Assessment of Diversity of Marine Organisms among Natural and Transplanted Seagrass Meadows
Abstract
:1. Introduction
2. Materials and Methods
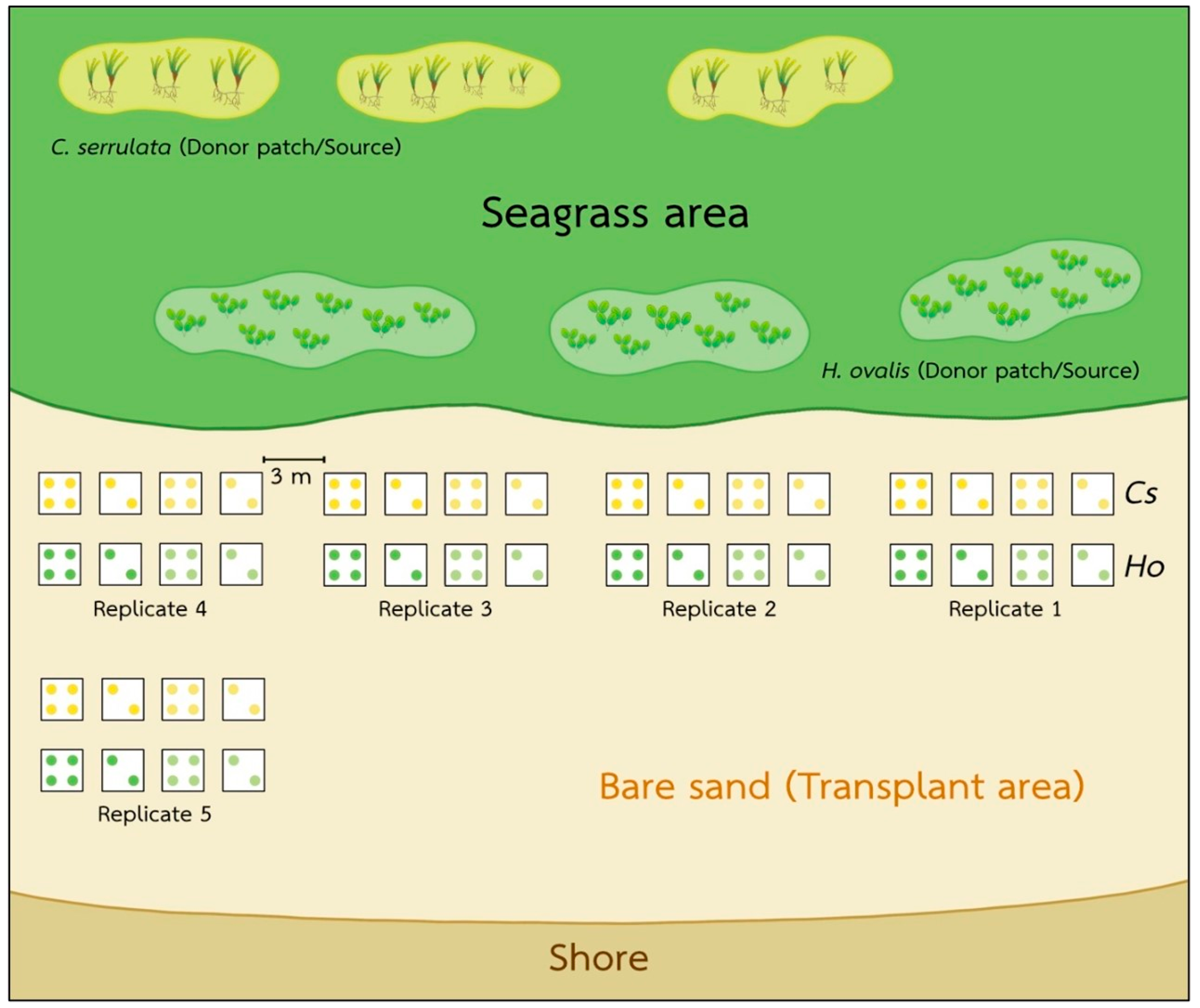
2.1. Study Site and Sample Collections
2.2. DNA Extraction and Sequencing
2.3. Data Processing and Statistics
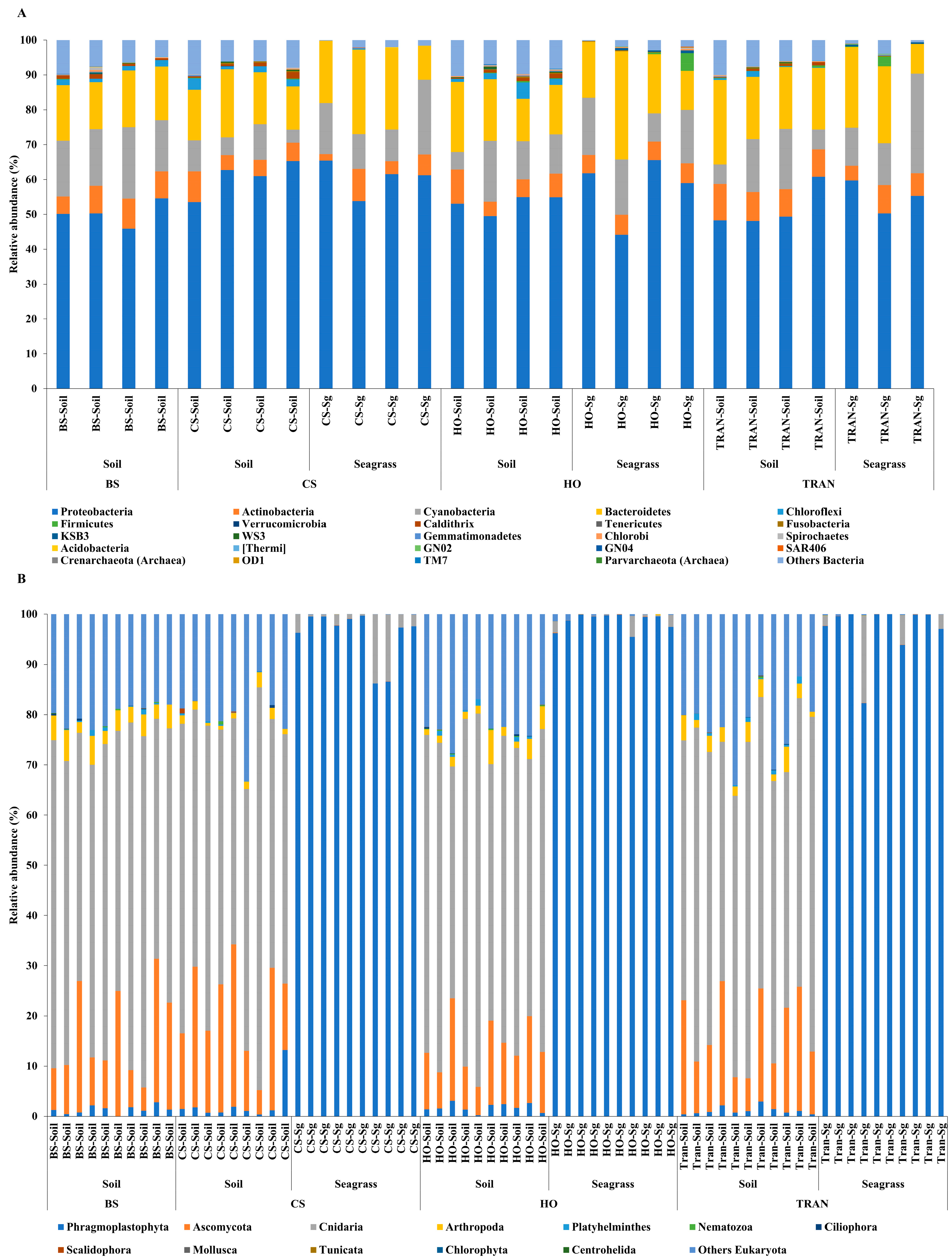
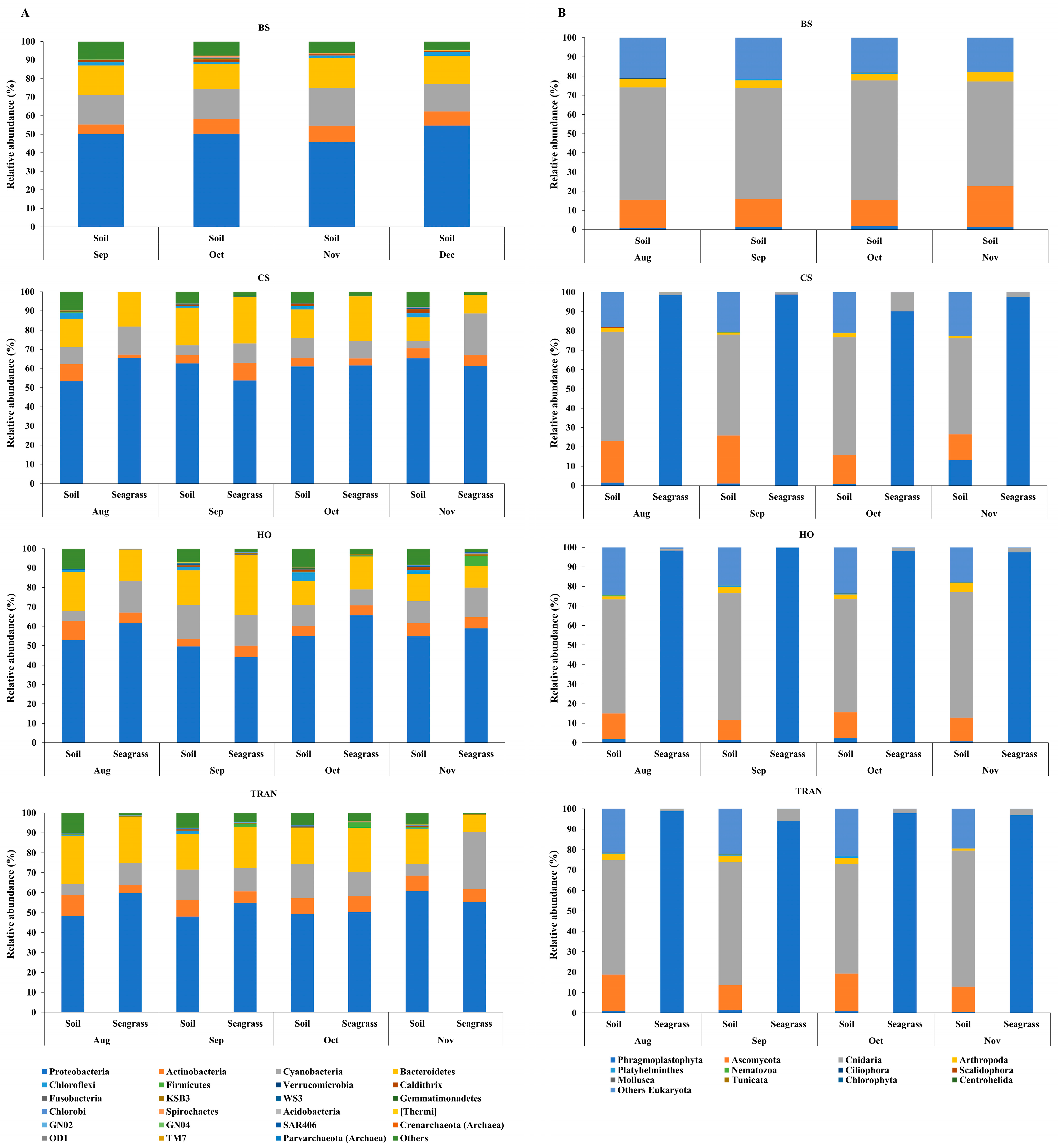
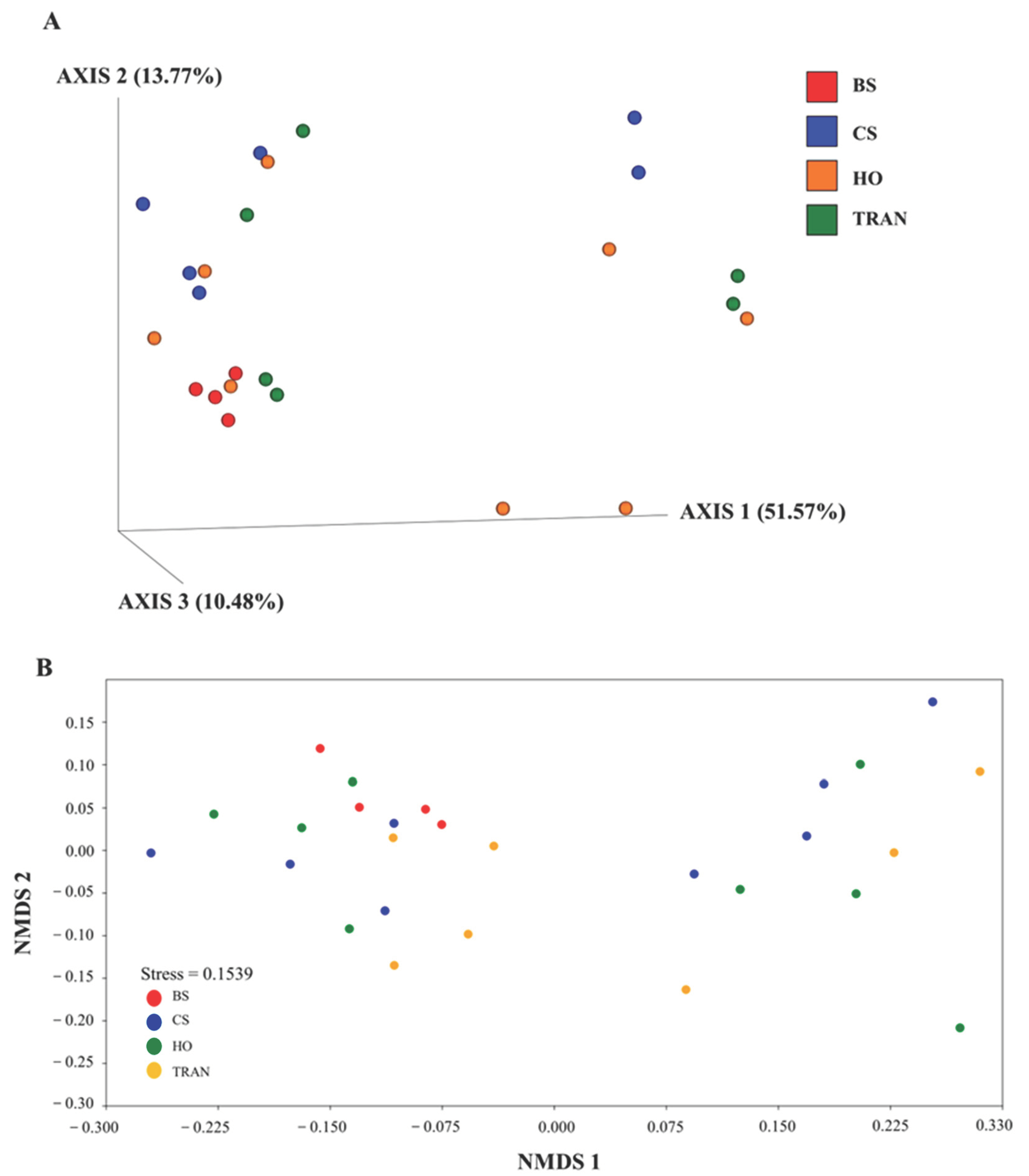
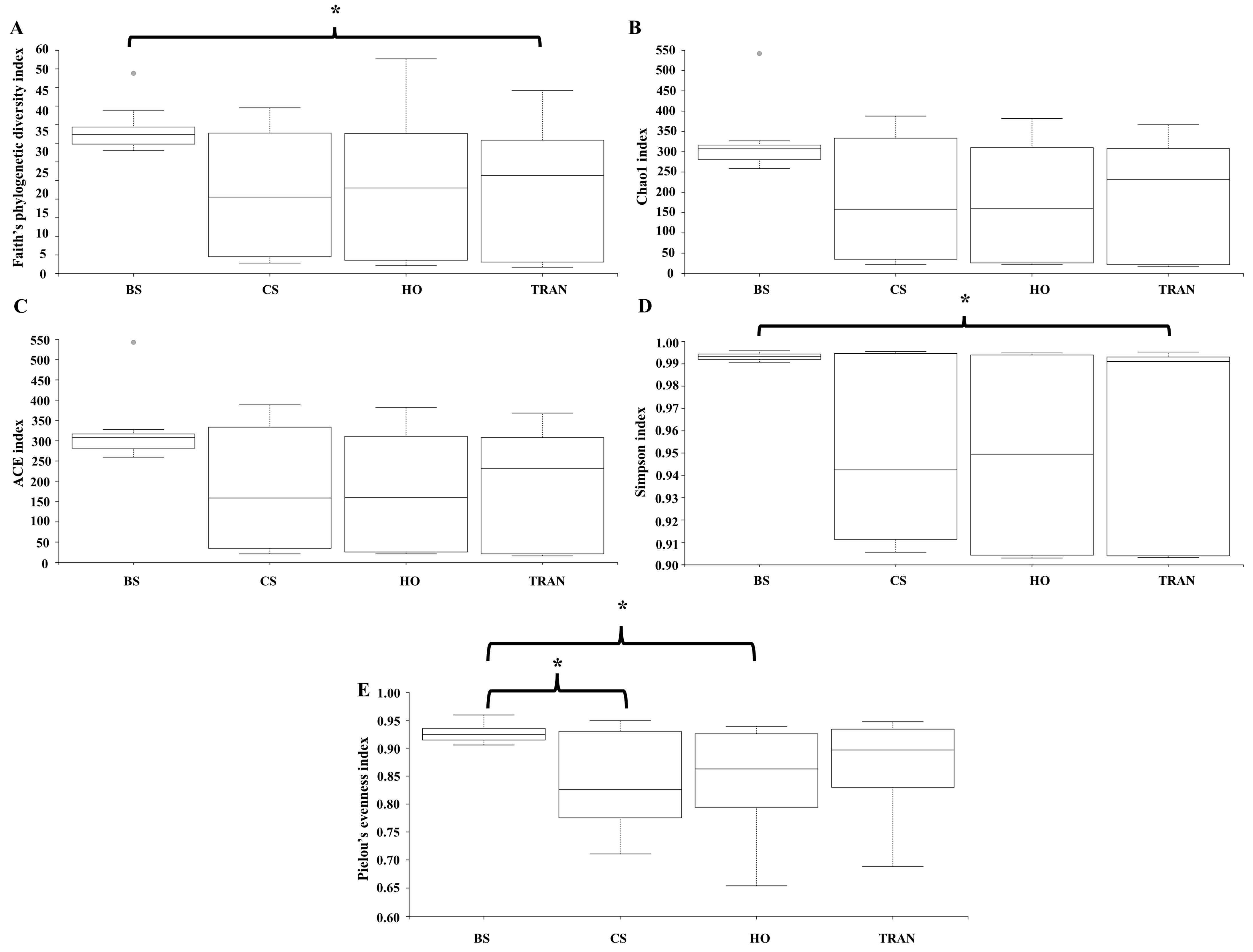
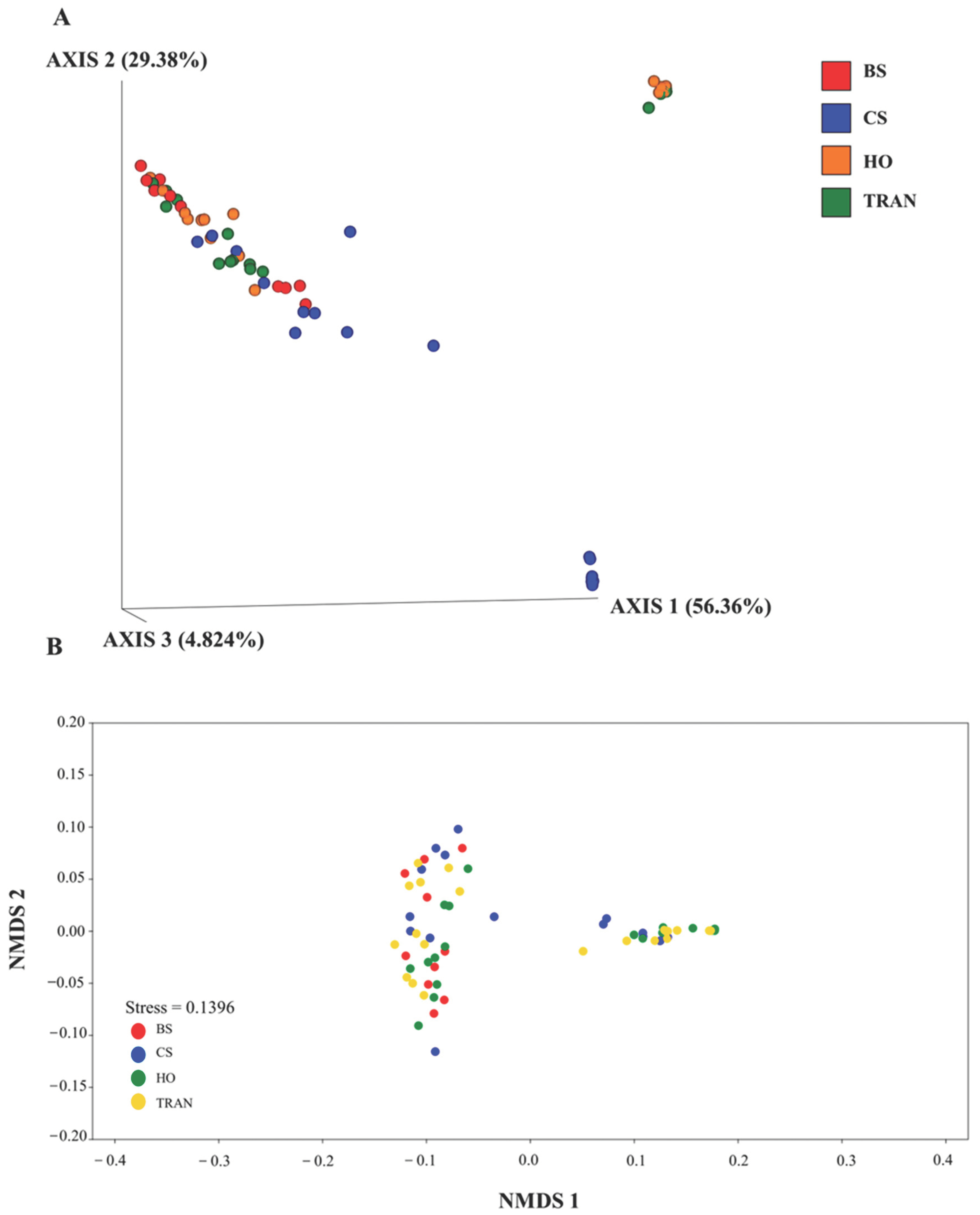
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cullen-Unsworth, L.; Unsworth, R. Seagrass meadows, ecosystem services, and sustainability. Environ. Sci. Policy 2013, 55, 14–28. [Google Scholar] [CrossRef]
- Ruiz-Frau, A.; Gelcich, S.; Hendriks, I.E.; Duarte, C.M.; Marbà, N. Current state of seagrass ecosystem services: Research and policy integration. Ocean. Coast. Manag. 2017, 149, 107–115. [Google Scholar] [CrossRef]
- Reusch, T.B.; Schubert, P.R.; Marten, S.M.; Gill, D.; Karez, R.; Busch, K.; Hentschel, U. Lower Vibrio spp. abundances in Zostera marina leaf canopies suggest a novel ecosystem function for temperate seagrass beds. Mar. Biol. 2021, 168, 149. [Google Scholar] [CrossRef]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K., Jr.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe Threatens coastal ecosystems. Proc. Natl. Acad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef]
- Robert, J.; Wong, P.P. Coastal systems and low-lying areas. In Climate Change 2007: Impacts, Adaptation and Vulnerability; Parry, M., Canziani, O., Eds.; Cambridge University Press: New York, NY, USA, 2007; pp. 326–367. [Google Scholar]
- Sala, E.; Knowlton, N. Global marine biodiversity trends. Annu. Rev. Environ. Resour. 2006, 31, 93–122. [Google Scholar] [CrossRef]
- Dunlop, M. Biodiversity: Strategy conservation. Nat. Clim. Chang. 2013, 3, 1019–1020. [Google Scholar] [CrossRef]
- Koch, M.; Bowes, G.; Ross, C.; Zhang, X.H. Climate change and ocean acidification effects on seagrasses and marine macroalgae. Glob. Chang. Biol. 2013, 19, 103–132. [Google Scholar] [CrossRef]
- Rasheed, M.; McKenna, S.A.; Carter, A.B.; Coles, R. Contrasting recovery of shallow and deep-water seagrass communities following climate associated losses in tropical north Queensland, Australia. Mar. Pollut. Bull. 2014, 83, 491–499. [Google Scholar] [CrossRef]
- Reed, B.J.; Hovel, K.A. Seagrass habitat disturbance: How loss and fragmentation of, eelgrass Zostera marina influences epifaunal abundance and diversity. Mar. Ecol. Prog. Ser. 2006, 326, 133–143. [Google Scholar] [CrossRef]
- Irlandi, E.A.; Orlando, B.A.; Ambrose, G.C., Jr. Influence of seagrass habitat patch size on growth and survival of juvenile bay scallops, Argopecten irradians concentricus (Say). J. Exp. Mar. Biol. Ecol. 1999, 235, 21–43. [Google Scholar] [CrossRef]
- Spivak, A.C.; Canuel, E.A.; Duffy, J.E.; Richardson, J.P. Top-down and bottom-up controls on sediment organic matter composition in an experimental seagrass ecosystem. Limnol. Oceanogr. 2007, 52, 2595–2607. [Google Scholar] [CrossRef]
- Bryars, C.; Neverauskas, V. Natural recolonization of seagrasses at a disused sewage sludge outfall. Aqua. Bot. 2004, 80, 283–289. [Google Scholar] [CrossRef]
- Riemann, B.; Carstensen, J.; Dahl, K.; Fossing, H.; Hansen, J.W.; Jakobsen, H.H.; Josefson, A.B.; Krause-Jensen, D.; Markager, S.; Stæhr, P.A.; et al. Recovery of Danish coastal ecosystems after reductions in nutrient loading: A holistic ecosystem approach. Estuaries Coasts 2016, 39, 82–97. [Google Scholar] [CrossRef]
- Orth, R.J.; McGlathery, J.K. Eelgrass recovery in the coastal bays of the Virginia Coast Reserve, USA. Mar. Ecol. Prog. Ser. 2012, 448, 173–176. [Google Scholar] [CrossRef]
- Matheson, F.E.; Reed, J.; Dos Santos, V.M.; Mackay, G.; Cummings, V.J. Seagrass rehabilitation: Successful transplants and evaluation of methods at different spatial scales. N. Z. J. Mar. Freshw. Res. 2016, 51, 96–109. [Google Scholar] [CrossRef]
- Blandon, A.; Zu Ermgassen, P.S. Quantitative estimate of commercial fish enhancement by seagrass habitat in southern Australia. Coast. Shelf Sci. 2014, 141, 1–8. [Google Scholar] [CrossRef]
- Lefcheck, J.S.; Marion, S.R.; Orth, R.J. Restored Eelgrass (Zostera marina L.) as a refuge for epifaunal biodiversity in mid-western Atlantic coastal bays. Estuaries Coast 2017, 40, 200–212. [Google Scholar] [CrossRef]
- Ondiviela, B.; Losada, I.J.; Lara, J.L.; Maza, M.; Galván, C.; Bouma, T.I.; van Belzen, J. The role of seagrasses in coastal protection in a changing climate. Coast. Eng. 2014, 87, 158–168. [Google Scholar] [CrossRef]
- Reynolds, L.K.; Waycott, M.; McGlathery, K.J.; Orth, R.J. Ecosystem services returned through seagrass restoration. Restor. Ecol. 2016, 24, 583–588. [Google Scholar] [CrossRef]
- Orth, R.J.; Lefcheck, J.S.; McGlathery, K.S.; Aoki, L.; Luckenbach, M.W.; Moore, K.A.; Oreska, M.; Snyder, R.; Wilcox, D.J.; Lusk, B. Restoration of seagrass habitat leads to rapid recovery of coastal ecosystem services. Sci. Adv. 2020, 6, eabc6434. [Google Scholar] [CrossRef] [PubMed]
- van Katwijk, M.M.; Thorhaug, A.; Marbà, N.; Orth, R.J.; Duarte, C.M.; Kendrick, G.A.; Althuizen, I.H.; Balestri, E.; Bernard, G.; Cambridge, M.L.; et al. Global analysis of seagrass restoration: The importance of large-scale planting. J. Appl. Ecol. 2016, 53, 567–578. [Google Scholar] [CrossRef]
- Valdemarsen, T.; Canal-Vergés, P.; Kristensen, E.; Holmer, M.; Kristiansen, M.; Flindt, M.V.R. Vulnerability of Zostera marina seedlings to physical stress. Mar. Ecol. Prog. Ser. 2010, 418, 119–130. [Google Scholar] [CrossRef]
- Carstensen, J.; Krause-Jensen, D.; Markager, S.; Timmermann, K.; Windolf, J. Water clarity and eelgrass responses to nitrogen reductions in the eutrophic Skive Fjord, Denmark. Hydrobiologia 2013, 704, 293–309. [Google Scholar] [CrossRef]
- He, H.; Stanley, R.R.E.; Rubidge, E.M.; Jeffery, N.W.; Hamilton, L.C.; Westfall, K.M.; Gilmore, S.R.; Roux, L.M.D.; Gale, K.S.P.; Heaslip, S.G.; et al. Fish community surveys in eelgrass beds using both eDNA metabarcoding and seining: Implications for biodiversity monitoring in the coastal zone. Can. J. Fish. Aquat. Sci. 2022, 79, 1335–1346. [Google Scholar] [CrossRef]
- Stankovic, M.; Hayashizaki, K.I.; Tuntiprapas, P.; Rattanachot, E.; Prathep, A. Two decades of seagrass area change: Organic carbon sources and stock. Mar. Pollut. Bull. 2021, 163, 111913. [Google Scholar] [CrossRef]
- Kaewsrikhaw, R.; Upanoi, T.; Prathep, A. Ecosystem services and vulnerability assessments of seagrass ecosystems: Basic tools for prioritizing conservation management actions using an example from Thailand. Water 2022, 14, 3650. [Google Scholar] [CrossRef]
- Tongnunui, P.; Tarangkoon, W.; Hukiew, P.; Kaeoprakan, P.; Horinouchi, M.; Rojchanaprasart, N.; Ponpai, W. Seagrass restoration: An update from Trang province, Southwestern Thailand. In Proceedings of the International Conference “Managinag Risks to Coastal Regions and Communities in a Changinag World” (EMECS’11—SeaCoasts XXVI), Saint-Petersburg, Russia, 22–27 August 2017; Academus Publishing: Redwood City, CA, USA, 2017; pp. 10–12. [Google Scholar]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Guardiola, M.; Uriz, M.J.; Taberlet, P.; Coissac, E.; Wangensteen, O.S.; Turon, X. Correction: Deep-sea, deep-sequencing: Metabarcoding extracellular DNA from sediments of marine canyons. PLoS ONE 2016, 11, e0153836. [Google Scholar] [CrossRef]
- Ortega, A.; Geraldi, N.R.; Duarte, C.M. Environmental DNA identifies marine macrophyte contributions to blue carbon sediments. Limnol. Oceanogr. 2013, 65, 3139–3149. [Google Scholar] [CrossRef]
- Closek, C.J.; Santora, J.A.; Starks, H.A.; Schroeder, I.D.; Andruszkiewicz, E.A.; Sakuma, K.M.; Bograd, S.J.; Hazen, E.L.; Field, J.C.; Boehm, A.B. Marine vertebrate biodiversity and distribution within the Central California current using environmental DNA (eDNA) metabarcoding and ecosystem surveys. Front. Mar. Sci. 2019, 6, 732. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids. Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Pielou, E.C. The Measurement of diversity in different types of biological collections. J. Theor. Biol. 1996, 13, 131–144. [Google Scholar] [CrossRef]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef]
- Somboonna, N.; Assawamakin, A.; Wilantho, A.; Tangphatsornruang, S.; Tongsima, S. Structural and functional diversity of free-living microorganisms in reef surface, Kra island, Thailand. BMC Genom. 2014, 15, 607. [Google Scholar] [CrossRef]
- Liang, J.; Yu, K.; Wang, Y.; Huang, X.; Huang, W.; Qin, Z.; Pan, Z.; Yao, Q.; Wang, W.; Wu, Z. Distinct bacterial communities associated with massive and branching Scleractinian corals and potential linkages to coral susceptibility to thermal or cold stress. Front. Microbiol. 2017, 8, 979. [Google Scholar] [CrossRef]
- Pootakham, W.; Mhuantong, W.; Yoocha, T.; Putchim, L.; Sonthirod, C.; Naktang, C.; Thongtham, N.; Tangphatsornruang, S. High resolution profiling of coral-associated bacterial communities using full-length 16S rRNA sequence data from PacBio SMRT sequencing system. Sci. Rep. 2017, 7, 2774. [Google Scholar] [CrossRef]
- Somboonna, N.; Wilantho, A.; Monanunsap, S.; Chavanich, S.; Tangphatsornruang, S.; Tongsima, S. Microbial communities in the reef water at Kham Island, lower Gulf of Thailand. PeerJ 2017, 5, e3625. [Google Scholar] [CrossRef]
- Mhuantong, W.; Nuryadi, H.; Trianto, A.; Sabdono, A.; Tangphatsornruang, S.; Eurwilaichitr, L.; Kanokkratana, P.; Champreda, V. Comparative analysis of bacterial communities associated with healthy and diseased corals in the Indonesian sea. PeerJ 2019, 7, e8137. [Google Scholar] [CrossRef]
- Pollock, F.J.; Lamb, J.B.; van de Water, J.A.; Smith, H.A.; Schaffelke, B.; Willis, B.L.; Bourne, D.G. Reduced diversity and stability of coral-associated bacterial communities and suppressed immune function precedes disease onset in corals. R. Soc. Open Sci. 2019, 6, 190355. [Google Scholar] [CrossRef]
- Nimnoi, P.; Pongsilp, N. Marine bacterial communities in the upper gulf of Thailand assessed by Illumina next-generation sequencing platform. BMC Microbiol. 2020, 20, 19. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Q.; Ling, J.; Long, L.; Huang, H.; Yin, J.; Wu, M.; Tang, X.; Lin, X.; Zhang, Y.; et al. Shifting the microbiome of a coral holobiont and improving host physiology by inoculation with a potentially beneficial bacterial consortium. BMC Microbiol. 2021, 21, 130. [Google Scholar] [CrossRef]
- Titioatchasai, J.; Surachat, K.; Kim, J.H.; Mayakun, J. Diversity of microbial communities associated with epilithic macroalgae in different coral reef conditions and damselfish territories of the Gulf of Thailand. J. Mar. Sci. Eng. 2023, 11, 514. [Google Scholar] [CrossRef]
- Jensen, S.I.; Kühl, M.; Priemé, A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 2007, 62, 108–117. [Google Scholar] [CrossRef]
- Marhaeni, B.; Radjasa, O.K.; Bengen, D.G.; Kaswadji, R.F. Screening of bacterial symbionts of seagrass Enhalus sp. against biofilm-forming bacteria. J. Coast. Dev. 2010, 13, 126–132. [Google Scholar]
- Ugarelli, K.; Laas, P.; Stingl, U. The microbial communities of leaves and roots associated with turtle Grass (Thalassia testudinum) and manatee grass (Syringodium filliforme) are distinct from seawater and sediment communities, but are similar between species and sampling sites. Microorganisms 2018, 7, 4. [Google Scholar] [CrossRef]
- Martin, B.C.; Alarcon, M.S.; Gleeson, D.; Middleton, J.A.; Fraser, M.W.; Ryan, M.H.; Holmer, M.; Kendrick, G.A.; Kilminster, K. Root microbiomes as indicators of seagrass health. FEMS Microbiol. Ecol. 2020, 96, fiz201. [Google Scholar] [CrossRef]
- Iqbal, M.M.; Nishimura, M.; Haider, M.N.; Sano, M.; Ijichi, M.; Kogure, K.; Yoshizawa, S. Diversity and composition of microbial communities in an eelgrass (Zostera marina) bed in Tokyo Bay, Japan. Microbes Environ. 2021, 36, ME21037. [Google Scholar] [CrossRef]
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Bier, E.J.; Sun, S.; Howard, E.C. Prokaryotic genomes and diversity in surface ocean waters: Interrogating the Global ocean sampling metagenome. Appl. Environ. Microbiol. 2009, 75, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, C.; Zhang, X.; Gong, J. Community structure and abundance of Archaea in a Zostera marina Meadow: A Comparison between seagrass-colonized and bare sediment sites. Archaea 2019, 2019, 5108012. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.L.; Wang, X.L.; Li, Y.C. The influence of eefgrass (Zostera marina L.) on the environmental factors and the microbial communities in sediment from Shuangdao bay of China. Appl. Ecol. Environ. Res. 2019, 9, 10767–11077. [Google Scholar]
- Banister, R.B.; Schwarz, M.T.; Fine, M.; Ritchie, K.B.; Muller, E.M. Instability and stasis among the microbiome of seagrass leaves, roots and rhizomes, and nearby sediments within a natural pH gradient. Microb. Ecol. 2022, 84, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Tarquinio, F.; Attlan, O.; Vanderklift, M.A.; Berry, O.; Bissett, A. Distinct endophytic bacterial communities inhabiting seagrass seeds. Front. Microbiol. 2021, 12, 703014. [Google Scholar] [CrossRef] [PubMed]
- Conte, C.; Rotini, A.; Manfra, L.; D’Andrea, M.M.; Winters, G.; Migliore, L. The seagrass holobiont: What we know and what we still need to disclose for its possible use as an ecological indicator. Water 2021, 13, 406. [Google Scholar] [CrossRef]
- Suzuki, D.; Ueki, A.; Amaishi, A.; Ueki, K. Desulfopila aestuarii gen. nov., sp. nov., a Gram-negative, rod-like, sulfate-reducing bacterium isolated from an estuarine sediment in Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 520–526. [Google Scholar] [CrossRef]
- Sundarakrishnan, B.; Pushpanathan, M.; Jayashree, S.; Rajendhran, J.; Sakthivel, N.; Jayachandran, S.; Gunasekaran, P. Assessment of microbial richness in pelagic sediment of Andaman Sea by bacterial Tag Encoded FLX Titanium Amplicon Pyrosequencing (bTEFAP). Indian J. Microbiol. 2012, 52, 544–550. [Google Scholar] [CrossRef]
- Guevara, R.; Ikenaga, M.; Dean, A.L.; Pisani, C.; Boyer, J.N. Changes in sediment bacterial community in response to long-term nutrient enrichment in a subtropical seagrass-dominated estuary. Microb. Ecol. 2014, 68, 427–440. [Google Scholar] [CrossRef]
- Spring, S.; Scheuner, C.; Göker, M.; Klenk, H.P. A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front. Microbiol. 2015, 6, 281. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, G.; Yan, B.C.; Lee, N.L.Y.; Ooi, J.L.S.; Lee, J.N.; Huang, D.; Wainwright, B.J. Spatial and structural factors shape seagrass-associated bacterial communities in Singapore and Peninsular Malaysia. Front. Mar. Sci. 2021, 8, 659180. [Google Scholar] [CrossRef]
- Carini, P.; Marsden, P.J.; Leff, J.W.; Morgan, E.E.; Strickland, M.S.; Fierer, N. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2017, 2, 16242. [Google Scholar] [CrossRef] [PubMed]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Vohník, M.; Borovec, O.; Kolarík, M. Communities of cultivable root mycobionts of the seagrass Posidonia oceanica in the northwest Mediterranean Sea are dominated by a hitherto undescribed pleosporalean dark septate endophyte. Microb. Ecol. 2016, 71, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Orfali, R.; Perveen, S.; Khan, M.F.; Ahmed, A.F.; Wadaan, M.A.; Al-Taweel, A.M.; Alqahtani, A.S.; Nasr, F.A.; Tabassum, S.; Luciano, P.; et al. Antiproliferative Illudalane Sesquiterpenes from the Marine Sediment Ascomycete Aspergillus oryzae. Mar. Drugs 2021, 19, 333. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Draeger, S.; Mitchell, J.I. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008, 74, 931–941. [Google Scholar] [CrossRef]
- Wesselmann, M.; Geraldi, N.R.; Marbà, N.; Hendriks, I.E.; Díaz-Rúa, R.; Duarte, C.M. eDNA reveals the associated metazoan diversity of mediterranean seagrass sediments. Diversity 2022, 14, 549. [Google Scholar] [CrossRef]
- Barnes, R.F. Patterns of benthic invertebrate biodiversity in intertidal seagrass in Moreton Bay, Queensland. Reg. Stud. Mar. Sci. 2017, 15, 17–25. [Google Scholar] [CrossRef]
- Hassenrück, C.; Hofmann, L.C.; Bischof, K.; Ramette, A. Seagrass biofilm communities at a naturally CO2-rich vent. Environ. Microbiol. Rep. 2015, 7, 516–525. [Google Scholar] [CrossRef]
- Tsioli, S.; Vasillis, P.; Anastasia, R.; Maria, K.; Christos, K.; Eva, P.; Küpper, F.C.; Sotiris, O. Diversity and composition of algal epiphytes on the Mediterranean seagrass Cymodocea nodosa: A scale-based study. Bot. Mar. 2021, 64, 101–118. [Google Scholar] [CrossRef]
- Gagnon, K.; Bocoum, E.H.; Chen, C.Y.; Baden, S.P.; Moksnes, P.O.; Infantes, E. Rapdi faunal colonization and recovery of biodiversity and functional diversity following eelgrass restoration. Restor. Ecol. 2023, 21, e13887. [Google Scholar] [CrossRef]


| Kruskal-Wallis | Group 1 | Group 2 | H | p-Value |
|---|---|---|---|---|
| Species richness | ||||
| Faith’s phylogenetic diversity | All | 0.408 | 0.817 | |
| Bare sand | Cymodocea serrulata | 0.045 | 0.831 | |
| Halophila ovalis | 0.461 | 0.497 | ||
| Transplant | 0.182 | 0.670 | ||
| Cymodocea serrulata | Halophila ovalis | 0.067 | 0.796 | |
| Transplant | 0.026 | 0.872 | ||
| Halophila ovalis | Transplant | 0.067 | 0.796 | |
| Chao1 | All | 2.034 | 0.565 | |
| Bare sand | Cymodocea serrulata | 1.038 | 0.308 | |
| Halophila ovalis | 0.007 | 0.932 | ||
| Transplant | 0.892 | 0.345 | ||
| Cymodocea serrulata | Halophila ovalis | 0.893 | 0.344 | |
| Transplant | 0.120 | 0.728 | ||
| Halophila ovalis | Transplant | 1.085 | 0.517 | |
| ACE | All | 1.832 | 0.607 | |
| Bare sand | Cymodocea serrulata | 1.038 | 0.308 | |
| Halophila ovalis | 0.003 | 0.865 | ||
| Transplant | 1.286 | 0.257 | ||
| Cymodocea serrulata | Halophila ovalis | 0.706 | 0.400 | |
| Transplant | 0.013 | 0.908 | ||
| Halophila ovalis | Transplant | 0.656 | 0.418 | |
| Simpson | All | 0.866 | 0.834 | |
| Bare sand | Cymodocea serrulata | 0.721 | 0.396 | |
| Halophila ovalis | 0.029 | 0.865 | ||
| Transplant | 0.574 | 0.449 | ||
| Cymodocea serrulata | Halophila ovalis | 0.397 | 0.529 | |
| Transplant | 0.054 | 0.817 | ||
| Halophila ovalis | Transplant | 0.054 | 0.817 | |
| Species evenness | ||||
| Pielou’s evenness | All | 0.039 | 0.998 | |
| Bare sand | Cymodocea serrulata | 0.045 | 0.831 | |
| Halophila ovalis | 0.029 | 0.865 | ||
| Transplant | 0.000 | 1.000 | ||
| Cymodocea serrulata | Halophila ovalis | 0.017 | 0.897 | |
| Transplant | 0.025 | 0.873 | ||
| Halophila ovalis | Transplant | 0.017 | 0.897 |
| Kruskal–Wallis | Group 1 | Group 2 | H | p-Value |
|---|---|---|---|---|
| Species richness | ||||
| Faith’s phylogenetic diversity | All | 6.060 | 0.108 | |
| Bare sand | Cymodocea serrulata | 3.579 | 0.058 | |
| Halophila ovalis | 3.414 | 0.065 | ||
| Transplant | 5.802 | 0.016 | ||
| Cymodocea serrulata | Halophila ovalis | 0.124 | 0.725 | |
| Transplant | 0.360 | 0.548 | ||
| Halophila ovalis | Transplant | 0.392 | 0.531 | |
| Chao1 | All | 5.041 | 0.169 | |
| Bare sand | Cymodocea serrulata | 1.982 | 0.159 | |
| Halophila ovalis | 3.256 | 0.071 | ||
| Transplant | 3.782 | 0.052 | ||
| Cymodocea serrulata | Halophila ovalis | 0.534 | 0.465 | |
| Transplant | 1.144 | 0.285 | ||
| Halophila ovalis | Transplant | 0.300 | 0.583 | |
| ACE | All | 5.057 | 0.168 | |
| Bare sand | Cymodocea serrulata | 1.982 | 0.159 | |
| Halophila ovalis | 3.256 | 0.071 | ||
| Transplant | 3.781 | 0.052 | ||
| Cymodocea serrulata | Halophila ovalis | 0.534 | 0.465 | |
| Transplant | 1.172 | 0.279 | ||
| Halophila ovalis | Transplant | 0.300 | 0.584 | |
| Simpson | All | 6.184 | 0.103 | |
| Bare sand | Cymodocea serrulata | 3.414 | 0.064 | |
| Halophila ovalis | 3.097 | 0.078 | ||
| Transplant | 4.464 | 0.035 | ||
| Cymodocea serrulata | Halophila ovalis | 2.055 | 0.152 | |
| Transplant | 0.882 | 0.348 | ||
| Halophila ovalis | Transplant | 0.010 | 0.917 | |
| Species evenness | ||||
| Pielou’s evenness | All | 7.109 | 0.068 | |
| Bare sand | Cymodocea serrulata | 5.436 | 0.020 | |
| Halophila ovalis | 4.278 | 0.039 | ||
| Transplant | 3.457 | 0.063 | ||
| Cymodocea serrulata | Halophila ovalis | 0.073 | 0.787 | |
| Transplant | 1.503 | 0.220 | ||
| Halophila ovalis | Transplant | 0.931 | 0.334 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Titioatchasai, J.; Surachat, K.; Rattanachot, E.; Tuntiprapas, P.; Mayakun, J. Assessment of Diversity of Marine Organisms among Natural and Transplanted Seagrass Meadows. J. Mar. Sci. Eng. 2023, 11, 1928. https://doi.org/10.3390/jmse11101928
Titioatchasai J, Surachat K, Rattanachot E, Tuntiprapas P, Mayakun J. Assessment of Diversity of Marine Organisms among Natural and Transplanted Seagrass Meadows. Journal of Marine Science and Engineering. 2023; 11(10):1928. https://doi.org/10.3390/jmse11101928
Chicago/Turabian StyleTitioatchasai, Jatdilok, Komwit Surachat, Ekkalak Rattanachot, Piyalap Tuntiprapas, and Jaruwan Mayakun. 2023. "Assessment of Diversity of Marine Organisms among Natural and Transplanted Seagrass Meadows" Journal of Marine Science and Engineering 11, no. 10: 1928. https://doi.org/10.3390/jmse11101928
APA StyleTitioatchasai, J., Surachat, K., Rattanachot, E., Tuntiprapas, P., & Mayakun, J. (2023). Assessment of Diversity of Marine Organisms among Natural and Transplanted Seagrass Meadows. Journal of Marine Science and Engineering, 11(10), 1928. https://doi.org/10.3390/jmse11101928









