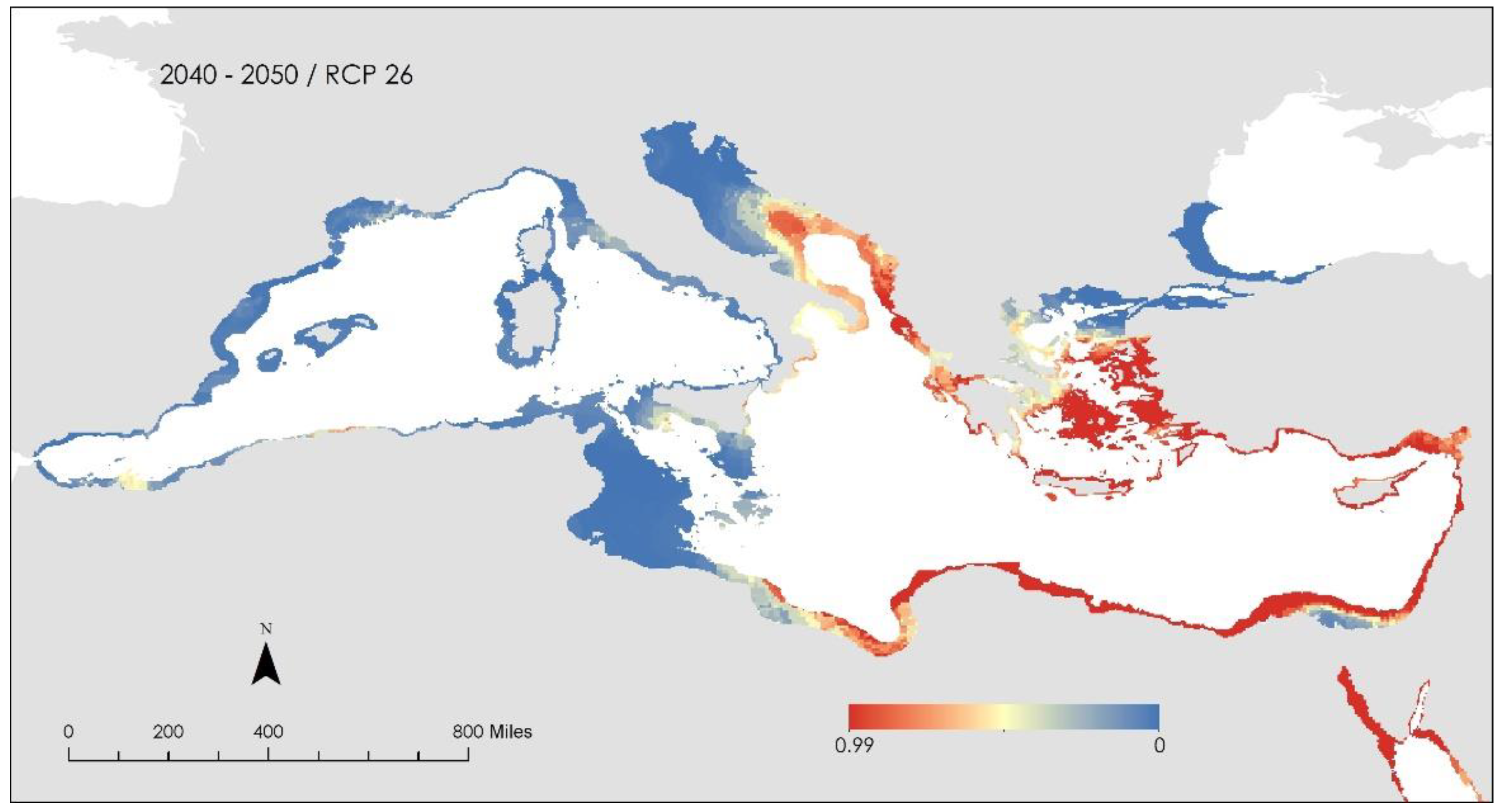
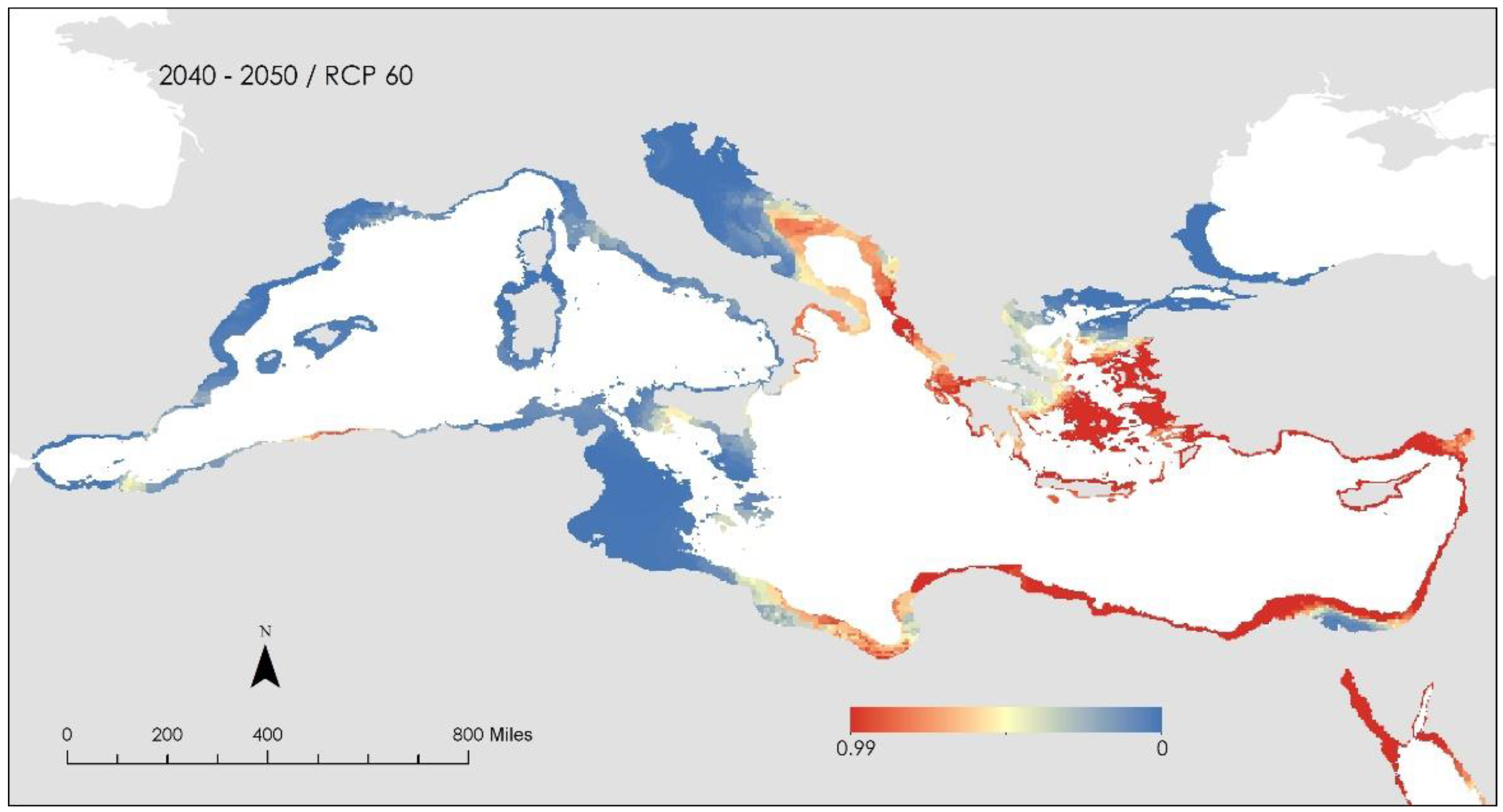
3. Results and Discussion
- i.
- the Patraikos Gulf, Greece
- ii.
- the Saronikos Gulf extending to the Northern Cyclades, Greece
- iii.
- the area surrounding Chios and Skyros islands, Greece
- iv.
- the area surrounding Corfu Island, Greece
- v.
- South of Sicily, Italy
- vi.
- the Gulf of Lion, France and
- vii.
- Certain scattered areas along the North African coast.
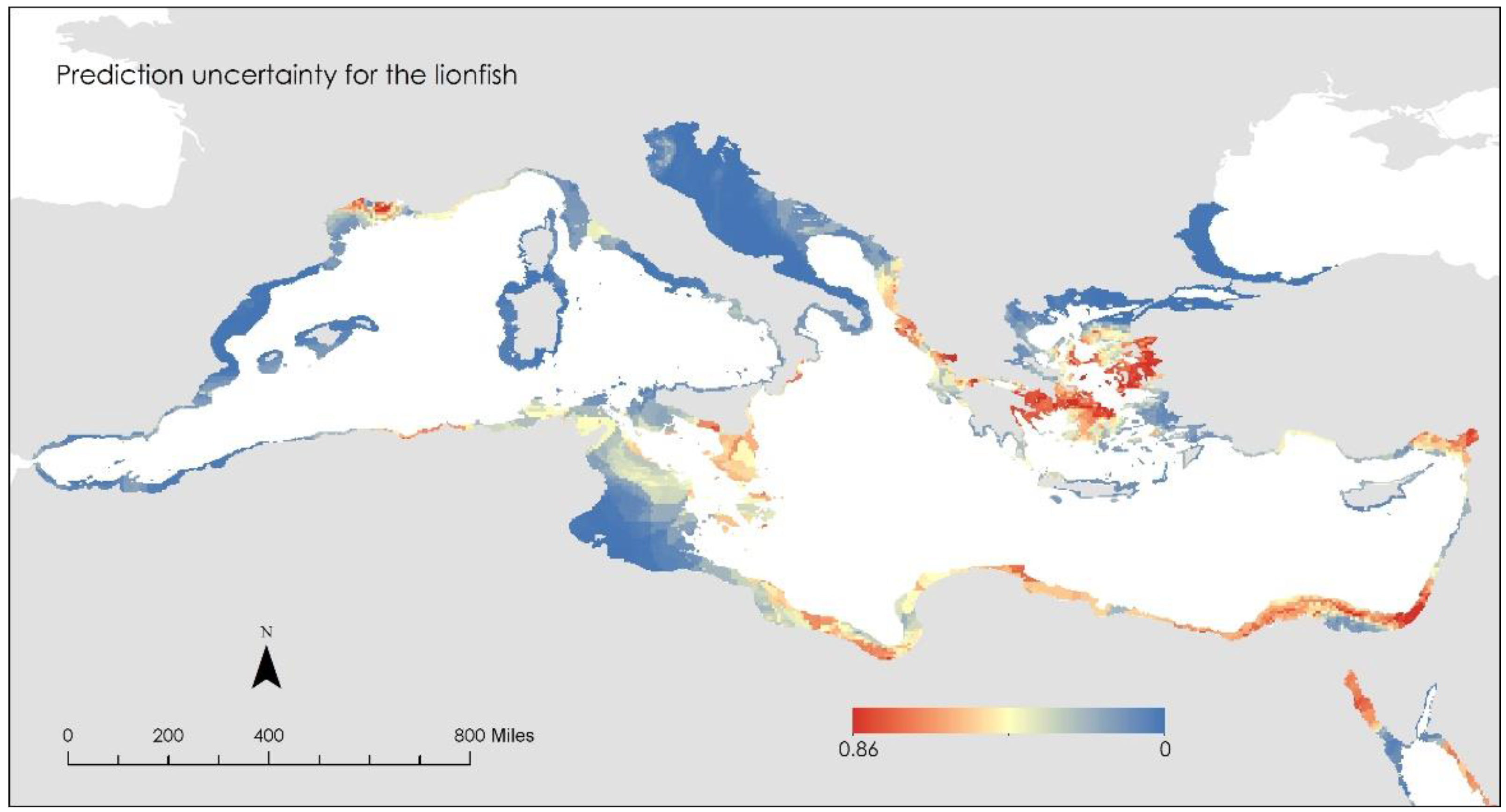
- i.
- The area around Lemnos Island, Greece
- ii.
- The area around the Nile Delta, Egypt
- iii.
- The area around Benghazi, Libya
- iv.
- The area around Algeria
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katsanevakis, S.; Moustakas, A. Uncertainty in Marine Invasion Science. Front. Mar. Sci. 2018, 5, 38. [Google Scholar] [CrossRef] [Green Version]
- Kalogirou, S. Alien Fish Species in the Eastern Mediterranean Sea: Invasion Biology in Coastal Ecosystems. Ph.D. Thesis, University of Gothenburg, Gothenburg, Sweden, 2011. [Google Scholar]
- Ballew, N.G.; Bacheler, N.M.; Kellison, G.T.; Schueller, A.M. Invasive lionfish reduce native fish abundance on a regional scale. Sci. Rep. 2016, 6, 32169. [Google Scholar] [CrossRef] [PubMed]
- Kalogirou, S. Ecological characteristics of the invasive pufferfish Lagocephalus sceleratus (Gmelin, 1789) in Rhodes, Eastern Mediterranean Sea. A case study from Rhodes. Mediterr. Mar. Sci. 2013, 14, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Ulman, A.; Kalogirou, S.; Pauly, D. The Dynamics of Maximum Lengths for the Invasive Silver-Cheeked Toadfish (Lagocephalus sceleratus) in the Eastern Mediterranean Sea. J. Mar. Sci. Eng. 2022, 10, 387. [Google Scholar] [CrossRef]
- CAR-SPAW-RA. 2022. Available online: https://www.car-spaw-rac.org/?Lionfish-The-lionfish-in-its-native-range-brief-description-of-its-biology-and#:~:text=Although%20lionfish%20is%20a%20food,endangered%20in%20their%20native%20range (accessed on 23 March 2022).
- Kulbicki, M.; Beets, J.; Chabanet, P.; Cure, K.; Darling, E.; Floeter, S.R.; Galzin, R.; Green, A.; Harmelin-Vivien, M.; Hixon, M.; et al. Distributions of Indo-Pacific lionfishes Pterois spp. in their native ranges: Implications for the Atlantic invasion. Mar. Ecol. Prog. Ser. 2012, 446, 189–205. [Google Scholar] [CrossRef] [Green Version]
- Albins, M.A.; Hixon, M.A. Invasive Indo-Pacific lionfish Pterois volitans reduce recruitment of Atlantic coral-reef fishes. Mar. Ecol. Prog. Ser. 2008, 367, 233–238. [Google Scholar] [CrossRef]
- Hixon, M.A.; Green, S.J.; Albins, M.A.; Akins, J.L.; Morris, J.A., Jr. Lionfish: A major marine invasion. Mar. Ecol. Prog. Ser. 2016, 558, 161–165. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, P.E.; Gardner, T.; Vives, S.P.; Gilligan, M.R.; Courtenay, W.R., Jr.; Ray, G.C.; Hare, J.A. Biological invasion of the Indo-Pacific lionfish Pterois volitans along the Atlantic coast of North America. Mar. Ecol. Prog. Ser. 2002, 235, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Semmens, B.X.; Buhle, E.R.; Salomon, A.K.; Pattengill-Semmens, C.V. A hotspot of non-native marine fishes: Evidence for the aquarium trade as an invasion pathway. Mar. Ecol. Prog. Ser. 2004, 266, 239–244. [Google Scholar] [CrossRef] [Green Version]
- Johnston, S.J.; Purkis, M.W. Are lionfish set for a Mediterranean invasion? Modelling explains why this is unlikely to occur. Mar. Pollut. Bull. 2014, 88, 138–147. [Google Scholar] [CrossRef]
- Morris, J.A., Jr. (Ed.) Invasive Lionfish: A Guide to Control and Management; Gulf and Caribbean Fisheries Institute: Marathon, FL, USA, 2012. [Google Scholar]
- Albins, M.A.; Hixon, M.A. Worst case scenario: Potential long-term effects of invasive predatory lionfish (Pterois volitans) on Atlantic and Caribbean coral-reef communities. Environ. Biol. Fish 2013, 96, 1151–1157. [Google Scholar] [CrossRef]
- Côté, I.M.; Green, S.; Hixon, M.A. Predatory fish invaders: Insights from the Indo-Pacific lionfish in the western Atlantic and Caribbean. Biol. Conserv. 2013, 164, 50–61. [Google Scholar] [CrossRef]
- Albins, M.A. Invasive Pacific lionfish Pterois volitans reduce abundance and species richness of native Bahamian coral-reef fishes. Mar. Ecol. Prog. Ser. 2015, 522, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Andradi-Brown, D.A.; Vermeij, M.; Slattery, M.; Lesser, M.; Bejarano, I.; Appeldoorn, R.; Goodbody-Gringley, G.; Chequer, A.D.; Pitt, J.M.; Eddy, C.; et al. Large-scale invasion of western Atlantic mesophotic reefs by invasive lionfish potentially undermines culling-based management. Biol. Invasions 2017, 19, 939–954. [Google Scholar] [CrossRef]
- Bariche, M.; Torres, M.; Azzurro, E. The presence of the invasive lionfish Pterois miles in the Mediterranean Sea Mediterr. Mar. Sci. 2013, 14, 292–294. [Google Scholar] [CrossRef]
- Kletou, D.; Hall-Spencer, J.M.; Kleitou, P. Kleitou A lionfish (Pterois miles) invasion has begun in the Mediterranean Sea. Mar. Biodivers. Rec. 2016, 9, 305. [Google Scholar] [CrossRef] [Green Version]
- Dailianis, T.; Akyol, O.; Babali, N.; Bariche, M.; Crocetta, F.; Gerovasileiou, V.; Chanem, R.; Gökoğlu, M.; Hasiotis, T.; Izquierdo-Muñoz, A.; et al. New Mediterranean biodiversity records (July 2016). Mediterr. Mar. Sci. 2016, 17, 608–626. [Google Scholar] [CrossRef] [Green Version]
- Azzurro, E.; Stancanelli, B.; Dimartino, V.; Bariche, M. Range expansion of the common lionfish Pterois miles (Bennett, 1828) in the Mediterranean Sea: An unwanted new guest for Italian waters. BioInvasions. Rec. 2017, 6, 95–98. [Google Scholar] [CrossRef]
- Bariche, M.; Kleitou, P.; Kalogirou, S.; Bernardi, G. Genetics reveal the identity and origin of the lionfish invasion in the Mediterranean Sea. Sci. Rep. 2017, 7, 6782. [Google Scholar] [CrossRef] [Green Version]
- Dimitriou, N.; Chartosia, J.M.; Hall-Spencer, P.; Kleitou, C.; Jimenez, C.; Antoniou, L.; Hadjioannou, D.; Kletou, S. Sfenthourakis Genetic data suggest multiple introductions of the lionfish (Pterois miles) into the Mediterranean Sea. Diversity 2019, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- Coll, M.; Piroddi, C.; Steenbeek, J.; Kaschner, K.; Ben Rais Lasram, F.; Aguzzi, J.; Ballesteros, E.; Bianchi, C.N.; Corbera, J.; Dailianis, T.; et al. The biodiversity of the Mediterranean Sea: Estimates, patterns, and threats. PLoS ONE 2010, 5, e11842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelist, G.; Rilov, D.; Golani, J.T.; Carlton, E. Spanier Restructuring the sea: Profound shifts in the world’s most invaded marine ecosystem. Divers. Distrib. 2013, 19, 69–77. [Google Scholar] [CrossRef]
- Hellmann, J.J.; Bierwagen, B.G.; Dukes, J.S.; Byers, J.E. Five Potential Consequences of Climate Change for Invasive Species. Conserv. Biol. 2008, 22, 534–543. [Google Scholar] [CrossRef]
- Walther, G.-R.; Roques, A.; Hulme, P.E.; Sykes, M.T.; Pyšek, P.; Kühn, I.; Zobel, M.; Bacher, S.; Botta-Dukát, Z.; Bugmann, H.; et al. Alien species in a warmer world: Risks and opportunities. Trends Ecol. Evol. 2009, 24, 686–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Occhipinti-Ambrogi, A. Global change and marine communities: Alien species and climate change. Mar. Pollut. Bull. 2007, 55, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Bellard, C.; Thuiller, W.; Leroy, B.; Genovesi, P.; Bakkenes, M.; Courchamp, F. Will climate change promote future invasions? Glob. Chang. Biol. 2013, 19, 3740–3748. [Google Scholar] [CrossRef] [PubMed]
- Ben Rais Lasram, F.; Guilhaumon, F.; Mouillot, D. Fish diversity patterns in the Mediterranean Sea: Deviations from a mid-domain model. Mar. Ecol. Prog. Ser. 2009, 376, 253–267. [Google Scholar] [CrossRef] [Green Version]
- Arndt, E.; Schembri, P.J. Common traits associated with establishment and spread of Lessepsian fishes in the Mediterranean Sea. Mar. Biol. 2015, 162, 2141–2153. [Google Scholar] [CrossRef]
- Hiddink, J.G.; Ben Rais Lasram, F.; Cantrill, J.; Davies, A.J. Keeping pace with climate change: What can we learn from the spread of Lessepsian migrants? Glob. Chang. Biol. 2012, 18, 2161–2172. [Google Scholar] [CrossRef]
- Poursanidis, D.; Kalogirou, S.; Azzurro, E.; Parravicini, V.; Bariche, M.; Dohna, H. Habitat suitability, niche unfilling and the potential spread of Pterois miles in the Mediterranean Sea. Mar. Pollut. Bull. 2020, 154, 111054. [Google Scholar] [CrossRef]
- Uusitalo, L.; Lehikoinen, A.; Helle, I.; Myrberg, K. An overview of methods to evaluate uncertainty of deterministic models in decision support. Environmental Modelling and Software 2015, 63, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Guisan, A.; Thuiller, W.; Zimmermann, N. Habitat Suitability and Distribution Models: With Applications in R (Ecology, Biodiversity and Conservation); Cambridge University Press: Cambridge, UK, 2017. [Google Scholar] [CrossRef]
- Carlson, C.J. embarcadero: Species distribution modelling with Bayesian additive regression trees in r. Methods Ecol. Evol. 2020, 11, 850–858. [Google Scholar] [CrossRef]
- Chipman, H.A.; George, E.I.; McCulloch, R.E. BART: Bayesian additive regression trees. Ann. Appl. Stat. 2010, 4, 266–298. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Liu, C.; Newell, G.; White, M. The effect of sample size on the accuracy of species distribution models: Considering both presences and pseudo-absences or background sites. Ecography 2019, 42, 535–548. [Google Scholar] [CrossRef] [Green Version]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Fielding, A.H.; Bell, J.F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 1997, 24, 38–49. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Le Lay, G.; Helfer, V.; Randin, C.; Guisan, A. Evaluating the ability of habitat suitability models to predict species presences. Ecol. Modell. 2006, 199, 142–152. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Measuring and comparing the accuracy of species distribution models with presence-absence data. Ecography 2011, 34, 232–243. [Google Scholar] [CrossRef]
- Liu, C.; Newell, G.; White, M. On the selection of thresholds for predicting species occurrence with presence-only data. Ecol. Evol. 2016, 6, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Sofaer, H.R.; Hoeting, J.A.; Jarnevich, C.S. The area under the precision-recall curve as a performance metric for rare binary events. Methods Ecol. Evol. 2019, 10, 565–577. [Google Scholar] [CrossRef]
- Leroy, B.; Delsol, R.; Hugueny, B.; Meynard, C.N.; Barhoumi, C.; Barbet-Massin, M.; Bellard, C. Without quality presence–absence data, discrimination metrics such as TSS can be misleading measures of model performance. J. Biogeogr. 2018, 45, 1994–2002. [Google Scholar] [CrossRef]
- Hammer, B.; Frasco, M. Metrics: Evaluation Metrics for Machine Learning, R Package Version 0.1.4 2018. 2018. Available online: https://cran.r-project.org/web/packages/Metrics/index.html (accessed on 25 March 2022).
- Schwarz, J.; Heider, D. GUESS: Projecting machine learning scores to well-calibrated probability estimates for clinical decision-making. Bioinformatics 2019, 35, 2458–2465. [Google Scholar] [CrossRef] [PubMed]
- Signorell, A.; Aho, K.; Anderegg, N.; Aragon, T.; Arppe, A.; Baddeley, A.; Bolker, B.; Caeiro, F.; Champely, S.; Chessel, D. DescTools: Tools for Descriptive Statistics, R Package Version 0.99-40 2021. 2021. Available online: https://cran.r-project.org/web/packages/Metrics/index.html (accessed on 25 March 2022).
- Smith, A.B. enmSdm: Tools for Modeling Species Niches and Distributions, R Package Version 0.5.1.5 2020. 2020. Available online: https://cran.r-project.org/web/packages/Metrics/index.html (accessed on 25 March 2022).
- Yan, Y. MLmetrics: Machine Learning Evaluation Metrics, R Package Version 1.1.1 2016. 2016. Available online: https://cran.r-project.org/web/packages/Metrics/index.html (accessed on 25 March 2022).
- Assis, J.; Tyberghein, L.; Bosch, S.; Verbruggen, H.; Serrão, E.A.; De Clerck, O.; Tittensor, D. Bio-ORACLE v2.0: Extending marine data layers for bioclimatic modelling. Global Ecol. Biogeogr. 2018, 27, 277–284. [Google Scholar] [CrossRef]
- D’Amen, M.; Azzurro, E. Lessepsian fish invasion in Mediterranean marine protected areas: A risk assessment under climate change scenarios. ICES J. Mar. Sci. 2020, 77, 388–397. [Google Scholar] [CrossRef]
- Dimitriadis, C.; Galanidi, M.; Zenetos, A.; Corsini-Foka, M.; Giovos, I.; Karachle, P.K.; Fournari-Konstantinidoy, I.; Kytinou, E.; Issaris, Y.; Azzurro, E.; et al. Updating the occurrences of Pterois miles in the Mediterranean Sea, with considerations on the thermal boundaries and future range expansion. Mediterr. Mar. Sci. 2020, 21, 62–69. [Google Scholar] [CrossRef]
- GBIF. 2021. GBIF.org (5 March 2021) GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.gmy9g2 (accessed on 5 March 2021).
- Dragičević, B.; Ugarković, P.; Krželj, M.; Zurub, D.; Dulčić, J. New record of Pterois cf. miles (Actinopterygii: Scorpaeniformes: Scorpaenidae) from the eastern middle Adriatic Sea (Croatian waters): Northward expansion. Acta Ichthyol. Piscat. 2021, 51, 379–383. [Google Scholar] [CrossRef]
- Coro, G.; Vilas, L.G.; Magliozzi, C.; Ellenbroek, A.; Scarponi, P.; Pagano, P. Forecasting the ongoing invasion of Lagocephalus sceleratus in the Mediterranean Sea. Ecol. Model. 2018, 371, 37–49. [Google Scholar] [CrossRef]
- Schmid, C.H.; Griffith, J.L. Multivariate classification rules: Calibration and discrimination. Encycl. Biostat. 2005. [Google Scholar] [CrossRef]
- IPCC. 2021: Summary for Policymakers. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Masson-Delmotte, V.P., Zhai, A., Pirani, S.L., Connors, C., Péan, S., Berger, N., Caud, Y., Chen, L., Goldfarb, M.I., Gomis, M., et al., Eds.; Cambridge University Press: Cambridge, UK, 2021; in press. [Google Scholar]
- Pörtner, H.-O.; Roberts, D.C.; Poloczanska, M.; Mintenbeck, E.S.; Tignor, K.; Alegría, A.; Craig, M.; Langsdorf, S.; Löschke, S.; Möller, V.; et al. (Eds.) IPCC, 2022: Summary for Policymakers. In Climate Change 2022: Impacts, Adaptation, and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2022; in press. [Google Scholar]
- Juza, M.; Fernández-Mora, À.; Tintoré, J. Sub-Regional Marine Heat Waves in the Mediterranean Sea from Observations: Long-Term Surface Changes, Sub-Surface and Coastal Responses. Front. Mar. Sci. 2022, 9, 785771. [Google Scholar] [CrossRef]
- Grodsky, S.A.; Reul, N.; Bentamy, A.; Vandemark, D.; Guimbard, S. Eastern Mediterranean salinification observed in satellite salinity from SMAP mission. J. Mar. Syst. 2019, 198, 103190. [Google Scholar] [CrossRef]
- Jarnevich, C.S.; Young, N.E.; Thomas, C.C.; Grissom, P.; Backer, D.; Frid, L. Assessing ecological uncertainty and simulation model sensitivity to evaluate an invasive plant species’ potential impacts to the landscape. Sci. Rep. 2020, 10, 19069. [Google Scholar] [CrossRef] [PubMed]
- Sniazhko, S.; Muralidharan, E. Uncertainty in decision-making: A review of the international business literature. Cogent Bus. Manag. 2019, 6, 1650692. [Google Scholar] [CrossRef]
- Sofaer, H.R.; Jarnevich, C.S.; Pearse, I.S.; Smyth, R.L.; Auer, S.; Cook, G.; Edwards, T.C.; Guala, G.F.; Howard, T.G.; Morisette, J.T.; et al. Development and Delivery of Species Distribution Models to Inform Decision-Making. BioScience 2019, 69, 544–557. [Google Scholar] [CrossRef]



| Metric | Value |
|---|---|
| TSS | 0.98 |
| AUC | 0.99 |
| Kappa | 0.94 |
| Boyce | 0.97 |
| Sorensen’s index | 0.75 |
| Nagelkerke | 0.94 |
| McFadden | 0.89 |
| Brier | 0.008 |
| bias | −0.0004 |
| MAE | 0.04 |
| RMSE | 0.11 |
| ECE | 0.02 |
| MCE | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poursanidis, D.; Kougioumoutzis, K.; Minasidis, V.; Chartosia, N.; Kletou, D.; Kalogirou, S. Uncertainty in Marine Species Distribution Modelling: Trying to Locate Invasion Hotspots for Pterois miles in the Eastern Mediterranean Sea. J. Mar. Sci. Eng. 2022, 10, 729. https://doi.org/10.3390/jmse10060729
Poursanidis D, Kougioumoutzis K, Minasidis V, Chartosia N, Kletou D, Kalogirou S. Uncertainty in Marine Species Distribution Modelling: Trying to Locate Invasion Hotspots for Pterois miles in the Eastern Mediterranean Sea. Journal of Marine Science and Engineering. 2022; 10(6):729. https://doi.org/10.3390/jmse10060729
Chicago/Turabian StylePoursanidis, Dimitris, Kostas Kougioumoutzis, Vasileios Minasidis, Niki Chartosia, Demetris Kletou, and Stefanos Kalogirou. 2022. "Uncertainty in Marine Species Distribution Modelling: Trying to Locate Invasion Hotspots for Pterois miles in the Eastern Mediterranean Sea" Journal of Marine Science and Engineering 10, no. 6: 729. https://doi.org/10.3390/jmse10060729
APA StylePoursanidis, D., Kougioumoutzis, K., Minasidis, V., Chartosia, N., Kletou, D., & Kalogirou, S. (2022). Uncertainty in Marine Species Distribution Modelling: Trying to Locate Invasion Hotspots for Pterois miles in the Eastern Mediterranean Sea. Journal of Marine Science and Engineering, 10(6), 729. https://doi.org/10.3390/jmse10060729










