Technological Advancements in Field Investigations of Marine Microorganisms: From Sampling Strategies to Molecular Analyses
Abstract
:1. Introduction
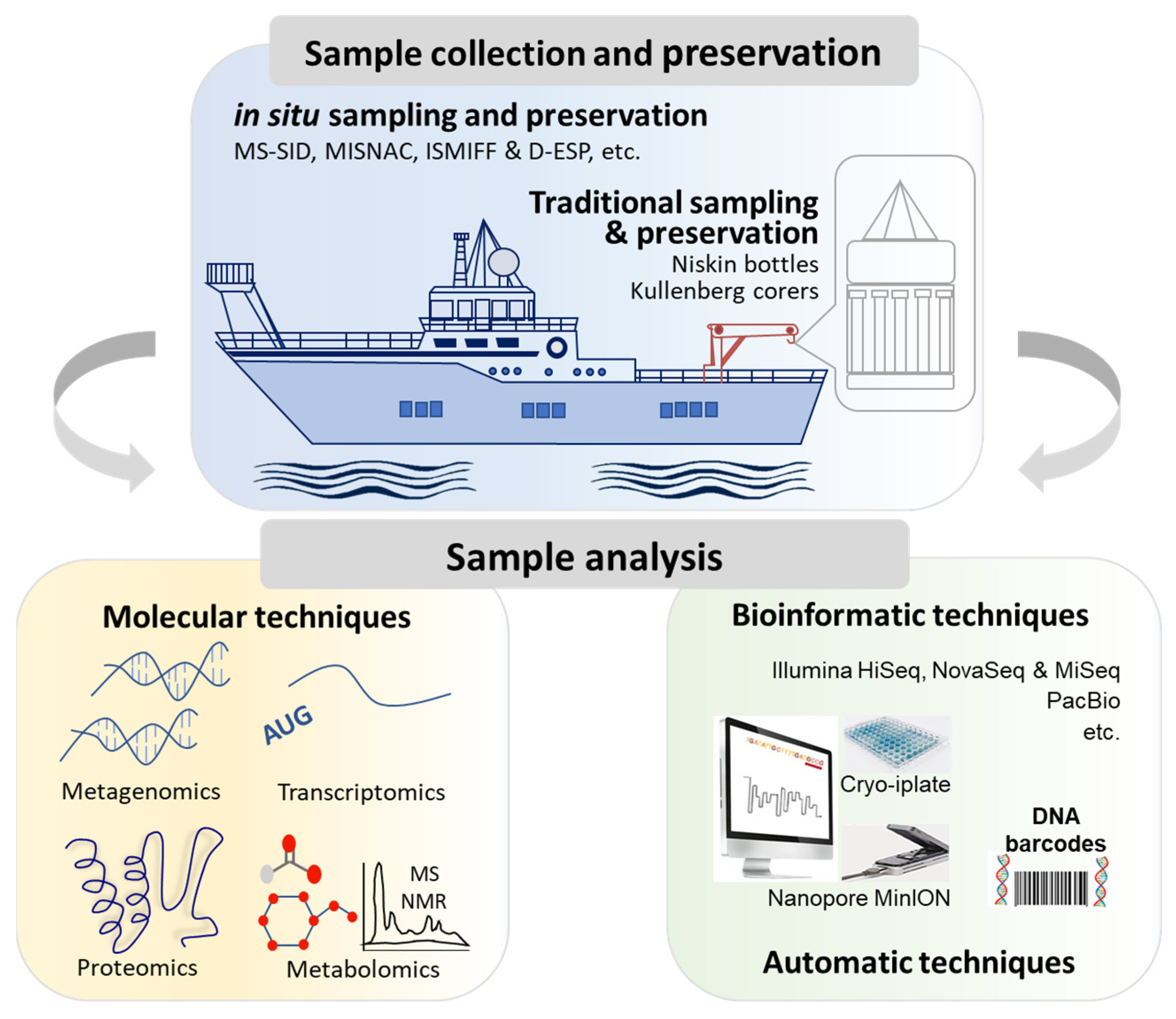
2. Field Investigation of Marine Microorganisms
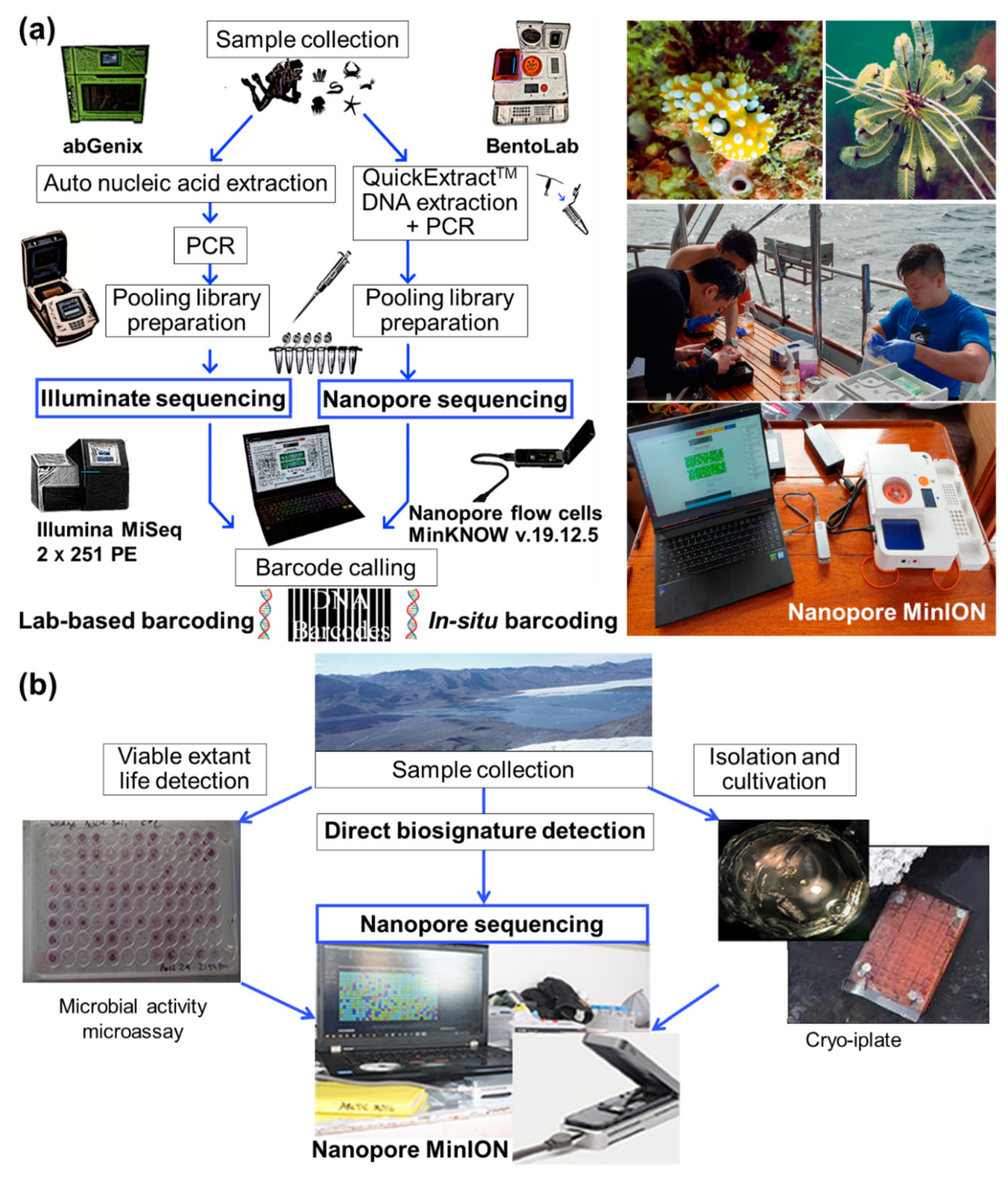
2.1. Collection of Marine Microbial Samples
2.2. Analyses of Marine Microbial Samples
2.2.1. Molecular Analyses for Molecular Ecology
2.2.2. Molecular Analyses by Omics Technologies
Marine Metagenomics
Marine Transcriptomics and Proteomics
Marine Metabolomics
3. Conclusions and Challenges
3.1. Conclusions
3.2. Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bickle, M.; Arculus, R.; Barrett, P.; DeConto, R.; Camoin, G.; Edwards, K.; Fisher, A.; Inagaki, F.; Kodaira, S.; Ohkouchi, N.; et al. Illuminating Earth’s Past, Present, and Future: The Science Plan for the International Ocean Discovery Program 2013–2023; I.O.D.P.M. International: Washington, DC, USA, 2011. [Google Scholar]
- Kumar, N.; Raven, K.E.; Blane, B.; Leek, D.; Brown, N.M.; Bragin, E.; Rhodes, P.A.; Parkhill, J.; Peacock, S.J. Evaluation of a fully automated bioinformatics tool to predict antibiotic resistance from MRSA genomes. J. Antimicrob. Chemother. 2020, 75, 1117–1122. [Google Scholar] [CrossRef]
- Sun, Y.H.; Liu, Y.; Pan, J.; Wang, F.P.; Li, M. Perspectives on cultivation strategies of archaea. Microb. Ecol. 2020, 79, 770–784. [Google Scholar]
- Cario, A.; Oliver, G.C.; Rogers, K.L. Exploring the deep marine biosphere: Challenges, innovations, and opportunities. Front. Earth Sci. 2019, 7, 225. [Google Scholar]
- Garel, M.; Bonin, P.; Martini, S.; Guasco, S.; Roumagnac, M.; Bhairy, N.; Armougom, F.; Tamburini, C. Pressure-retaining sampler and high-pressure systems to study deep-sea microbes under in situ conditions. Front. Microbiol. 2019, 10, 453. [Google Scholar]
- Imachi, H.; Nobu, M.K.; Miyazaki, M.; Tasumi, E.; Saito, Y.; Sakai, S.; Ogawara, M.; Ohashi, A.; Takai, K. Cultivation of previously uncultured microorganisms with a continuous-flow down-flow hanging sponge (DHS) bioreactor, using a syntrophic archaeon culture obtained from deep marine sediment as a case study. Nat. Protoc. 2022, 17, 2784–2814. [Google Scholar]
- Liu, W.; Zheng, X.W.; Dai, X.; Zhang, Z.F.; Zhang, W.Y.; Xiao, T.; Huang, L. Isolation and characterization of the first temperate virus infecting Psychrobacillus from marine sediments. Viruses 2022, 14, 108. [Google Scholar] [CrossRef]
- Pope, E.; Cartmell, C.; Haltli, B.; Ahmadi, A.; Kerr, R.G. Microencapsulation and in situ incubation methodology for the cultivation of marine bacteria. Front. Microbiol. 2022, 13, 958660. [Google Scholar]
- Wang, F.P.; Li, M.; Huang, L.; Zhang, X.H. Cultivation of uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 117–120. [Google Scholar]
- Caron, D.A.; Alexander, H.; Allen, A.E.; Archibald, J.M.; Armbrust, E.V.; Bachy, C.; Bell, C.J.; Bharti, A.; Dyhrman, S.T.; Guida, S.M.; et al. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 2017, 15, 6–20. [Google Scholar]
- Kodama, T.; Taniuchi, Y.; Kasai, H.; Yamaguchi, T.; Nakae, M.; Okumura, Y. Empirical estimation of marine phytoplankton assemblages in coastal and offshore areas using an in situ multi-wavelength excitation fluorometer. PLoS ONE 2022, 17, e0257258. [Google Scholar]
- Hiraoka, S.; Sumida, T.; Hirai, M.; Toyoda, A.; Kawagucci, S.; Yokokawa, T.; Nunoura, T. Diverse DNA modification in marine prokaryotic and viral communities. Nucleic Acids Res. 2022, 50, 1531–1550. [Google Scholar] [PubMed]
- Bruns, S.; Wienhausen, G.; Scholz-Boettcher, B.; Wilkes, H. Simultaneous quantification of all B vitamins and selected biosynthetic precursors in seawater and bacteria by means of different mass spectrometric approaches. Anal. Bioanal. Chem. 2022, 414, 7839–7854. [Google Scholar] [PubMed]
- Tuit, C.B.; Wait, A.D. A review of marine sediment sampling methods. Environ. Forensics 2020, 21, 291–309. [Google Scholar] [CrossRef]
- Feike, J.; Juergens, K.; Hollibaugh, J.T.; Krueger, S.; Jost, G.; Labrenz, M. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 2012, 6, 461–470. [Google Scholar] [CrossRef]
- Lunne, T.; Long, M. Review of long seabed samplers and criteria for new sampler design. Mar. Geol. 2006, 226, 145–165. [Google Scholar]
- Wei, Z.-F.; Li, W.-L.; Li, J.; Chen, J.; Xin, Y.-Z.; He, L.-S.; Wang, Y. Multiple in situ nucleic acid collections (MISNAC) from deep-sea waters. Front. Mar. Sci. 2020, 7, 81. [Google Scholar] [CrossRef]
- Taylor, C.D.; Edgcomb, V.P.; Doherty, K.W.; Engstrom, I.; Shanahan, T.; Pachiadaki, M.G.; Molyneaux, S.J.; Honjo, S. Fixation filter, device for the rapid in situ preservation of particulate samples. Deep-Sea Res. Part I 2015, 96, 69–79. [Google Scholar] [CrossRef]
- Hendricks, A.; Mackie, C.M.; Luy, E.; Sonnichsen, C.; Smith, J.; Grundke, I.; Tavasoli, M.; Furlong, A.; Beiko, R.G.; LaRoche, J.; et al. Compact and automated eDNA sampler for in situ monitoring of marine environments. Sci. Rep. 2023, 13, 5210. [Google Scholar] [CrossRef]
- He, S.; Peng, Y.; Jin, Y.; Wan, B.; Liu, G. Review and analysis of key techniques in marine sediment sampling. Chin. J. Mech. Eng. 2020, 33, 66. [Google Scholar] [CrossRef]
- Case, D.H.; Ijiri, A.; Morono, Y.; Tavormina, P.; Orphan, V.J.; Inagaki, F. Aerobic and anaerobic methanotrophic communities associated with methane hydrates exposed on the seafloor: A high-pressure sampling and stable isotope-incubation experiment. Front. Microbiol. 2017, 8, 2569. [Google Scholar]
- He, S.; Qiu, S.; Tang, W.; Peng, Y.; Jin, Y. A novel submersible-mounted sediment pressure-retaining sampler at full ocean depth. Front. Mar. Sci. 2023, 10, 1154269. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Taylor, C.; Pachiadaki, M.G.; Honjo, S.; Engstrom, I.; Yakimov, M. Comparison of Niskin vs. in situ approaches for analysis of gene expression in deep Mediterranean Sea water samples. Deep-Sea Res. Part II 2016, 129, 213–222. [Google Scholar] [CrossRef]
- Li, J.; Xin, Y.; Cai, D.; Chen, J.; Li, W.; Wang, S.; Wei, Z.; Zhang, A. Development and scientific application of deep-sea multiple in situ nucleic acid collections (MISNAC) apparatus. In Proceedings of the Global Oceans 2020: Singapore—U.S. Gulf Coast, Biloxi, MS, USA, 5–30 October 2020; pp. 1–5. [Google Scholar]
- Wang, Y.; Gao, Z.-M.; Li, J.; He, L.-S.; Cui, G.-J.; Li, W.-L.; Chen, J.; Xin, Y.-Z.; Cai, D.-S.; Zhang, A.-Q. Hadal water sampling by in situ microbial filtration and fixation (ISMIFF) apparatus. Deep-Sea Res. Part I 2019, 144, 132–137. [Google Scholar] [CrossRef]
- Teague, J.; Scott, T.B.; Sharma, S.; Graham, G.; Allen, M.J. An Alternative Method to Niskin Sampling for Molecular Analysis of the Marine Environment. J. Mar. Sci. Eng. 2017, 5, 22. [Google Scholar] [CrossRef]
- He, Z.; Deng, Y.; Van Nostrand, J.D.; Tu, Q.; Xu, M.; Hemme, C.L.; Li, X.; Wu, L.; Gentry, T.J.; Yin, Y.; et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010, 4, 1167–1179. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, H.L.; Chen, M.J.; Song, X.X.; Yu, C.X.; Zhao, Y.; Wu, Y.J. Reference gene selection for quantitative real-time pcr of mycelia from lentinula edodes under high-temperature stress. Biomed Res. Int. 2018, 2018, 1670328. [Google Scholar] [CrossRef]
- Tu, Q.C.; Yu, H.; He, Z.L.; Deng, Y.; Wu, L.Y.; Van Nostrand, J.D.; Zhou, A.F.; Voordeckers, J.; Lee, Y.J.; Qin, Y.J.; et al. GeoChip 4: A functional gene-array-based high-throughput environmental technology for microbial community analysis. Mol. Ecol. Resour. 2014, 14, 914–928. [Google Scholar] [CrossRef]
- Chu, Y.H.; Heather, H.; Zhang, R.R.; Guo, Z.Y.; Ricardo, V.L. In situ hybridization: Introduction to techniques, applications and pitfalls in the performance and interpretation of assays. Semin. Diagn. Pathol. 2019, 36, 336–341. [Google Scholar] [CrossRef]
- Li, Z.H.; Rui, J.P.; Li, X.Z.; Li, J.B.; Dong, L.; Huang, Q.L.; Huang, C.; Wang, Z.P.; Li, L.; Xuan, P.; et al. Bacterial community succession and metabolite changes during doubanjiang-meju fermentation, a Chinese traditional fermented broad bean (Vicia faba L.) paste. Food Chem. 2017, 218, 534–542. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef]
- Rybalka, N.; Blanke, M.; Tzvetkova, A.; Noll, A.; Roos, C.; Boy, J.; Boy, D.; Nimptsch, D.; Godoy, R.; Friedl, T. Unrecognized diversity and distribution of soil algae from Maritime Antarctica (Fildes Peninsula, King George Island). Front. Microbiol. 2023, 14, 1118747. [Google Scholar]
- Pendergraft, M.A.; Belda-Ferre, P.; Petras, D.; Morris, C.K.; Mitts, B.A.; Aron, A.T.; Bryant, M.; Schwartz, T.; Ackermann, G.; Humphrey, G.; et al. Bacterial and chemical evidence of coastal water pollution from the Tijuana river in sea spray aerosol. Environ. Sci. Technol. 2023, 57, 4071–4081. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Soto, M.F.; Cerqueda-Garcia, D.; Aguirre-Macedo, M.L.; Garcia-Maldonado, J.Q. Spatiotemporal dynamics of benthic bacterial communities in the Perdido Fold Belt, Northwestern Gulf of Mexico. Front. Mar. Sci. 2023, 10, 1070548. [Google Scholar]
- Savi, D.C.; Aluizio, R.; Galli-Terasawa, L.; Kava, V.; Glienke, C. 16S-gyrB-rpoB multilocus sequence analysis for species identification in the genus Microbispora. Anton. Leeuw. 2016, 109, 801–815. [Google Scholar]
- Guo, S.L.; Yang, Q.H.; Feng, J.J.; Duan, L.H.; Zhao, J.P. Phylogenetic analysis of the pathogenic genus Aeromonas spp. isolated from diseased eels in China. Microb. Pathog. 2016, 101, 12–23. [Google Scholar] [PubMed]
- Ogier, J.C.; Pages, S.; Galan, M.; Barret, M.; Gaudriault, S. rpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiol. 2019, 19, 171. [Google Scholar]
- Peixoto, R.S.; Coutinho, H.L.D.; Rumjanek, N.G.; Macrae, A.; Rosado, A.S. Use of rpoB and 16S rRNA genes to analyse bacterial diversity of a tropical soil using PCR and DGGE. Lett. Appl. Microbiol. 2002, 35, 316–320. [Google Scholar] [CrossRef]
- Ki, J.-S.; Zhang, W.; Qian, P.-Y. Discovery of marine Bacillus species by 16S rRNA and rpoB comparisons and their usefulness for species identification. J. Microbiol. Methods 2009, 77, 48–57. [Google Scholar] [CrossRef]
- Freitas-Silva, J.; de Oliveira, B.F.R.; Vigoder, F.d.M.; Muricy, G.; Dobson, A.D.W.; Laport, M.S. Peeling the layers away: The genomic characterization of Bacillus pumilus 64-1, an isolate with antimicrobial activity from the marine sponge Plakina cyanorosea (Porifera, homoscleromorpha). Front. Microbiol. 2021, 11, 592735. [Google Scholar]
- Zhang, Y.; Wang, X.; Zhen, Y.; Mi, T.; He, H.; Yu, Z. Microbial diversity and community structure of sulfate-reducing and sulfur-oxidizing bacteria in sediment cores from the East China Sea. Front. Microbiol. 2017, 8, 2133. [Google Scholar]
- Sanchez-Soto, M.F.; Cerqueda-Garcia, D.; Alcantara-Hernandez, R.J.; Falcon, L.I.; Pech, D.; arcega-Cabrera, F.; Aguirre-Macedo, M.L.; Garcia-Maldonado, J.Q. Assessing the diversity of benthic sulfate-reducing microorganisms in Northwestern Gulf of Mexico by Illumina sequencing of dsrB gene. Microb. Ecol. 2021, 81, 908–921. [Google Scholar] [PubMed]
- Winkel, M.; Mitzscherling, J.; Overduin, P.P.; Horn, F.; Winterfeld, M.; Rijkers, R.; Grigoriev, M.N.; Knoblauch, C.; Mangelsdorf, K.; Wagner, D.; et al. Anaerobic methanotrophic communities thrive in deep submarine permafrost. Sci. Rep. 2018, 8, 1291. [Google Scholar] [PubMed]
- Hinsa-Leasure, S.M.; Bhavaraju, L.; Rodrigues, J.L.M.; Bakermans, C.; Gilichinsky, D.A.; Tiedje, J.M. Characterization of a bacterial community from a Northeast Siberian seacoast permafrost sample. FEMS Microbiol. Ecol. 2010, 74, 103–113. [Google Scholar] [PubMed]
- Rani, S.; Koh, H.W.; Rhee, S.K.; Fujitani, H.; Park, S.J. Detection and diversity of the nitrite oxidoreductase alpha subunit (nxrA) gene of Nitrospina in marine sediments. Microb. Ecol. 2017, 73, 111–122. [Google Scholar]
- Lau, E.; Fisher, M.C.; Steudler, P.A.; Cavanaugh, C.M. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PLoS ONE 2013, 8, e56993. [Google Scholar]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar]
- Cao, S.; Zhang, W.; Ding, W.; Wang, M.; Fan, S.; Yang, B.; McMinn, A.; Wang, M.; Xie, B.-b.; Qin, Q.-L.; et al. Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics. Microbiome 2020, 8, 47. [Google Scholar]
- Goldberg, S.M.D.; Johnson, J.; Busam, D.; Feldblyum, T.; Ferriera, S.; Friedman, R.; Halpern, A.; Khouri, H.; Kravitz, S.A.; Lauro, F.M.; et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11240–11245. [Google Scholar] [CrossRef]
- Wolf, C.; Kilias, E.S.; Metfies, K. Evaluating the potential of 18S rDNA clone libraries to complement pyrosequencing data of marine protists with near full-length sequence information. Mar. Biol. Res. 2014, 10, 771–780. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Rodriguez-Ezpeleta, N. Environmental status assessment using DNA metabarcoding: Towards a genetics based marine biotic index (gAMBI). PLoS ONE 2014, 9, e90529. [Google Scholar]
- Singer, G.A.C.; Fahner, N.A.; Barnes, J.G.; McCarthy, A.; Hajibabaei, M. Comprehensive biodiversity analysis via ultra-deep patterned flow cell technology: A case study of eDNA metabarcoding seawater. Sci. Rep. 2019, 9, 5991. [Google Scholar] [PubMed]
- Haro-Moreno, J.M.; López-Pérez, M.; Rodríguez-Valera, F. Long read metagenomics, the next step? BioRxiv 2020. [Google Scholar] [CrossRef]
- Haro-Moreno, J.M.; Lopez-Perez, M.; Rodriguez-Valera, F. Enhanced recovery of microbial genes and genomes from a marine water column using long-read metagenomics. Front. Microbiol. 2021, 12, 708782. [Google Scholar]
- Singer, E.; Bushnell, B.; Coleman-Derr, D.; Bowman, B.; Bowers, R.M.; Levy, A.; Gies, E.A.; Cheng, J.-F.; Copeland, A.; Klenk, H.-P.; et al. High-resolution phylogenetic microbial community profiling. ISME J. 2016, 10, 2020–2032. [Google Scholar] [PubMed]
- Wang, S.; Su, X.; Cui, H.; Wang, M.; Hu, X.; Ding, W.; Zhang, W. Microbial richness of marine biofilms revealed by sequencing full-length 16s rRNA genes. Genes 2022, 13, 1050. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Leung, A.W.-S.; Zheng, Z.; Zhang, D.; Xiao, C.; Luo, R.; Luo, M.; Zhang, S. Applications and potentials of nanopore sequencing in the (epi)genome and (epi)transcriptome era. Innovation 2021, 2, 100153. [Google Scholar] [PubMed]
- Marsay, K.S.; Koucherov, Y.; Davidov, K.; Iankelevich-Kounio, E.; Itzahri, S.; Salmon-Divon, M.; Oren, M. High-resolution screening for marine prokaryotes and eukaryotes with selective preference for polyethylene and polyethylene terephthalate surfaces. Front. Microbiol. 2022, 13, 845144. [Google Scholar]
- Curren, E.; Yoshida, T.; Kuwahara, V.S.; Leong, S.C.Y. Rapid profiling of tropical marine cyanobacterial communities. Reg. Stud. Mar. Sci. 2019, 25, 100485. [Google Scholar]
- Chang, J.J.; Ip, Y.C.; Ng, C.S.; Huang, D. Takeaways from mobile DNA barcoding with BentoLab and MinION. Genes 2020, 11, 1121. [Google Scholar] [CrossRef]
- Goordial, J.; Altshuler, I.; Hindson, K.; Chan-Yam, K.; Marcolefas, E.; Whyte, L.G. In situ field sequencing and life detection in remote (79 degrees 26′ N) Canadian high Arctic permafrost ice wedge microbial communities. Front. Microbiol. 2017, 8, 2594. [Google Scholar] [CrossRef]
- Hassan, S.; Sabreena; Khurshid, Z.; Bhat, S.A.; Kumar, V.; Ameen, F.; Ganai, B.A. Marine bacteria and omic approaches: A novel and potential repository for bioremediation assessment. J. Appl. Microbiol. 2022, 133, 2299–2313. [Google Scholar] [PubMed]
- Dong, X.; You, Y.; Wu, J.Q. Building an RNA sequencing transcriptome of the central nervous system. Neuroscientist 2016, 22, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Li, W.V.; Chen, Y.; Li, J.J. TROM: A testing-based method for finding transcriptomic similarity of biological samples. Stat. Biosci. 2017, 9, 105–136. [Google Scholar] [PubMed]
- Page, T.M.; Lawley, J.W. The next generation is here: A review of transcriptomic approaches in marine ecology. Front. Mar. Sci. 2022, 9, 757921. [Google Scholar]
- Rachinger, N.; Fischer, S.; Bohme, I.; Linck-Paulus, L.; Kuphal, S.; Kappelmann-Fenzl, M.; Bosserhoff, A.K. Loss of gene information: Discrepancies between RNA sequencing, cDNA microarray, and qRT-PCR. Int. J. Mol. Sci. 2021, 22, 9349. [Google Scholar] [PubMed]
- Veluchamy, A.; Lin, X.; Maumus, F.; Rivarola, M.; Bhavsar, J.; Creasy, T.; O’Brien, K.; Sengamalay, N.A.; Tallon, L.J.; Smith, A.D.; et al. Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 2013, 4, 2091. [Google Scholar] [CrossRef]
- Lu, Y.; Lv, Y.X.; Zhang, Y.; Liu, Q.; Xu, X.W.; Xiao, X.; Xu, J. Metatranscriptomes reveal the diverse responses of Thaumarchaeota ecotypes to environmental variations in the northern slope of the South China Sea. Environ. Microbiol. 2023, 25, 410–427. [Google Scholar] [CrossRef]
- Geoghegan, J.L.; Giallonardo, F.D.; Wille, M.; Ortiz-Baez, A.S.; Costa, V.A.; Ghaly, T.; Mifsud, J.C.O.; Turnbull, O.M.H.; Bellwood, D.R.; Williamson, J.E.; et al. Erratum: Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 2021, 7, veab005. [Google Scholar]
- Ren, L.; Hu, X.; Zhao, X.; Chen, S.; Wu, Y.; Li, D.; Yu, Y.; Geng, L.; Ji, X.; Huang, H. Transcriptomic analysis of the regulation of lipid fraction migration and fatty acid biosynthesis in Schizochytrium sp. Sci. Rep. 2017, 7, 3562. [Google Scholar]
- Matilda, C.S.; Madhusudan, I.; Isola, R.G.; Shanthi, C. Potential of proteomics to probe microbes. J. Basic Microbiol. 2020, 60, 471–483. [Google Scholar] [CrossRef]
- Zhang, S.F.; Han, B.B.; Wu, F.X.; Huang, H.H. Quantitative proteomic analysis provides insights into the algicidal mechanism of Halobacillus sp. P1 against the marine diatom Skeletonema costatum. Sci. Total Environ. 2020, 717, 137048. [Google Scholar] [PubMed]
- Muthusamy, S.; Lundin, D.; Branca, R.M.M.; Baltar, F.; Gonzalez, J.M.; Lehtio, J.; Pinhassi, J. Comparative proteomics reveals signature metabolisms of exponentially growing and stationary phase marine bacteria. Environ. Microbiol. 2017, 19, 2301–2319. [Google Scholar] [PubMed]
- Gregson, B.H.; Metodieva, G.; Metodiev, M.V.; Golyshin, P.N.; McKew, B.A. Protein expression in the obligate hydrocarbon-degrading psychrophile Oleispira antarctica RB-8 during alkane degradation and cold tolerance. Environ. Microbiol. 2020, 22, 1870–1883. [Google Scholar] [PubMed]
- Lin, C.Y.; Huang, F.P.; Ling, Y.S.; Liang, H.J.; Lee, S.H.; Hu, M.Y.; Tsao, P.N. Use of nuclear magnetic resonance-based metabolomics to characterize the biochemical effects of naphthalene on various organs of tolerant mice. PLoS ONE 2015, 10, e0120429. [Google Scholar]
- Utermann, C.; Echelmeyer, V.A.; Oppong-Danquah, E.; Bluemel, M.; Tasdemir, D. Diversity, bioactivity profiling and untargeted metabolomics of the cultivable gut microbiota of Ciona intestinalis. Mar. Drugs 2021, 19, 6. [Google Scholar] [CrossRef]
- Emwas, A.-H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. In Metabonomics: Methods in Molecular Biology; Bjerrum, J., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1277, pp. 161–193. [Google Scholar]
- Poho, P.; Lipponen, K.; Bespalov, M.M.; Sikanen, T.; Kotiaho, T.; Kostiainen, R. Comparison of liquid chromatography-mass spectrometry and direct infusion microchip electrospray ionization mass spectrometry in global metabolomics of cell samples. Eur. J. Pharm. Sci. 2019, 138, 104991. [Google Scholar] [CrossRef]
- Mueller, C.; Kremb, S.; Gonsior, M.; Brack-Werner, R.; Voolstra, C.R.; Schmitt-Kopplin, P. Advanced identification of global bioactivity hotspots via screening of the metabolic fingerprint of entire ecosystems. Sci. Rep. 2020, 10, 1319. [Google Scholar]
- Kizhakkekalam, V.K.; Chakraborty, K. Pharmacological properties of marine macroalgae-associated heterotrophic bacteria. Arch. Microbiol. 2019, 201, 505–518. [Google Scholar]
- Baran, R.; Bowen, B.P.; Northen, T.R. Untargeted metabolic footprinting reveals a surprising breadth of metabolite uptake and release by Synechococcus sp. PCC 7002. Mol. Biosyst. 2011, 7, 3200–3206. [Google Scholar]
- Bae, M.; Chung, B.; Oh, K.B.; Shin, J.; Oh, D.C. Hormaomycins B and C: New antibiotic cyclic depsipeptides from a marine mudflat-derived Streptomyces sp. Mar. Drugs 2015, 13, 5187–5200. [Google Scholar] [CrossRef]
- Wang, W.J.; Li, D.Y.; Li, Y.C.; Hua, H.M.; Ma, E.L.; Li, Z.L. Caryophyllene sesquiterpenes from the marine-derived fungus Ascotricha sp. ZJ-M-5 by the one strain-many compounds strategy. J. Nat. Prod. 2014, 77, 1367–1371. [Google Scholar] [CrossRef] [PubMed]
- Ueoka, R.; Shinzato, N.; Kagaya, N.; Suenaga, H.; Shin-ya, K. Pseudoalteropeptide A, a novel lipopeptide from the marine bacterium Pseudoalteromonas piscicida SWA4_PA4 isolated from marine seaweed. J. Antibiot. 2021, 74, 105–110. [Google Scholar]
- Zhang, D.; Shu, C.Y.; Lian, X.Y.; Zhang, Z.Z. New antibacterial bagremycins F and G from the marine-derived Streptomyces sp. ZZ745. Mar. Drugs 2018, 16, 330. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.M.; Nicoletti, R.; Salvatore, F.; Naviglio, D.; Andolfi, A. GC–MS approaches for the screening of metabolites produced by marine-derived Aspergillus. Mar. Chem. 2018, 206, 19–33. [Google Scholar]
- Al-Dhabi, N.A.; Mohammed Ghilan, A.K.; Esmail, G.A.; Valan Arasu, M.; Duraipandiyan, V.; Ponmurugan, K. Bioactivity assessment of the Saudi Arabian Marine Streptomyces sp. Al-Dhabi-90, metabolic profiling and its in vitro inhibitory property against multidrug resistant and extended-spectrum beta-lactamase clinical bacterial pathogens. J. Infect. Public Health 2019, 12, 549–556. [Google Scholar] [CrossRef]
- Vinale, F.; Salvatore, M.M.; Nicoletti, R.; Staropoli, A.; Manganiello, G.; Venneri, T.; Borrelli, F.; DellaGreca, M.; Salvatore, F.; Andolfi, A. Identification of the main metabolites of a marine-derived strain of Penicillium brevicompactum using LC and GC MS techniques. Metabolites 2020, 10, 55. [Google Scholar] [CrossRef]
- Felline, S.; Del Coco, L.; Kaleb, S.; Guarnieri, G.; Fraschetti, S.; Terlizzi, A.; Fanizzi, F.P.; Falace, A. The response of the algae Fucus virsoides (Fucales, Ochrophyta) to Roundup® solution exposure: A metabolomics approach. Environ. Pollut. 2019, 254, 112977. [Google Scholar]
- Bose, U.; Hewavitharana, A.K.; Ng, Y.K.; Shaw, P.N.; Fuerst, J.A.; Hodson, M.P. LC-MS-based metabolomics study of marine bacterial secondary metabolite and antibiotic production in Salinispora arenicola. Mar. Drugs 2015, 13, 249–266. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Amin, M.; Xu, X.Y.; Qi, S.H. Antifouling potentials and metabolite profiles of two marine derived fungal isolates. Nat. Prod. Commun. 2018, 13, 423–426. [Google Scholar] [CrossRef]
- Teame, T.; Zhang, Z.; Ran, C.; Zhang, H.; Yang, Y.; Ding, Q.; Xie, M.; Gao, C.; Ye, Y.; Duan, M.; et al. The use of zebrafish (Danio rerio) as biomedical models. Anim. Front. 2019, 9, 68–77. [Google Scholar]
- Danouche, M.; El Ghachtouli, N.; El Arroussi, H. Phycoremediation mechanisms of heavy metals using living green microalgae: Physicochemical and molecular approaches for enhancing selectivity and removal capacity. Heliyon 2021, 7, e07609. [Google Scholar] [PubMed]
- Wang, S.C.; Liu, G.Z.; Liu, F.F. Physiological and metabolic toxicity of polystyrene microplastics to Dunaliella salina. Environ. Pollut. 2023, 316, 120544. [Google Scholar] [PubMed]
- Villar, E.; Farrant, G.K.; Follows, M.; Garczarek, L.; Speich, S.; Audic, S.; Bittner, L.; Blanke, B.; Brum, J.R.; Brunet, C.; et al. Environmental characteristics of Agulhas rings affect interocean plankton transport. Science 2015, 348, 1261447. [Google Scholar] [PubMed]
- Bork, P.; Bowler, C.; de Vargas, C.; Gorsky, G.; Karsenti, E.; Wincker, P. Tara Oceans studies plankton at planetary scale. Science 2015, 348, 873. [Google Scholar] [CrossRef]


| Methods | Advantages | Disadvantages | Application | |
|---|---|---|---|---|
| Traditional methods | Niskin bottle [26] |
|
| Water |
| Kullenberg pistone corer [16] | Water/sediments | |||
| In situ high-fidelity collection methods [20] | Pressure core barrel |
|
| Water/sediments |
| Pressure core sampler |
| Water/sediments | ||
| Advanced piston corer |
|
| Water/sediments | |
| Pressure temperature core sampler |
|
| Water/sediments | |
| Fugro pressure corer |
|
| Water/sediments | |
| Rotary corer |
| Water/sediments | ||
| Methods | Advantages | Limitations |
|---|---|---|
| DGGE/TGGE [31] |
|
|
| qRT-PCR [32] |
|
|
| CARD-FISH [30] |
|
|
| Geochip [29] |
|
|
| Clone Library [33] |
|
|
| Gene Markers | Marine Microorganisms | Molecular Techniques | Marine Ecosystems |
|---|---|---|---|
| rpoB [40] | Bacillus species | PCR | Intertidal areas, fish farms, biofilms, water, and sediments |
| gyrB [41] | Bacillus pumilus | Illumina sequencing | Marine sponge |
| dsrB/soxB [42,43] | Sulfate-reducing/sulfur-oxidizing bacteria | qPCR, Illumina sequencing, and 454 pyrosequencing | Marine sediments |
| mcrA [44] | Anaerobic methanotrophic (ANME-2a/b, ANME-2d) archaea | qPCR, CARD-FISH | Deep submarine permafrost |
| icl [45] | Firmicutes, Proteobacteria, and Actinobacteria | Geochip 2.0 | Seacoast permafrost |
| nxrA [46] | Nitrospina | 454 pyrosequencing and qPCR | Marine sediments |
| mxaF [47] | Methylococcaceae | PCR | Hydrothermal vent |
| Categories | Metabolites | Marine Microorganisms | Techniques |
|---|---|---|---|
| Depsipeptides | Hormaomycins B and C | Streptomyces sp. | NMR [83] |
| Sesquiterpenes | Caryophyllene derivatives | Ascotricha sp. | NMR [84] |
| Ketones | 1,3,6-Trihydroxy-8-methylxanthone | ||
| Lipopeptides | Pseudoalteropeptide A | Pseudoalteromonas piscicida | NMR [85] |
| Antibiotics | Bagremycins F and G | Streptomyces sp. | NMR [86] |
| Dicarboxylic acids | Succinic acid, fumaric acid, malic acid, 2-methylglutaconic acid, and citric acid | Aspergillus sp. | GC-MS [87] |
| Alcohols | 7-Hexadecanoleicosane | ||
| Alkanes | Eicosane | ||
| Ketones | 7-Methyl-oxa-cyclododeca-6 and 10-dien-2-one | ||
| Pyridazines | 3-Methylpyridazine and indazol-4-one | Streptomyces sp. | GC-MS [88] |
| Alkanoic acids | n-Hexadecanoic acid and octadecanoic acid | ||
| Ketones | Indazol-4-one | ||
| Tetrapeptides | Cis-bis(methylthio)silvatin, 6-oxo-methylthiosilvatin and deprenyl-bis(methylthio)silvatin | Penicillium brevicompactum | GC-MS [89] |
| Immunosuppressants | Mycophenolic acid | ||
| Diketopiperazines | Fusaperazine A/E/F, bilain B, and saroclazine A/B | ||
| Alkaloids, antibiotics | Brevianamide A/B | ||
| Alkaloids | Pyrrole-derived alkaloids | Micromonospora sp. | LC-MS [90] |
| Antibiotics | Rifamycins and staurosporine | Salinispora arenicola | LC-MS [91] |
| Polyketides | Saliniketals | ||
| Alkaloids | α-Methoxyroquefortine C, roquefortine C, and isoroquefortine C | Aspergillus sydowii and Penicillium chrysogenum | LC-MS [92] |
| Antibiotics | Meleagrin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Fang, F.; Ding, L.; Yu, K.; Zhang, L.; Lu, H. Technological Advancements in Field Investigations of Marine Microorganisms: From Sampling Strategies to Molecular Analyses. J. Mar. Sci. Eng. 2023, 11, 1981. https://doi.org/10.3390/jmse11101981
Huang Z, Fang F, Ding L, Yu K, Zhang L, Lu H. Technological Advancements in Field Investigations of Marine Microorganisms: From Sampling Strategies to Molecular Analyses. Journal of Marine Science and Engineering. 2023; 11(10):1981. https://doi.org/10.3390/jmse11101981
Chicago/Turabian StyleHuang, Zhishan, Fang Fang, Lingyun Ding, Ke Yu, Lijuan Zhang, and Hailong Lu. 2023. "Technological Advancements in Field Investigations of Marine Microorganisms: From Sampling Strategies to Molecular Analyses" Journal of Marine Science and Engineering 11, no. 10: 1981. https://doi.org/10.3390/jmse11101981
APA StyleHuang, Z., Fang, F., Ding, L., Yu, K., Zhang, L., & Lu, H. (2023). Technological Advancements in Field Investigations of Marine Microorganisms: From Sampling Strategies to Molecular Analyses. Journal of Marine Science and Engineering, 11(10), 1981. https://doi.org/10.3390/jmse11101981







