Complex Plastids and the Evolution of the Marine Phytoplankton
Abstract
:1. Origin of Photosynthesis
2. Origin of Plastids
2.1. Primary Endosymbiosis

2.2. Geologic and Atmospheric Chemistry Context of Primary Endosymbiosis
2.3. Geochemical Consequences of Primary Endosymbiosis
2.4. Genome Reduction of Symbiont, Transfer of Genes to the Nucleus and Retention of Genes in the Plastid
2.5. Theories for the Relationship between Host and Symbiont
2.6. Ecological and Diversity Consequences of Endosymbiosis
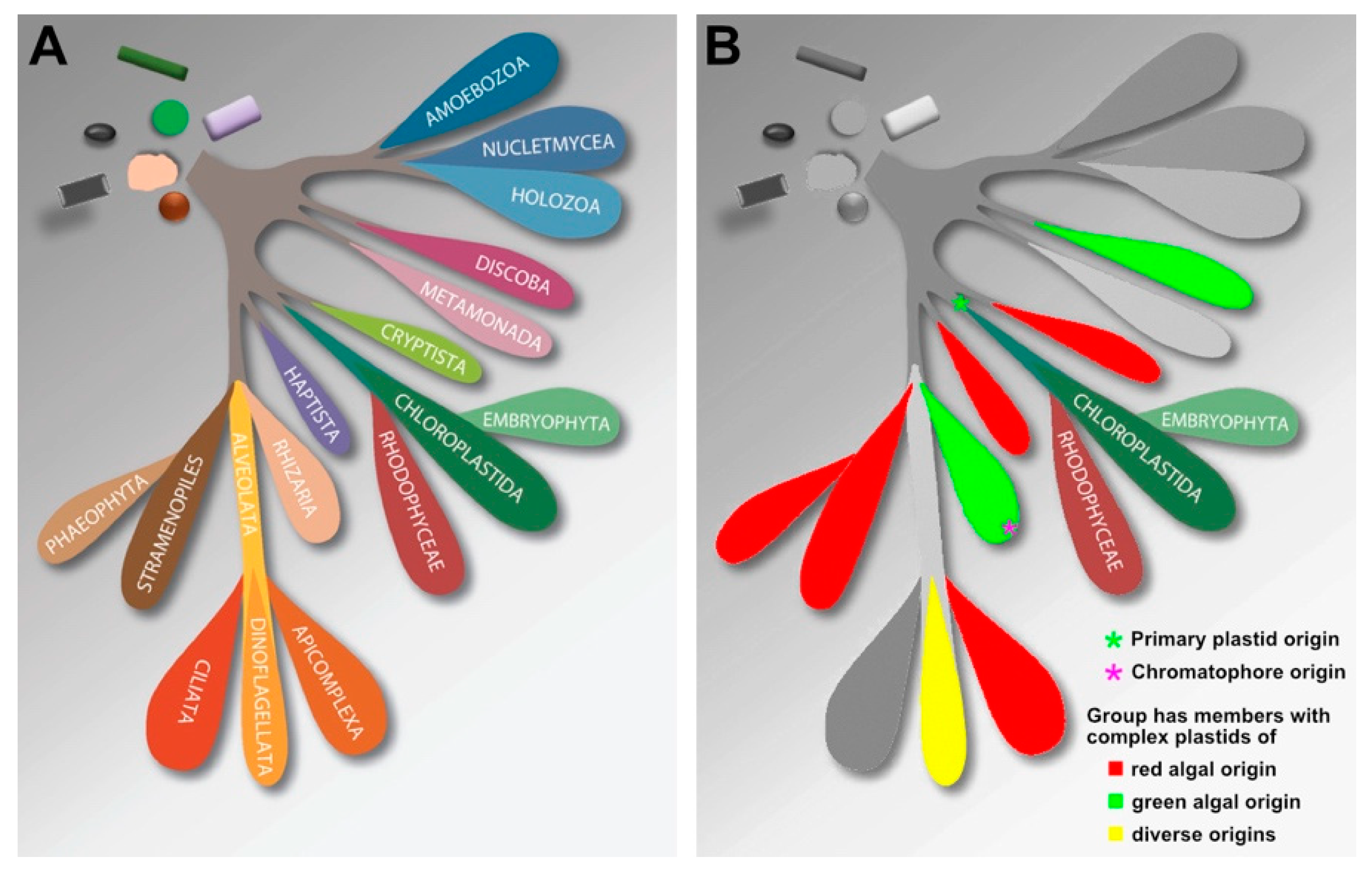
2.7. Diversification of Archaeplastida and Relationship to Other Eukaryotic Groups
2.8. Ecological Niches and Roles of Red vs Green
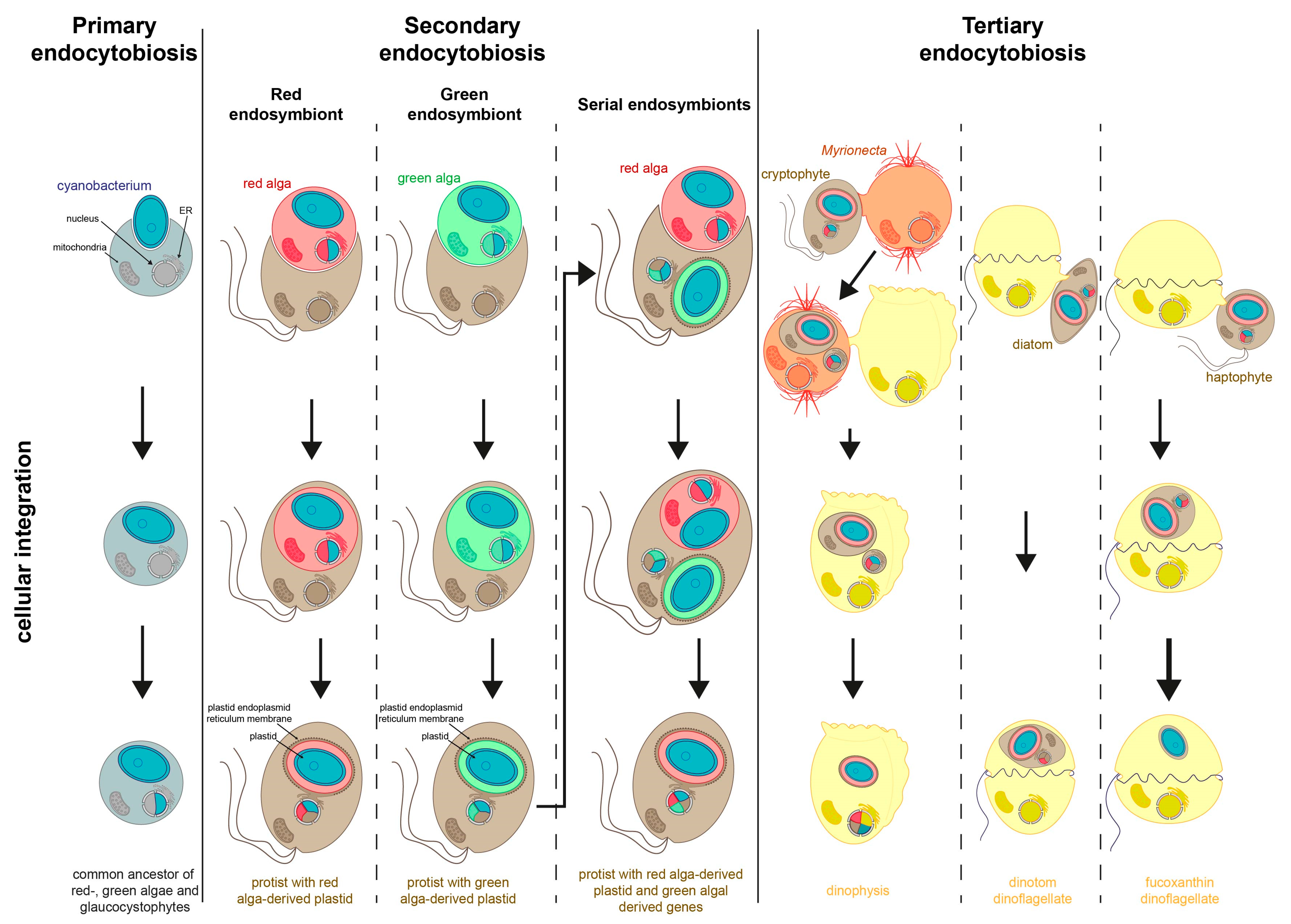
3. Secondary Endosymbiosis
3.1. Diversification of Secondary Plastids
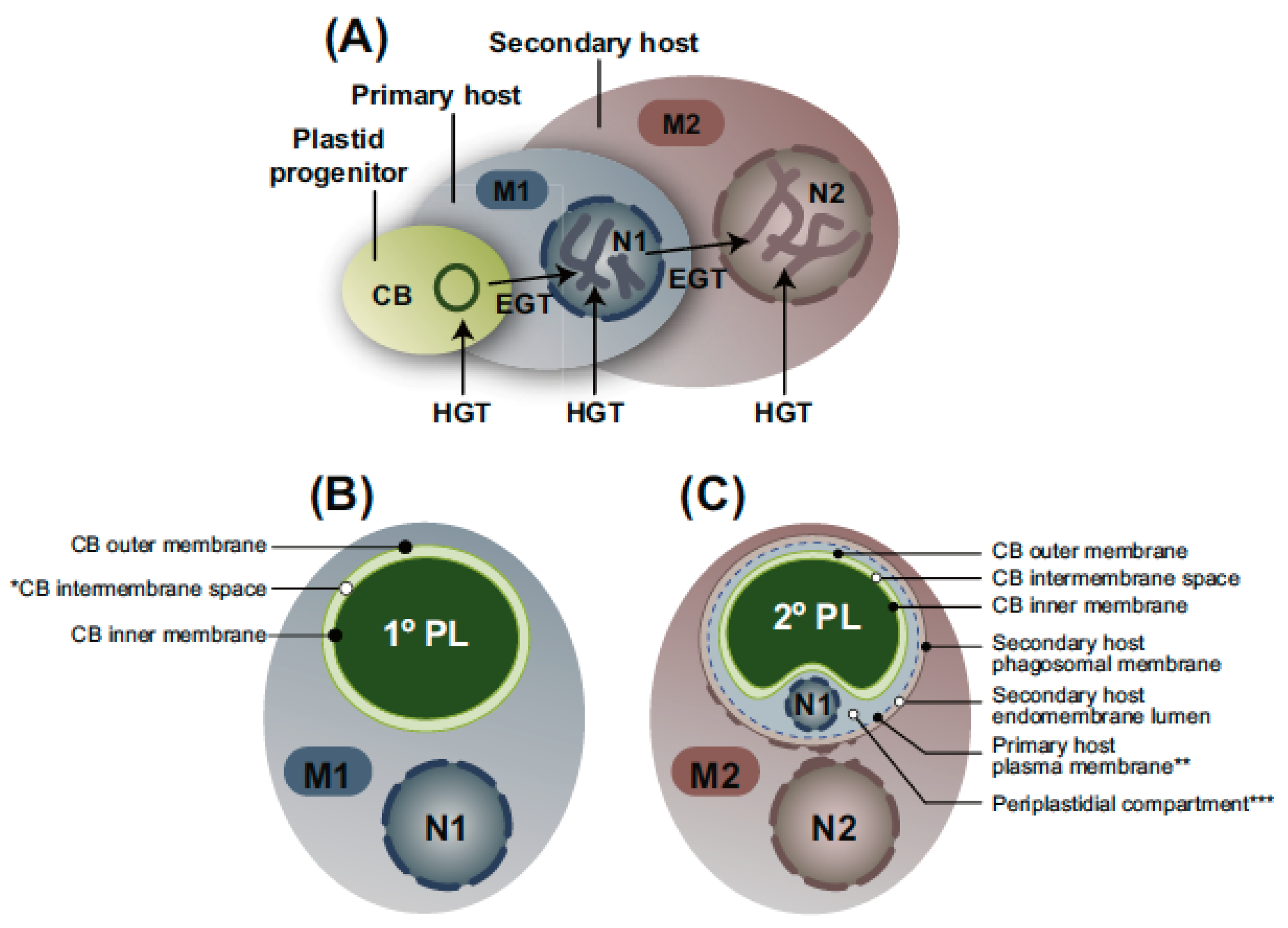
3.2. Discussion of Number and Timing of Secondary Endosymbiosis
3.3. Ancient Green Endosymbiosis
3.4. Permian/Triassic (P/T) Extinction and the Green vs. Red Switch in the Ocean
3.5. Eukaryotic Nuclei Reduction and Gene Transfer
3.6. Genome Evolution Consequences of Higher Order Endosymbioses
3.7. Convergent Evolution of Nucleomorph Structure in Chloroarachniophytes and Cryptophytes
3.8. Relationship of Membrane Number to Protein Targeting
4. Tertiary Endosymbiosis
4.1. Theories for Relationships of Hosts and Symbionts (Symbiosis vs. Predation)
4.2. Kleptoplasts as Intermediate States during Organelle Evolution
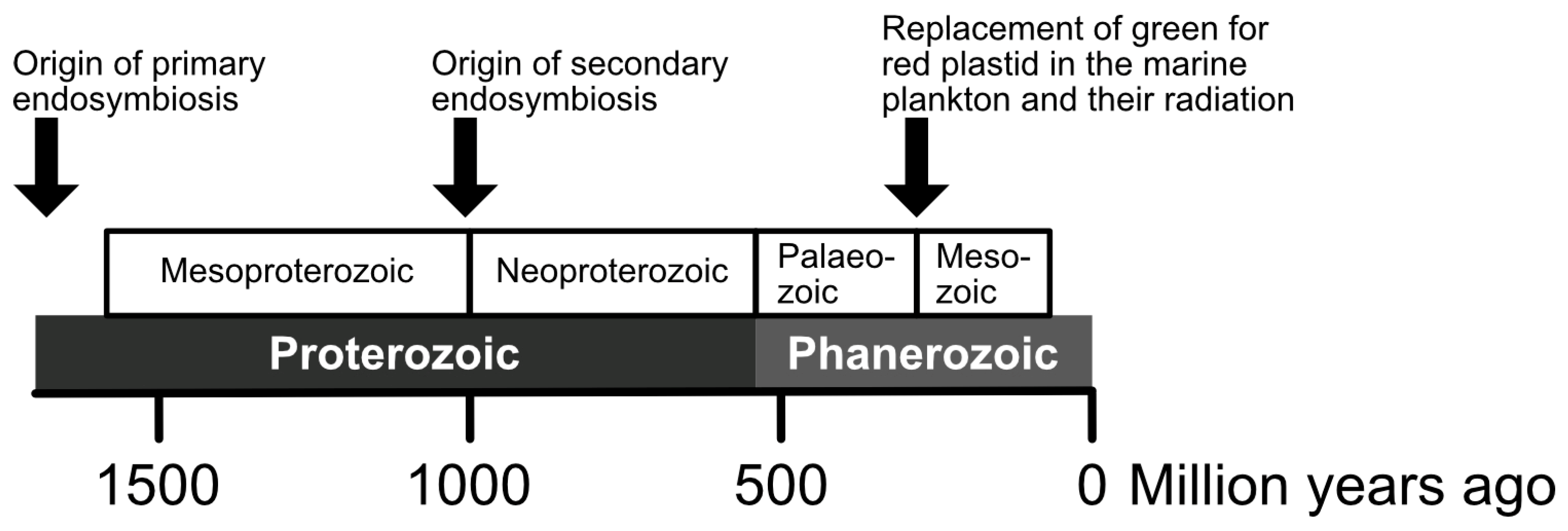
5. Molecular Clocks for the Timing and Diversification of Complex Plastids
6. Multigene Trees Showing How Many Groups Are in Plankton
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hohmann-Marriott, M.F.; Blankenship, R.E. Evolution of photosynthesis. Annu. Rev. Plant Biol. 2011, 62, 515–548. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.W.; Hemp, J.; Johnson, J.E. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci. 2016, 44, 647–683. [Google Scholar] [CrossRef]
- Holland, H.D. The oxygenation of the atmosphere and oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209. [Google Scholar] [CrossRef]
- Rasmussen, B.; Fletcher, I.R.; Brocks, J.J.; Kilburn, M.R. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 2008, 455, 1101–1104. [Google Scholar] [CrossRef]
- Geider, R.J.; Delucia, E.H.; Falkowski, P.G.; Finzi, A.C.; Grime, J.P.; Grace, J.; Kana, T.M.; La Roche, J.; Long, S.P.; Osborne, B.A.; et al. Primary productivity of planet earth: Biological determinants and physical constraints in terrestrial and aquatic habitats. Glob. Change Biol. 2001, 7, 849–882. [Google Scholar] [CrossRef]
- Bendall, D.S.; Howe, C.J.; Nisbet, E.G.; Nisbet, R.E. Photosynthetic and atmospheric evolution. Introduction. Philosophical transactions of the Royal Society of London. Series B, Biol. Sci. 2008, 363, 2625–2628. [Google Scholar] [CrossRef]
- Cardona, T. Early Archean origin of heterodimeric photosystem I. Heliyon 2018, 4, e00548. [Google Scholar] [CrossRef]
- Elsevier. Photosynthesis Originated a Billion Years Earlier than We Thought, Study Shows. Available online: www.sciencedaily.com/releases/2018/03/180306093304.htm (accessed on 14 July 2023).
- Baymann, F.; Brugna, M.; Mühlenhoff, U.; Nitschke, W. Daddy, where did (PS)I come from? Biochim. Et Biophys. Acta 2001, 1507, 291–310. [Google Scholar] [CrossRef]
- Raymond, J. The role of horizontal gene transfer in photosynthesis, oxygen production, and oxygen tolerance. Methods Mol. Biol. 2009, 532, 323–338. [Google Scholar] [CrossRef]
- Soo, R.M.; Hemp, J.; Parks, D.H.; Fischer, W.W.; Hugenholtz, P. On the origins of oxygenic photosynthesis and aerobic respiration in cyanobacteria. Science 2017, 355, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Deusch, O.; Landan, G.; Roettger, M.; Gruenheit, N.; Kowallik, K.V.; Allen, J.F.; Martin, W.; Dagan, T. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol. Biol. Evol. 2008, 25, 748–761. [Google Scholar] [CrossRef]
- McFadden, G.I. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2014, 6, a016105. [Google Scholar] [CrossRef] [PubMed]
- Domman, D.; Horn, M.; Embley, T.M.; Williams, T.A. Plastid establishment did not require a chlamydial partner. Nat. Commununications 2015. [CrossRef] [PubMed]
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef]
- Howe, C.J.; Barbrook, A.C.; Nisbet, R.E.; Lockhart, P.J.; Larkum, A.W. The origin of plastids. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 2675–2685. [Google Scholar] [CrossRef]
- Gruber, A. What’s in a name? How organelles of endosymbiotic origin can be distinguished from endosymbionts. Microb. Cell 2019, 6, 123–133. [Google Scholar] [CrossRef]
- Keeling, P.J.; Archibald, J.M. Organelle evolution: what’s in a name? Curr. Biol. 2008, 18, R345–R347. [Google Scholar] [CrossRef]
- Oborník, M. In the beginning was the word: How terminology drives our understanding of endosymbiotic organelles. Microb. Cell 2019, 6, 134–141. [Google Scholar] [CrossRef]
- Sun, J.; Rutherford, S.T.; Silhavy, T.J.; Huang, K.C. Physical properties of the bacterial outer membrane. Nat. Rev. Microbiol. 2022, 20, 236–248. [Google Scholar] [CrossRef]
- Höhr, A.I.C.; Lindau, C.; Wirth, C.; Qiu, J.; Stroud, D.A.; Kutik, S.; Guiard, B.; Hunte, C.; Becker, T.; Pfanner, N.; et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 2018, 359, eaah6834. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000, 5, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Huang, L.F.; Li, H.M.; Chen, Y.R.; Yu, S.M. Signal peptide-dependent targeting of a rice alpha-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 2004, 135, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Villarejo, A.; Buren, S.; Larsson, S.; Dejardin, A.; Monne, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Price, D.C.; Chan, C.X.; Yoon, H.S.; Yang, E.C.; Qiu, H.; Weber, A.P.; Schwacke, R.; Gross, J.; Blouin, N.A.; Lane, C.; et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 2012, 335, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Tanidokoro, K.; Shimizu, Y.; Kawarabayasi, Y.; Ohshima, T.; Sato, M.; Tadano, S.; Ishikawa, H.; Takio, S.; Takechi, K.; et al. Moss chloroplasts are surrounded by a peptidoglycan wall containing D-Amino acids. Plant Cell 2016, 28, 1521–1532. [Google Scholar] [CrossRef]
- Lauterborn, R. Protozoenstudien II. Paulinella chromatophora nov. gen., nov. spec., ein beschalter Rhizopode des Süßwassers mit blaugrünen chromatophorenartigen Einschlüssen. Z. Wiss. Zool. 1895, 59, 537–544. [Google Scholar]
- Marin, B.M.; Nowack, E.C.; Melkonian, M. A plastid in the making: evidence for a second primary endosymbiosis. Protist 2005, 156, 425–432. [Google Scholar] [CrossRef]
- Kies, L. Electron microscopical investigations on Paulinella chromatophora Lauterborn, a thecamoeba containing blue-green endosymbionts (Cyanelles). Protoplasma 1974, 80, 69–89. [Google Scholar] [CrossRef]
- Nowack, E.C.M.; Melkonian, M.; Glöckner, G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 2008, 18, 410–418. [Google Scholar] [CrossRef]
- Nowack, E.C.M.; Grossman, A.R. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc. Natl. Acad. Sci. USA 2012, 109, 5340–5345. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, S.; Kim, E. A modern descendant of early green algal phagotrophs. Curr. Biol. 2013, 23, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Gawryluk, R.M.R.; Tikhonenkov, D.V.; Hehenberger, E.; Husnik, F.; Mylnikov, A.P.; Keeling, P.J. Non-photosynthetic predators are sister to red algae. Nature 2019, 572, 240–243. [Google Scholar] [CrossRef]
- Irisarri, I.; Strassert, J.F.H.; Burki, F. Phylogenomic insights into the origin of primary plastids. Syst. Biol. 2021, 71, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Nakayama, T.; Reyes-Prieto, A.; Andersen, R.A.; Boo, S.M.; Ishida, K.; Bhattacharya, D. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol. Biol. 2009, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.M.; Löffelhardt, W. Protein translocation into and within cyanelles (Review). Mol. Membr. Biol. 2005, 22, 123–132. [Google Scholar] [CrossRef]
- Chan, C.X.; Gross, J.; Yoon, H.S.; Bhattacharya, D. Plastid origin and evolution: New models provide insights into old problems. Plant Physiol. 2011, 155, 1552–1560. [Google Scholar] [CrossRef]
- Rosing, M.T.; Bird, D.K.; Sleep, N.H.; Glassley, W.; Albarede, F. The rise of continents—An essay on the geologic consequences of photosynthesis. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2006, 232, 99–113. [Google Scholar] [CrossRef]
- Ozaki, K.; Tajika, E.; Hong, P.K.; Nakagawa, Y.; Reinhard, C.T. Effects of primitive photosynthesis on Earth’s early climate system. Nat. Geosci. 2018, 11, 55–59. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Godfrey, L.V. Electrons, life and the evolution of Earth's oxygen cycle. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 2705–2716. [Google Scholar] [CrossRef]
- Larkum, A.W.; Lockhart, P.J.; Howe, C.J. Shopping for plastids. Trends Plant Sci. 2007, 12, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Martin, W. Gene transfer from organelles to the nucleus: Frequent and in big chunks. Proc. Natl. Acad. Sci. USA 2003, 100, 8612–8614. [Google Scholar] [CrossRef]
- Martin, W.; Herrmann, R.G. Gene transfer from organelles to the nucleus: How much, what happens, and Why? Plant Physiol. 1998, 118, 9–17. [Google Scholar] [CrossRef]
- Allen, J.F. The function of genomes in bioenergetic organelles. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2003, 358, 19–38. [Google Scholar] [CrossRef] [PubMed]
- McFadden, G.I. Plastids and protein targeting. J. Eukaryot. Microbiol. 1999, 46, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.F. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. TIG 1998, 14, 307–311. [Google Scholar] [CrossRef] [PubMed]
- de Vries, J.; Gould, S.B. The monoplastidic bottleneck in algae and plant evolution. J. Cell Sci. 2018, 131, jcs203414. [Google Scholar] [CrossRef]
- Allen, J.F. The CoRR hypothesis for genes in organelles. J. Theor. Biol. 2017, 434, 50–57. [Google Scholar] [CrossRef]
- Gross, J.; Bhattacharya, D. Endosymbiont or host: Who drove mitochondrial and plastid evolution? Biol. Direct 2011, 6, 12. [Google Scholar] [CrossRef]
- Johnson, M.D. The acquisition of phototrophy: Adaptive strategies of hosting endosymbionts and organelles. Photosynth. Res. 2011, 107, 117–132. [Google Scholar] [CrossRef]
- Nowack, E.C.; Melkonian, M. Endosymbiotic associations within protists. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Hengeveld, R.; Fedonkin, M.A. Causes and consequences of eukaryotization through mutualistic endosymbiosis and compartmentalization. Acta Biotheor. 2004, 52, 105–154. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.F.; Garg, S.; Zimorski, V. Endosymbiotic theories for eukaryote origin. Philosophical Transactions of the Royal Society of London. Ser. B Biol. Sci. 2015, 370, 20140330. [Google Scholar] [CrossRef]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef]
- Rumpho, M.E.; Pelletreau, K.N.; Moustafa, A.; Bhattacharya, D. The making of a photosynthetic animal. J. Exp. Biol. 2011, 214, 303–311. [Google Scholar] [CrossRef]
- Stoecker, D.K.; Johnson, M.D.; Vargas, C.d.; Not, F. Acquired phototrophy in aquatic protists. Aquat. Microb. Ecol. 2009, 57, 279–310. [Google Scholar]
- Mishra, S.; Joshi, B.; Dey, P.; Pathak, H.; Pandey, N.; Kohra, A. CCM in photosynthetic bacteria and marine alga. J. Pharmacogn. Phytochem. 2018, 7, 928–937. [Google Scholar]
- He, S.; Crans, V.L.; Jonikas, M.C. The pyrenoid: The eukaryotic CO2-concentrating organelle. Plant Cell 2023, 35, 3236–3259. [Google Scholar] [CrossRef] [PubMed]
- Woodward, F.I.; Lomas, M.R.; Kelly, C.K. Global climate and the distribution of plant biomes. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2004, 359, 1465–1476. [Google Scholar] [CrossRef]
- Tan, J.; Slattery, M.R.; Yang, X.; Jiang, L. Phylogenetic context determines the role of competition in adaptive radiation. Proc. Biol. Sci. 2016, 283, 20160241. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J. The evolution of modern eukaryotic phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef]
- Gruber, A.; Oborník, M. Evolution of plastids and mitochondria in diatoms. In Diatom Photosynthesis: From Primary Production to High Value; Goessling, J.W., Serodio, J., Lavaud, J., Eds.; Scrivener Publishing LLC: Berverly, CA, USA, 2023. [Google Scholar]
- Archibald, J.M. The puzzle of plastid evolution. Curr. Biol. 2009, 19, R81–R88. [Google Scholar] [CrossRef]
- Archibald, J.M. Secondary endosymbiosis. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Oxford, UK, 2009; pp. 438–446. [Google Scholar] [CrossRef]
- Delwiche, C.F. Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 1999, 154, S164–S177. [Google Scholar] [CrossRef]
- Melkonian, M. Phylogeny of photosynthetic protists and their plastids. Verh. Dtsch. Zool. Ges. 1996, 89, 71–96. [Google Scholar]
- Gould, S.; Maier, U.-G.; Martin, W.F. Protein import and the origin of red complex plastids. Curr. Biol. 2015, 25, R515–R521. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.; Zauner, S.; Fraunholz, M.; Beaton, M.; Penny, S.; Deng, L.-T.; Wu, X.; Reith, M.; Cavalier-Smith, T.; Maier, U.-G. The highly reduced genome of an enslaved algal nucleus. Nature 2001, 410, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Gilson, P.R.; Su, V.; Slamovits, C.H.; Reith, M.E.; Keeling, P.J.; McFadden, G.I. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus. Proc. Natl. Acad. Sci. USA 2006, 103, 9566–9571. [Google Scholar] [CrossRef] [PubMed]
- Gagat, P.; Bodyl, A.; Mackiewicz, P.; Stiller, J.W. Tertiary plastid endosymbioses in dinoflagellates. In Endosymbiosis; Loeffelhardt, W., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 233–290. [Google Scholar]
- Green, B.R. After the primary endosymbiosis: An update on the chromalveolate hypothesis and the origins of algae with Chl c. Photosynth. Res. 2011, 107, 103–115. [Google Scholar] [CrossRef]
- Hackett, J.D.; Anderson, D.M.; Erdner, D.L.; Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 2004, 91, 1523–1534. [Google Scholar] [CrossRef]
- Archibald, J.M. Chapter Three—The evolution of algae by secondary and tertiary endosymbiosis. In Advances in Botanical Research; Piganeau, G., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 64, pp. 87–118. [Google Scholar]
- Cavalier-Smith, T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 1999, 46, 347–366. [Google Scholar] [CrossRef]
- Baurain, D.; Brinkmann, H.; Petersen, J.; Rodriguez-Ezpeleta, N.; Stechmann, A.; Demoulin, V.; Roger, A.J.; Burger, G.; Lang, B.F.; Philippe, H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol. Biol. Evol. 2010, 27, 1698–1709. [Google Scholar] [CrossRef]
- Burki, F.; Okamoto, N.; Pombert, J.F.; Keeling, P.J. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Biol. Sci./R. Soc. 2012, 279, 2246–2254. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Grant, J.; Tekle, Y.I.; Lasek-Nesselquist, E.; Morrison, H.G.; Sogin, M.L.; Patterson, D.J.; Katz, L.A. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst. Biol. 2010, 59, 518–533. [Google Scholar] [CrossRef]
- Strassert, J.F.H.; Irisarri, I.; Williams, T.A.; Burki, F. A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids. Nat. Commun. 2021, 12, 1879. [Google Scholar] [CrossRef]
- Moustafa, A.; Beszteri, B.; Maier, U.G.; Bowler, C.; Valentin, K.; Bhattacharya, D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 2009, 324, 1724–1726. [Google Scholar] [CrossRef] [PubMed]
- Archibald, J.M.; Rogers, M.B.; Toop, M.; Ishida, K.-i.; Keeling, P.J. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. 2003, 100, 7678–7683. [Google Scholar] [CrossRef] [PubMed]
- Minge, M.A.; Shalchian-Tabrizi, K.; Tørresen, O.K.; Takishita, K.; Probert, I.; Inagaki, Y.; Klaveness, D.; Jakobsen, K.S. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 2010, 10, 191. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Medlin, L. The Phylogeny of Plastids—A review based on comparisons of small-subunit ribosomal-rna coding regions. J. Phycol. 1995, 31, 489–498. [Google Scholar] [CrossRef]
- Medlin, L.K. The Permian–Triassic mass extinction forces the radiation of the modern marine phytoplankton. Phycologia 2011, 50, 684–693. [Google Scholar] [CrossRef]
- Chan, C.X.; Bhattacharya, D. The origin of plastids. Nat. Educ. 2010, 3, 84. [Google Scholar]
- Falkowski, P.G.; Oliver, M.J. Mix and match: How climate selects phytoplankton. Nat. Rev. Microbiol. 2007, 5, 813–819. [Google Scholar] [CrossRef]
- Allen, J.F.; Raven, J.A. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J. Mol. Evol. 1996, 42, 482–492. [Google Scholar] [CrossRef]
- Gruber, A.; Kroth, P.G. Intracellular metabolic pathway distribution in diatoms and tools for genome-enabled experimental diatom research. Philos. Trans. R. Soc. B-Biol. Sci. 2017, 372, 20160402. [Google Scholar] [CrossRef] [PubMed]
- Río Bártulos, C.; Rogers, M.B.; Williams, T.A.; Gentekaki, E.; Brinkmann, H.; Cerff, R.; Liaud, M.-F.; Hehl, A.B.; Yarlett, N.R.; Gruber, A.; et al. Mitochondrial glycolysis in a major lineage of eukaryotes. Genome Biol. Evol. 2018, 10, 2310–2325. [Google Scholar] [CrossRef] [PubMed]
- Fabris, M.; Matthijs, M.; Rombauts, S.; Vyverman, W.; Goossens, A.; Baart, G.J.E. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 2012, 70, 1004–1014. [Google Scholar] [CrossRef]
- Gruber, A.; Haferkamp, I. Nucleotide transport and metabolism in diatoms. Biomolecules 2019, 9, 761. [Google Scholar] [CrossRef]
- Curtis, B.A.; Tanifuji, G.; Burki, F.; Gruber, A.; Irimia, M.; Maruyama, S.; Arias, M.C.; Ball, S.G.; Gile, G.H.; Hirakawa, Y.; et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 2012, 492, 59–65. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Bodył, A.; Gagat, P. Nucleomorph genomes. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 128–133. [Google Scholar] [CrossRef]
- Kroth, P.G. Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int. Rev. Cytol. 2002, 221, 191–255. [Google Scholar] [CrossRef]
- Patron, N.J.; Waller, R.F. Transit peptide diversity and divergence: A global analysis of plastid targeting signals. BioEssays News Rev. Mol. Cell. Dev. Biol. 2007, 29, 1048–1058. [Google Scholar] [CrossRef]
- Apt, K.E.; Zaslavkaia, L.; Lippmeier, J.C.; Lang, M.; Kilian, O.; Wetherbee, R.; Grossman, A.R.; Kroth, P.G. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 2002, 115, 4061–4069. [Google Scholar] [CrossRef]
- Kilian, O.; Kroth, P.G. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005, 41, 175–183. [Google Scholar] [CrossRef]
- Gruber, A.; Vugrinec, S.; Hempel, F.; Gould, S.B.; Maier, U.G.; Kroth, P.G. Protein targeting into complex diatom plastids: Functional characterisation of a specific targeting motif. Plant Mol. Biol. 2007, 64, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.S.; Gould, S.B.; Lehmann, P.; Gruber, A.; Przyborski, J.M.; Maier, U.-G. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol. Biol. Evol. 2007, 24, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Stork, S.; Moog, D.; Przyborski, J.M.; Wilhelmi, I.; Zauner, S.; Maier, U.G. Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Eukaryot. Cell 2012, 11, 1472–1481. [Google Scholar] [CrossRef] [PubMed]
- Maier, U.G.; Zauner, S.; Hempel, F. Protein import into complex plastids: Cellular organization of higher complexity. Eur. J. Cell Biol. 2015, 94, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Kingdom Chromista and its eight phyla: A new synthesis emphasising periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 2018, 255, 297–357. [Google Scholar] [CrossRef]
- Novák Vanclová, A.M.G.; Nef, C.; Vancl, A.; Liu, F.; Zoltán Füssy, Z.; Bowler, C.; Dorrell, R.G. Divergent and diversified proteome content across a serially acquired plastid lineage. Biorxiv Prepr. 2022, 2022, 30.518497. [Google Scholar] [CrossRef]
- Hehenberger, E.; Burki, F.; Kolisko, M.; Keeling, P.J. Functional relationship between a dinoflagellate host and its diatom endosymbiont. Mol. Biol. Evol. 2016, 33, 2376–2390. [Google Scholar] [CrossRef]
- Yamada, N.; Bolton, J.J.; Trobajo, R.; Mann, D.G.; Dąbek, P.; Witkowski, A.; Onuma, R.; Horiguchi, T.; Kroth, P.G. Discovery of a kleptoplastic ‘dinotom’ dinoflagellate and the unique nuclear dynamics of converting kleptoplastids to permanent plastids. Sci. Rep. 2019, 9, 10474. [Google Scholar] [CrossRef]
- Yamada, N.; Sym, S.D.; Horiguchi, T. Identification of highly divergent diatom-derived chloroplasts in dinoflagellates, including a description of Durinskia kwazulunatalensis sp. nov. (Peridiniales, Dinophyceae). Mol. Biol. Evol. 2017, 34, 1335–1351. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Lee, J.J. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J. Protozool. 1985, 32, 376–379. [Google Scholar] [CrossRef]
- Hehenberger, E.; Gast, R.J.; Keeling, P.J. A kleptoplastidic dinoflagellate and the tipping point between transient and fully integrated plastid endosymbiosis. Proc. Natl. Acad. Sci. USA 2019, 116, 17934–17942. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.D.; Oldach, D.; Delwiche, C.F.; Stoecker, D.K. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature 2007, 445, 426–428. [Google Scholar] [CrossRef] [PubMed]
- Pelletreau, K.N.; Bhattacharya, D.; Price, D.C.; Worful, J.M.; Moustafa, A.; Rumpho, M.E. Sea slug kleptoplasty and plastid maintenance in a metazoan. Plant Physiol. 2011, 155, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.K.; Curtis, N.E. Cell biology of the chloroplast symbiosis in sacoglossan sea slugs. Int. Rev. Cell Mol. Biol. 2012, 293, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef]
- Sanchez-Puerta, M.V.; Lippmeier, J.C.; Apt, K.E.; Delwiche, C.F. Plastid genes in a non-photosynthetic dinoflagellate. Protist 2007, 158, 105–117. [Google Scholar] [CrossRef]
- Patron, N.J.; Waller, R.F.; Keeling, P.J. A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 2006, 357, 1373–1382. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Lahr, D.J.; Knoll, A.H.; Katz, L.A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. USA 2011, 108, 13624–13629. [Google Scholar] [CrossRef]
- Yoon, H.S.; Hackett, J.D.; Ciniglia, C.; Pinto, G.; Bhattacharya, D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004, 21, 809–818. [Google Scholar] [CrossRef]
- Shih, P.M.; Matzke, N.J. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 12355–12360. [Google Scholar] [CrossRef] [PubMed]
- Bryłka, K.; Alverson, A.J.; Pickering, R.A.; Richoz, S.; Conley, D.J. Uncertainties surrounding the oldest fossil record of diatoms. Sci. Rep. 2023, 13, 8047. [Google Scholar] [CrossRef] [PubMed]
- Brocks, J.J.; Nettersheim, B.J.; Adam, P.; Schaeffer, P.; Jarrett, A.J.M.; Güneli, N.; Liyanage, T.; van Maldegem, L.M.; Hallmann, C.; Hope, J.M. Lost world of complex life and the late rise of the eukaryotic crown. Nature 2023, 618, 767–773. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. Large-Scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol. Biol. Evol. 2011, 28, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Prieto, A.; Yoon, H.S.; Moustafa, A.; Yang, E.C.; Andersen, R.A.; Boo, S.M.; Nakayama, T.; Ishida, K.; Bhattacharya, D. Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 2010, 27, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, P.; Colleoni, C.; Nakamura, Y.; Suzuki, E.; Putaux, J.L.; Buléon, A.; Haebel, S.; Ritte, G.; Steup, M.; Falcón, L.I.; et al. Metabolic symbiosis and the birth of the plant kingdom. Mol. Biol. Evol. 2008, 25, 536–548. [Google Scholar] [CrossRef]
- Dagan, T.; Roettger, M.; Stucken, K.; Landan, G.; Koch, R.; Major, P.; Gould, S.B.; Goremykin, V.V.; Rippka, R.; Tandeau de Marsac, N.; et al. Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 2012, 5, 31–44. [Google Scholar] [CrossRef]
- Jaffe, A.L.; Castelle, C.J.; Dupont, C.L.; Banfield, J.F. Lateral gene transfer shapes the distribution of RuBisCO among candidate phyla radiation bacteria and DPANN Archaea. Mol. Biol. Evol. 2018, 36, 435–446. [Google Scholar] [CrossRef]
- Kerr, A.C. Oceanic plateau formation: A cause of mass extinction and black shale deposition around the Cenomanian–Turonian boundary? J. Geol. Soc. 1998, 155, 619–626. [Google Scholar] [CrossRef]
- Li, G.; Wang, Y.; Shi, G.R.; Liao, W.; Yu, L. Fluctuations of redox conditions across the Permian–Triassic boundary—New evidence from the GSSP section in Meishan of South China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2016, 448, 48–58. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruber, A.; Medlin, L.K. Complex Plastids and the Evolution of the Marine Phytoplankton. J. Mar. Sci. Eng. 2023, 11, 1903. https://doi.org/10.3390/jmse11101903
Gruber A, Medlin LK. Complex Plastids and the Evolution of the Marine Phytoplankton. Journal of Marine Science and Engineering. 2023; 11(10):1903. https://doi.org/10.3390/jmse11101903
Chicago/Turabian StyleGruber, Ansgar, and Linda K. Medlin. 2023. "Complex Plastids and the Evolution of the Marine Phytoplankton" Journal of Marine Science and Engineering 11, no. 10: 1903. https://doi.org/10.3390/jmse11101903
APA StyleGruber, A., & Medlin, L. K. (2023). Complex Plastids and the Evolution of the Marine Phytoplankton. Journal of Marine Science and Engineering, 11(10), 1903. https://doi.org/10.3390/jmse11101903





