Abstract
The oceanic lightfish Vinciguerria nimbaria is a mesopelagic species found in tropical and subtropical waters. In this study, we collected a total of 266 mixed fish egg samples from 78 stations in Korean waters of the Northwest Pacific Ocean from January to November 2021, and analysed these samples for V. nimbaria using cytochrome c oxidase I (COI) metabarcoding. We detected V. nimbaria eggs five times in May and once in August, with 20 V. nimbaria eggs to be estimated among the 266 mixed samples, which consisted of 68,844 eggs. To verify the accuracy of the metabarcoding results, two samples consisting of 1 and 6 eggs (diameter, 0.82 ± 0.07 mm; n = 5), respectively, that were identified as V. nimbaria were reanalysed using partial COI regions with the Sanger sequencing method. COI sequences obtained using both sequencing methods showed 100% identity in the overlapping regions. The mixed eggs formed one clade with V. nimbaria (average pairwise genetic distance, 0.002 ± 0.003; n = 7) in a phylogenetic ML tree based on the mitogenome (2 rRNAs and 13 protein-coding genes) of order Stomiiformes, including partial COIs from the mixed egg samples. The pairwise genetic distances in this clade were smaller than that of Stomiiformes (0.468 ± 0.081), except for V. nimbaria. These eggs represent direct evidence of the intrusion or distribution of adult V. nimbaria, an unrecorded species, in Korean waters.
1. Introduction
Vinciguerria nimbaria (Jordan and Williams 1895; Stomiiformes: Phosichthyidae), a mesopelagic species found abundantly at depths of 200–400 m in tropical and subtropical oceans [1,2], is distributed in the Western Central, Eastern, and Northwestern Atlantic Oceans [3,4,5] and Indo-Pacific waters, including the Southeastern Pacific Ocean and South China Sea [6,7] except for Korean waters [8]. Vinciguerria nimbaria feeds mainly on copepods and is a major prey of tuna [2,9]; it forms loose schools that occur in clusters during the daytime and dense schools in large aggregations during the night [10], organized through large vertical diel migrations [11].
The total life span of V. nimbaria is 6–7 months. Mature females (standard length, >30.6 mm; age, >85 days) spawn pelagic eggs (diameter, 0.65 mm) continuously throughout the year [5]. The lifetime fecundity of V. nimbaria is approximately 9000 eggs (109,000 eggs without considering mortality), and the maximum stock egg production of a theoretical cohort occurs at a standard length of 37 mm [5].
Many marine teleost fishes release large quantities of pelagic eggs as a spawning strategy [12,13]. The probability of finding pelagic eggs increases exponentially during the spawning period, exceeding that of adults [14,15]. The geographic distribution of these eggs is critical to locating spawning fish, as well as identifying their spawning grounds and periods. Because fish eggs are extremely sensitive to environmental changes, spawning conditions can be used as ecological indicators of climate change [16].
The identification of fish eggs to the species level using morphological traits alone is difficult because of their high morphological similarity [17,18]. DNA barcode analysis has become increasingly prevalent in egg identification for a limited number of species, including Lophius litulon [19], Anguilliformes [20], and Larimichthys polyactis [21]. Egg DNA barcoding is also useful for the long-term monitoring of various fish spawning grounds [22,23]. Recently, the intrusion of rare fish that were previously unreported in Korean waters has been detected through DNA metabarcoding of mixed fish eggs based on high-throughput sequencing (HTS) [24].
In this study, we applied DNA metabarcoding to mixed fish egg samples to test its efficacy in monitoring rare species. The Vinciguerria nimbaria barcode reads from these samples were confirmed using Sanger sequencing. Our results offer evidence of the distribution of unrecorded adult V. nimbaria in Korean waters based on DNA metabarcoding of pelagic fish eggs.
2. Materials and Methods
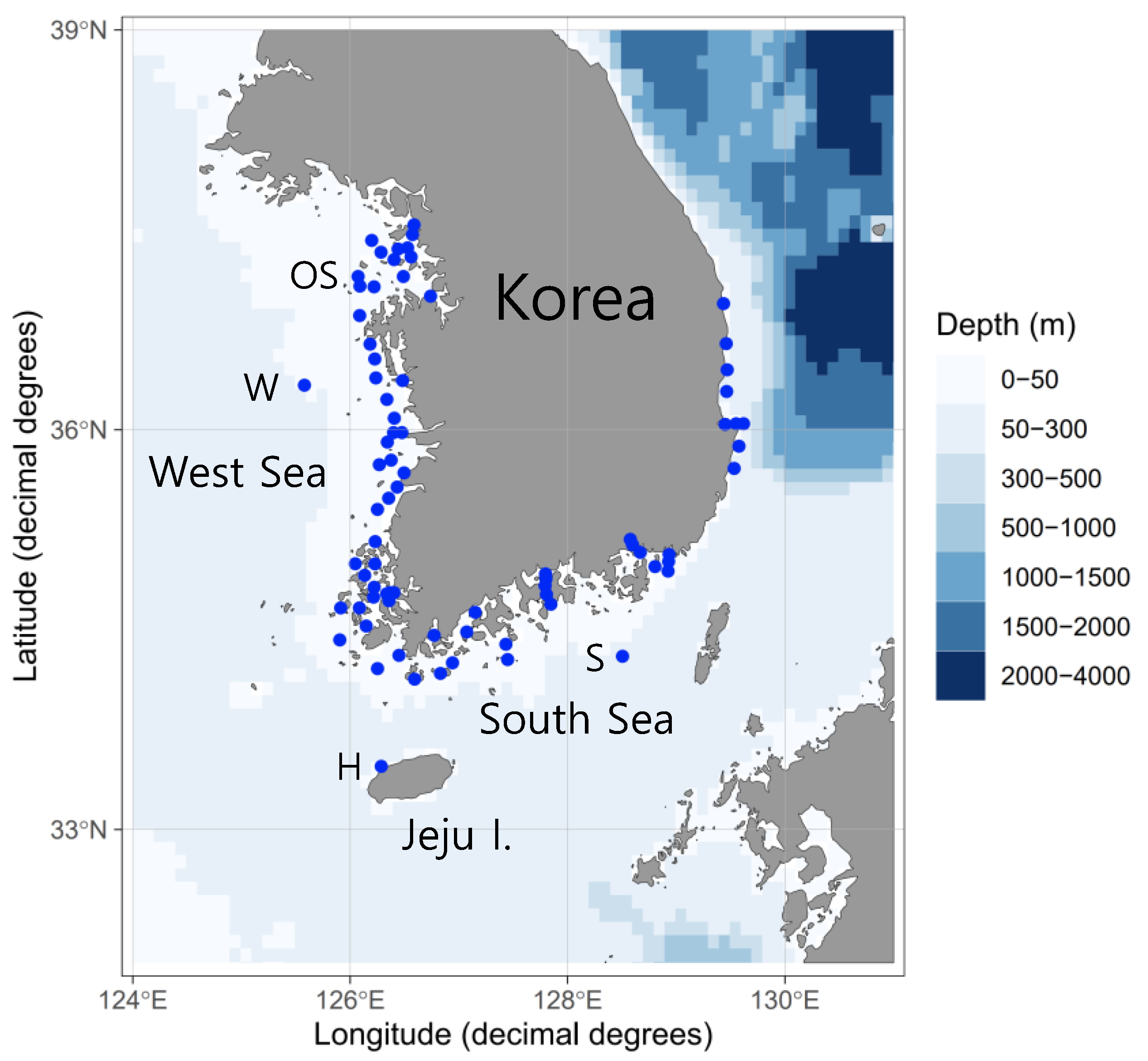
Fish egg samples were collected from 78 stations in the coastal waters of Korea between January and November 2021. A map of the sampling stations was created using ggOceanMaps software [25] (Figure 1). Egg sampling was conducted using two types of nets (mesh, 300 μm), with mouth diameters of 60 and 80 cm (net towing speed, 1 m/s; net trajectory; vertical, horizontal, and oblique; net towing time, <10 min). We collected a total of 266 specimens containing fish eggs; these were stored in 95% ethanol in the field on the research vessel (10% sample volume in ethanol). A total of 68,844 eggs were extracted from the raw samples under a dissecting microscope (Table 1). The water temperature and salinity of the study area were measured using a Sea-Bird SBE 9 or Sea-Bird SBE 19 Plus V2 instrument (Sea-Bird Electronics, Bellevue, WA, USA).
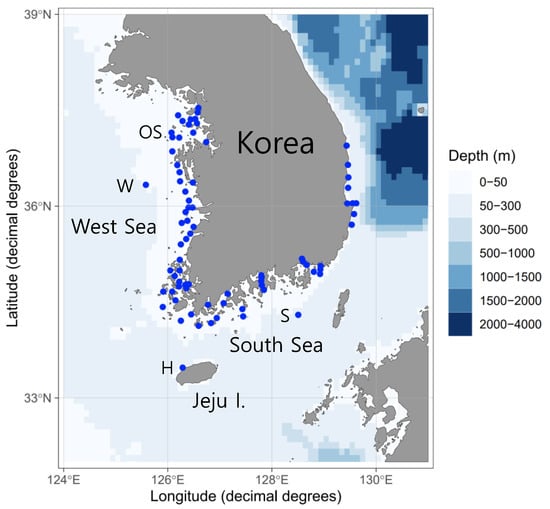
Figure 1.
Sampling stations where pelagic fish eggs were collected off the Korean coast.

Table 1.
Information on the survey area and high-throughput sequencing data for pelagic fish eggs.
We performed genomic DNA (gDNA) extraction from one egg to mixed eggs up to 250 ul or less per sample and two adult samples of Vinciguerria sp. (Indian Ocean; 4.097° N, 77.350° E) on March 23, 2018 (accession no.: OP983980, OP983981) using a MagListoTM 5M Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea) following the manufacturer’s protocols. Prior to gDNA extraction, all samples were photographed using a digital camera. A MiSeq next-generation sequencer (Illumina, San Diego, CA, USA) was used to analyse the cytochrome c oxidase I (COI) barcode region of mixed eggs. Sequencing library construction consisted of two steps: polymerase chain reaction (PCR) and amplicon sequencing [26]. The first PCR was performed using the COI region primers used for species identification, including the MiSeq adapter. The primers used for the first PCR were as follows: forward, TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGWACWGGWTGAACWGTWTAYCCYCC; reverse, GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTAIACYTCIGGRTGICCRAARAAYCA [26,27]. The PCR conditions were: initial denaturation at 95 °C for 3 min; 40 cycles of denaturation at 95 °C for 30 s, annealing at 46 °C for 30 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 5 min. The products were held at 4 °C.
For Sanger sequencing of two Vinciguerra sp. adults (Figure 2) identified followed by Smith and Heems [28], and Nakao [7], one egg (SM310), and six mixed eggs (SM332), PCR was performed using COI fish cocktail primers, as follows: forward, M13(20)-VF2_t1 GTAAACGACGGCCAGTCAACCAACCACAAAGACATTGGCAC and M13(20)-FishF2_t1 GTAAAACGACGGCCAGTCGACTAATCATAAAGATATCGGCAC; and reverse, M13(40)-FishR2_t1 CAGGAAACAGCTATGACACTTCAGGGTGACCGAAGAATCAGAA and M13(40)-FR1d_t1 CAGGAAACAGCTATGACACCTCAGGGTGTCCGAARAAYCARAA [29,30]. The PCR conditions were: initial denaturation at 94 °C for 3 min; 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 40 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 4 min. The products were held at 4 °C, and then sequenced using a DNA Analyzer (3730XL; Applied Biosystems, Waltham, MA, USA) with a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA).

Figure 2.
Vinciguerria sp. (IB18IM 4–8; total length, 32.2 mm) collected in the Indian Ocean on 23 March 2018.
Secondary PCR was performed for sample classification using the purified product of the first PCR, using the following primers: forward, AATGATACGGCGACCACCGAGATCTACAC-index2-TCGTCGGCAGCGT; and reverse, CAAGCAGAAGACGGCATACGAGAT-index1-GTCTCGTGGGCTCGG [26]. The secondary PCR conditions were: initial denaturation at 95 °C for 3 min; 8 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 1 min. The products were held at 4 °C. The products of the first and second PCR analyses were purified using a MagListoTM 5M PCR Purification Kit (Bioneer).
The concentration of the purified second PCR product was measured using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific), and then equal amounts of each sample were mixed. A total of 408 raw FASTQ datasets (≥1–14 segmentation analyses per sample; 68,844 eggs within 266 mixed fish egg samples) were obtained by MiSeq analysis (2 × 301 bp; Illumina), merged using the BBMerge tool (default, [31]), and mapped to the COI region of V. nimbaria reference sequences (GenBank accession no. AP012958; complete mitogenome) using Geneious v11.1.5 software.
The merged paired contig reads (MPCRs; >99% identity with V. nimbaria COI AP012956) were identified as V. nimbaria if the mapping ratio of MPCRs to MMPRs (mean merged paired reads) among sampling stations with >1000 MPRs (merged paired reads) was >80%. To validate the identification accuracy, six complete Stomiiformes genomes, including that of V. nimbaria (AP012958), were extracted from GenBank [32]. The six mitogens concatenated 2 rRNAs and 13 protein-coding genes (PCGs), and COI contig reads (including two Sanger sequences) constructed from six mixed egg samples and partial COI sequences from two Vinciguerria sp. adults were aligned using ClustalW [33]. The GTR + G + I model (1000 bootstraps) was used to construct an ML (maximum likelihood) tree of aligned sequences with MEGA-X software [34,35].
3. Results
A total of 79,928,854 reads were obtained through HTS of 266 mixed fish egg samples (68,844 eggs; 408 raw FASTQ dataset) collected from 78 stations along the Korean coast. A total of 12,272,357 MPRs were constructed from the 79,928,854 bidirectional raw FASTQ reads. Among the MPRs from the raw HTS reads, 79 samples had 1–9806 MPCRs with >99% identity with V. nimbaria COI (AP012958). Only six samples passed the quality control condition for identification as V. nimbaria, accounting for at least 1551–25,617 MPRs, was >85.7% ratio of MMPRs to MPCRs (three eggs; SM377) (Table 2). Two MPCRs, from one, single-egg sample (SM310) and one, six-egg mixed sample (SM332), that were identified as the same species were found to be 100% concordant in their overlapping regions according to Sanger sequencing (OP975709, OP975710).

Table 2.
Descriptions of reference mapping stations, samples, and merged paired contig reads from mixed pelagic fish egg samples.
Vinciguerria nimbaria eggs were found off the northwest coast of Jeju Island (H: May, one time) and in the Korea Strait (S: May, three times), south of the Korean Peninsula; and in the West Sea (W: once in May and once in August), west of the Korean Peninsula (Table 2). Eggs were collected using vertical, oblique, and surface net towing methods. Surface net towing was performed twice (Table 2). The water temperature of the surface layer was 16.8–17.4 °C, and the salinity was 35.2–35.5 PSU at the three sampling areas (H, S, and W) in May. In August, the water temperature was 27.8–28.1 °C at the surface and 11.4–11.5 °C at the bottom, and the salinity was 31.8–31.9 PSU at the surface and 32.3–32.4 PSU at the bottom at the W sampling area.
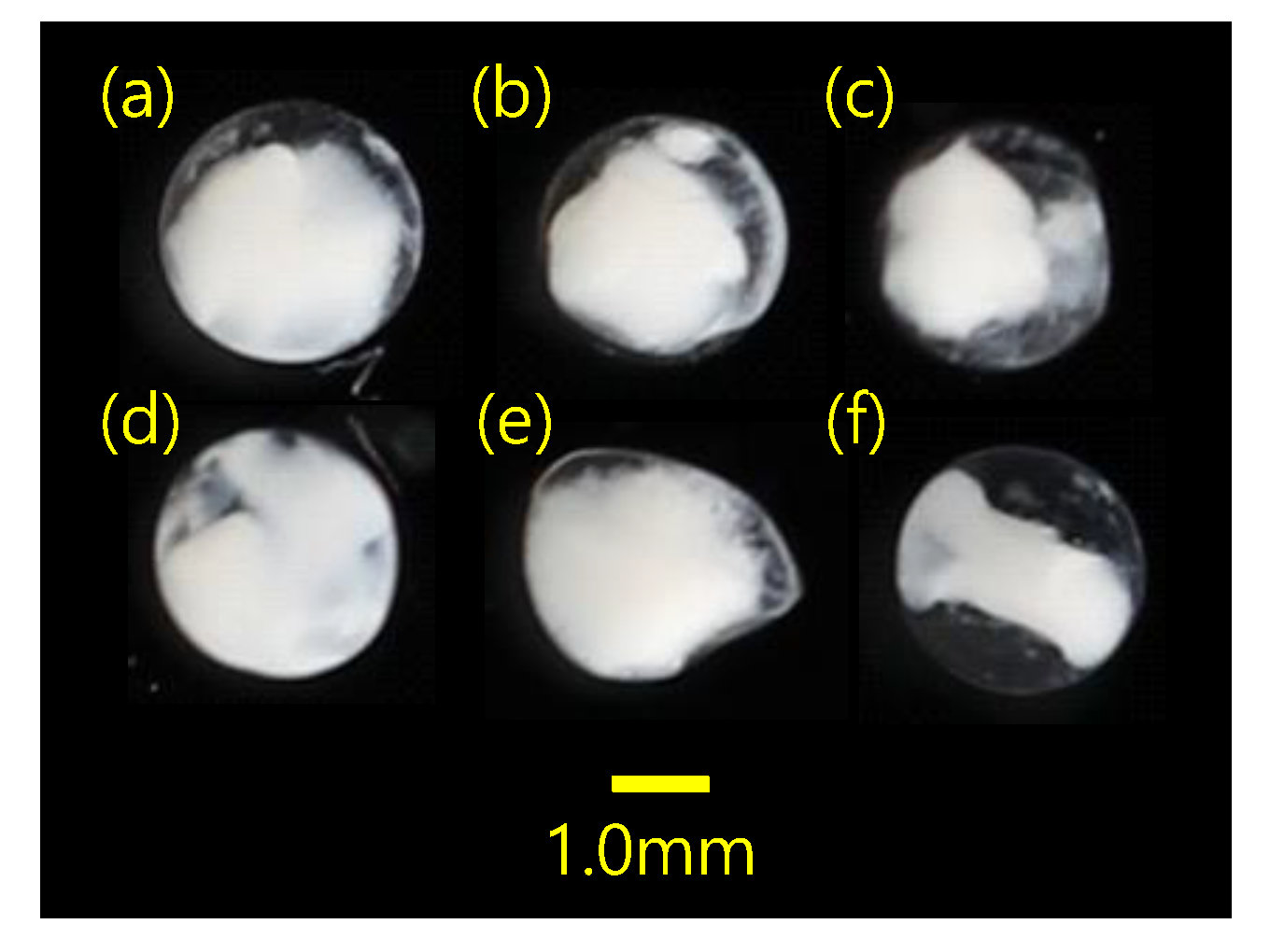
The proportion of MMPRs to MPCRs for two V. nimbaria eggs (SM310 and SM332) was 99.4 and 556.7% respectively. The proportion of MMPRs to MPCRs for the other four mixed eggs was 85.7–870.9%. If the ratio of MMPRs to MPCRs was >50%, it was estimated as one egg. There were one to nine V. nimbaria eggs per sample, for a total of 20 eggs (Table 2). The diameter of the V. nimbaria eggs was 0.71–0.90 mm (0.82 ± 0.07 mm; n = 5) in 95% ethanol. The eggs were spherical, with no oil globule, and were in the (a–e) early and (f) middle stages of development (Figure 3).
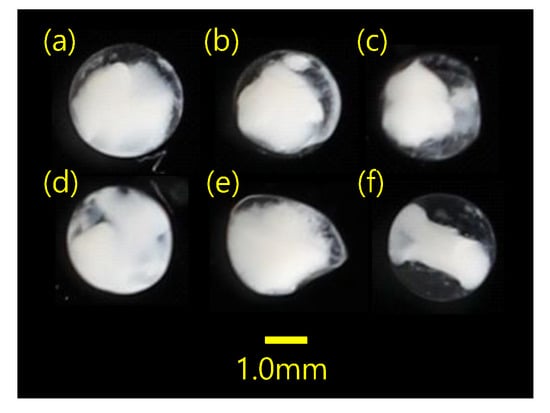
Figure 3.
Morphological characteristics of the sampled eggs, with diameters (mm) of (a) 0.90, (b) 0.84, (c) 0.79, (d) 0.71, (e) no data (not measured because of deformed egg) and (f) 0.84. The average egg diameter was 0.82 ± 0.07 mm (n = 5). All eggs were sampled from station W (sample SM332).
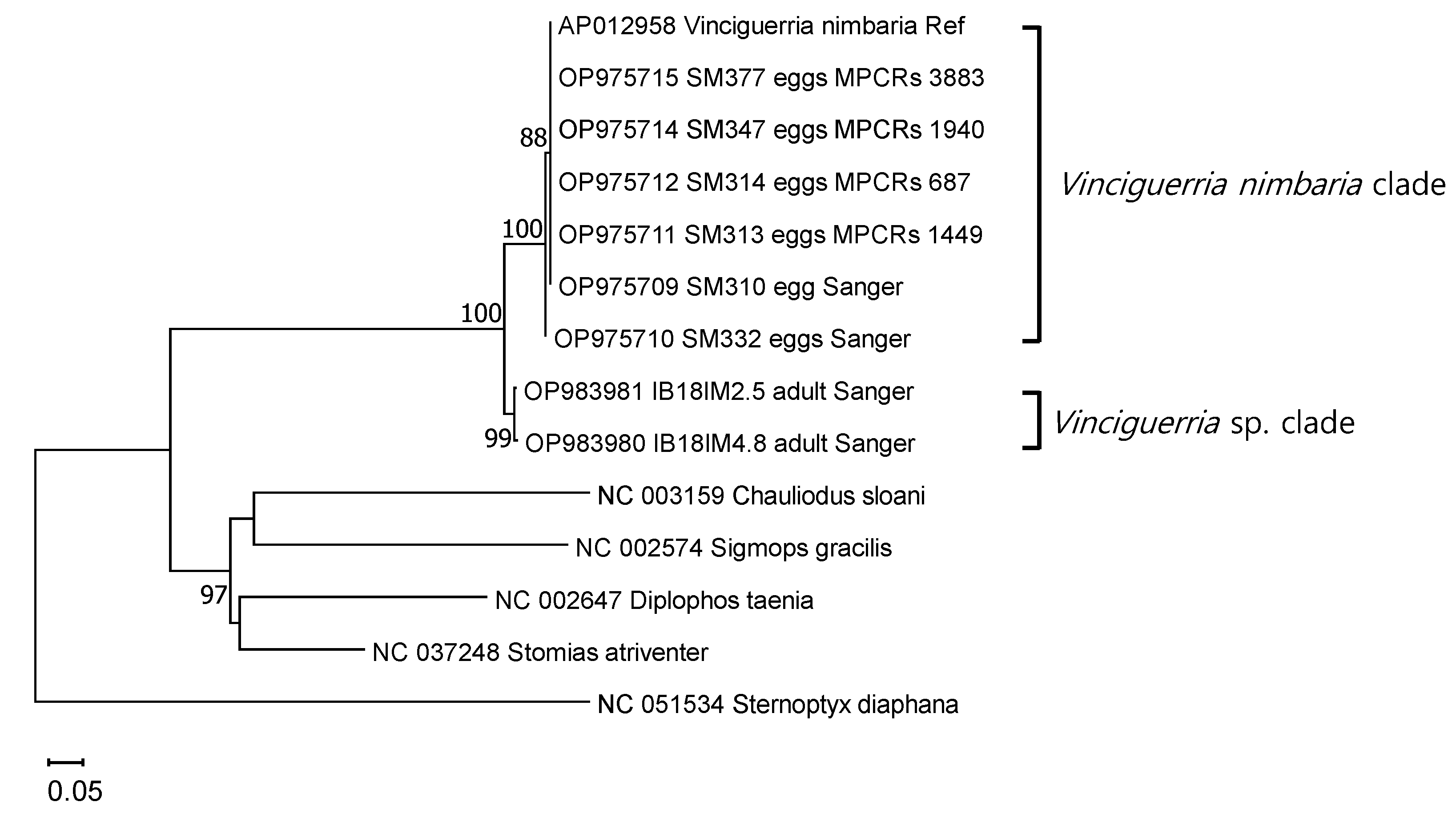
All six egg MPCRs formed a phylum with V. nimbaria (AP012958), which was used as a standard for species identification in the COI ML tree (Figure 4). Genetic distances within this clade ranged from 0.000 to 0.007 (average, 0.002 ± 0.003; n = 7). This clade was clearly distinguished from the Vinciguerria sp. clade, a similar taxon, with a genetic distance of 0.068–0.074 (average, 0.073 ± 0.002). The two lineages of genus Vinciguerria were distinctly separated from other taxa in Stomiiformes, with a genetic distance of 0.301–0.660 (average, 0.468 ± 0.081).
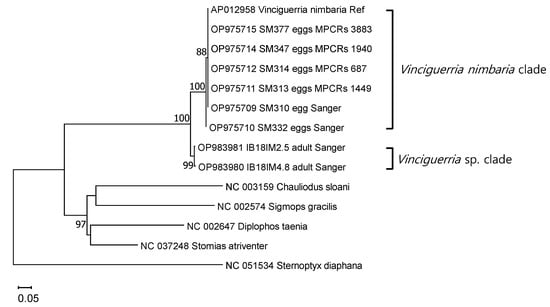
Figure 4.
Phylogenetic relationships determined using two rRNAs and 13 protein-coding genes, including partial cytochrome c oxidase I, from six V. nimbaria egg merged paired contig reads, two adult Vinciguerria sp., and five outgroups as shown in the ML tree. Ref, reference sequence; Sanger, Sanger sequencing; MPRCRs, merged paired read contig reads.
4. Discussion
Information on the geographical distribution of fish can be obtained from a wide variety of sample types, including eggs, adults, environmental DNA (eDNA; traces of organisms in their habitat), and photographs [23,36,37,38]. Among these, the easiest samples to attain are eDNA, which can be obtained through water sampling, and fish eggs, which can be collected using small nets or instruments such as the Continuous Underway Fish Egg Sampler [23,39,40]. Unlike eDNA, fish eggs are highly correlated with the distribution of adults during the spawning season, and are therefore useful for identifying spawning grounds through the detection of egg spatiotemporal distribution [41,42]. In this study, we examined the coastal distribution of V. nimbaria around the Korean Peninsula by analysing the gDNA of single or mixed fish egg samples by HTS and Sanger sequencing. To date, adults of this species have not been reported in the waters around the Korean Peninsula [8]. The V. nimbaria eggs identified by DNA barcoding in this study are the first to be discovered throughout the coastal waters of the Korean Peninsula, as well as in the Pacific Northwest. In a similar case, Trachipterus jacksonensis, which had been reported only in the Southern Hemisphere of the Pacific, was found in the East Sea, east of the Korean Peninsula through DNA barcoding of individual eggs [43].
Many marine teleost fishes release large amounts of pelagic eggs for reproduction [12,13]. These eggs have the highest natural mortality rates among teleost life stages. Nevertheless, the likelihood of finding eggs in the early stages of development immediately after spawning is higher than that of adults [14,15]. Thus, most of the eggs collected in this study were in the early stages of development (Figure 3).
Egg size is useful information for selecting DNA barcode target samples when specifying the study area [20,43]. Two species in Korean waters, Engraulis japonicus and Maurolicus japonicus, were identified as species level without DNA barcoding among lots of fish eggs [15,44,45]. In this study, the average egg size was 0.82 ± 0.07 mm (no oil globule) in 95% ethanol (Figure 3). When fish eggs are stored in 95% ethanol, shrinkage or deformation occurs due to dehydration. However, the V. nimbaria eggs collected in this study were approximately 30% larger than those collected in the Atlantic Ocean in a previous study (diameter, 0.65 mm) [5]. Neither the eggs collected in this study nor those collected in the Atlantic had oil globules. Thus, further study is needed to determine whether this large difference in egg size represents intraspecies variation or a potential interspecies difference. Despite these morphological differences, egg morphological traits remain useful for selecting study species.
The length of the DNA barcode region produced via Sanger sequencing was approximately 600 bp, which is a higher resolution than that of the short sequences used in HTS (~313 bp). Large numbers of individual fish egg DNA barcodes require substantial analysis time and cost. An alternative to this costly process is DNA metabarcoding, which is performed on large numbers of nucleotide sequences obtained by HTS based on gDNA extracted from large numbers of mixed fish egg samples. This method is used for various taxa such as zooplankton, and has been applied in eDNA-based fish species exploration [36,37,46] and species analyses of mock mixed fish egg samples or mixed fish larvae [47,48].
In this study, V. nimbaria was detected six times through reference mapping of a large number of reads obtained by HTS from a huge quantity of mixed fish egg samples. The two HTS analysis samples, one consisting of a single egg (SM377) and the other of six mixed eggs (SM332), were re-analysed using Sanger sequencing, and the results showed 100% identity of the nucleotide sequences analysed by the two methods (Table 2). These results suggest that our analysis strategy was useful for identifying species from fish eggs.
In fish egg DNA barcode analysis, the accuracy of the reference sequence as the criterion for species identification is important. In this study, the ML tree was created using the mitogenomes (two rRNAs and 13 PCGs) of six fish species from order Stomiiformes, including V. nimbaria, the partial COI of two adults of a Vinciguerria species, and partial COIs from individual and mixed egg reference mapping contig sequences (including those for Sanger sequencing) (Figure 2 and Figure 3). In the ML tree, the V. nimbaria clade formed a lineage with fish eggs that was closest to the adult Vinciguerria species clade. The genetic distance between the two clades was 0.068–0.074 (average, 0.073 ± 0.002). These results confirm that the reference sequence of V. nimbaria belongs to the genus Vinciguerria, indicating that it may be used as an identifying criterion.
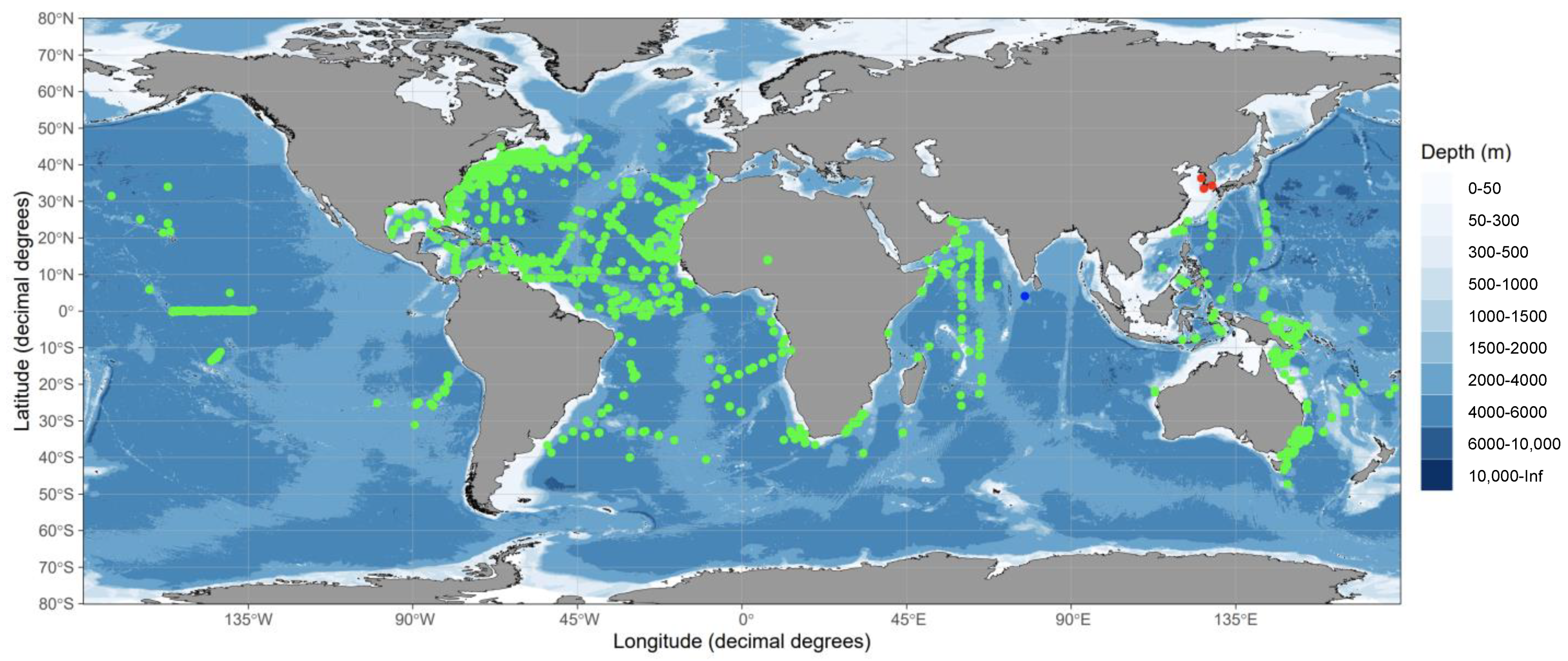
Fertilised eggs of vertebrates such as frogs and mice, including the zebrafish Danio rerio, do not replicate mitochondrial DNA during a certain period of development [49,50,51]. In sardine eggs, the 16S rRNA/18sRNA ratio decreases with embryogenesis, from the early stage to the immediately pre-hatching stage. The ratio of populations occupied by E. japonicus and M. japonicus in mixed egg samples and the ratio of reads obtained by HTS also showed a significant linear relationship [52]. The ratio of MPCRs to MMPRs for six samples detected V. nimbaria were 99.4% and 556.7%, respectively. Based on our results, a total of 20 possible eggs were identified from the ratio of MPCRs in V. nimbaria eggs to total MPRs in 151 mixed eggs, including individual eggs. Considering the number of V. nimbaria eggs found in three different sea areas, and that this species was found twice within one region after a time interval, and three times in another survey area, there appears to have been a continuous intrusion of V. nimbaria adults into the waters around the Korean Peninsula. Therefore, the waters around the Korean Peninsula may be a V. nimbaria habitat and the northern limit for spawning grounds based on the direct evidence of Vinciguerria nimbaria eggs as shown in the worldwide distribution of V. nimbaria (Figure 5).
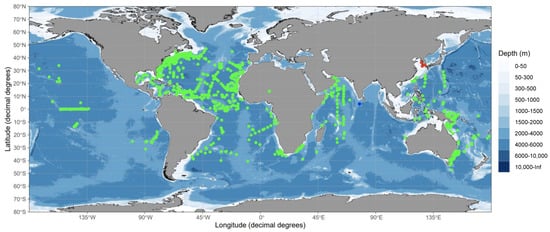
Figure 5.
Distribution map (by ggOceanMaps; 24) of Vinciguerria nimbaria in tropical and subtropical oceans, showing V. nimbaria eggs (red dots) and adult Vinciguerria sp. (blue dot) examined in this study, as well as V. nimbaria (green dots) from FishBase with permission from [53].
Author Contributions
Conceptualization, S.K.; methodology, S.K.; validation, S.K., B.-s.C. and S.-y.W.; formal analysis, S.K., B.-s.C. and S.-y.W.; investigation, B.-s.C. and S.-y.W.; data curation, B.-s.C. and S.-y.W.; writing— original draft preparation, S.K.; writing—review and editing, S.K., B.-s.C. and S.-y.W.; visualization, S.K.; supervision, S.K., B.-s.C. and S.-y.W.; project administration, S.K., B.-s.C. and S.-y.W.; funding acquisition, S.K., B.-s.C. and S.-y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Korea Institute of Ocean Science and Technology (PEA0012), the National Research Foundation of Korea (2016R1D1A1B03935737). Additionally, “Development of Advanced Science and Technology for Marine Environmental Impact Assessment” of the Korea Institute of Marine Science & Technology Promotion (KIMST-20210427), and the National Marine Ecosystem Comprehensive Survey supervised by the Korea Marine Environment Corporation, both funded by the Ministry of Oceans and Fisheries. We thank the many researchers who contributed to the field survey and sample analysis of this project and those who operated the survey vessel.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Sequence data supporting this study’s findings are deposited in NCBI/GenBank (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 6 January 2023) under accession numbers OP975709- OP975712, OP975714, OP975715, OP983980, and OP983981.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gjøsæter, J.; Kawaguchi, K. A review of the world resources of mesopelagic fish. FAO Fish. Tech. Pap. 1980, 193, 157. [Google Scholar]
- Badcock, J. Photichthyidae. In Fishes of the North-Eastern Atlantic and the Mediterranean; Whitehead, P.J.P., Bauchot, M.L., Hureau, J.C., Nielsen, J., Tortonese, E., Eds.; UNESCO: Paris, France, 1984; Volume 1, pp. 318–324. [Google Scholar]
- Scott, W.B.; Scott, M.G. Atlantic fishes of Canada. Can. Bull. Fish. Aquat. Sci. 1988, 219, 731. [Google Scholar]
- Claro, R. Características generales de la ictiofauna. In Ecología de Los Peces Marinos de Cuba; Claro, R., Ed.; Instituto de Oceanología Academia de Ciencias de Cuba: La Habana, Cuba; Centro de Investigaciones de Quintana Roo: Chetumal, Mexico, 1994; pp. 55–70. [Google Scholar]
- Stéquert, B.; Ménard, F.; Marchal, E. Reproductive biology of Vinciguerria nimbaria in the equatorial waters of the eastern Atlantic Ocean. J. Fish Biol. 2003, 62, 1116–1136. [Google Scholar] [CrossRef]
- Pequeño, G.; de Peces, C. Lista sistematica revisaday comentada. Rev. Biol. Mar. 1989, 24, 1–132. [Google Scholar]
- Nakabo, T. Fishes of Japan with Pictorial Keys to the Species; Tokai University Press: Tokyo, Japan, 2013. [Google Scholar]
- Youn, C.H.; Shim, J.H.; Kim, J.J. Pisces of Korea; Haksul Information Center: Seoul, Korea, 2021; p. 2148. [Google Scholar]
- Lebourges-Dhaussy, A.; Marchal, E.; Menkès, C.; Champalbert, G.; Biessy, B. Vinciguerria nimbaria (micronekton), environment and tuna: Their relationships in the Eastern Tropical Atlantic. Oceanol. Acta 2000, 23, 515–528. [Google Scholar] [CrossRef]
- Ménard, F.; Marchal, E. Foraging behaviour of tuna feeding on small schooling Vinciguerria nimbaria in the surface layer of the equatorial Atlantic Ocean. Aquat. Living Resour. 2003, 16, 231–238. [Google Scholar] [CrossRef]
- Marchal, E.; Lebourges, A. Acoustic evidence for unusual diel behaviour of a mesopelagic fish (Vinciguerria nimbaria) exploited by tuna. ICES J. Mar. Sci. 1996, 53, 443–447. [Google Scholar] [CrossRef]
- Kendall, A.W., Jr.; Ahlstrom, E.H.; Moser, H.G. Early life history stages of fishes and their characters. In Ontogeny and Systematics of Fishes; Moser, H.G., Richards, W.J., Cohen, D.M., Fahay, M.P., Kendall, A.W., Richardson, S.L., Eds.; American Society of Ichthyologists and Herpetologists: Kansas, MO, USA, 1984. [Google Scholar]
- Leiby, M.M. Life history and ecology of pelagic fish eggs and larvae. Mar. Plankton Life Cycle Strateg. 1984, 6, 121–140. [Google Scholar]
- Houde, E.D. Fish early life dynamics and recruitment variability. Am. Fish. Soc. Symp. 1987, 2, 17–29. [Google Scholar]
- Jung, S.; Hwang, S.D.; Kim, J. Fecundity and growth-dependent mortality of Pacific anchovy (Engraulis japonicus) in Korean coastal waters. Fish. Res. 2008, 93, 39–46. [Google Scholar] [CrossRef]
- Dahlke, F.T.; Wohlrab, S.; Butzin, M.; Pörtner, H.O. Thermal bottlenecks in the life cycle define climate vulnerability of fish. Science 2020, 369, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.T.; Yang, J.S.; Chen, K.C.; Lee, Y.S. An Identification Guide of Marine Fish Eggs from Taiwan; Institute of Zoology, Academia Sinica and Taiwan Power Company: New Taipei, Taiwan, 2001; p. 179. [Google Scholar]
- Ikeda, T.; Hirai, A.; Tabata, S.; Onishi, Y.; Mito, S. An Atlas of Early Stage Fishes in Japan, 2nd ed.; Okiyama, M., Ed.; Tokai University Press: Tokyo, Japan, 2014; p. 108. [Google Scholar]
- Oh, J.; Kim, S. Morphological and molecular characterization of separated pelagic eggs from Lophius litulon (Lophiiformes; Lophiidae). J. Fish Biol. 2015, 86, 1887–1891. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y.; Oh, J.; Kim, S. Genetic identification of eggs from four species of Ophichthidae and Congridae (Anguilliformes) in the northern East China Sea. PLoS ONE 2018, 13, e0195382. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.H.; Kim, J.K.; Ryu, J.H. First report on the occurrence of eggs of the small yellow croaker Larimichthys polyactis from Chilsan-do Island, Jeollanam-do, Korea. Korean J. Fish. Aquat. Sci. 2020, 53, 650–655. [Google Scholar]
- Han, S.H.; Kim, M.J.; Song, C.B. Molecular identification and distribution pattern of fish eggs collected around Jejudo Island. Korean J. Ichthyol. 2015, 27, 284–292. [Google Scholar]
- Harada, A.E.; Lindgren, E.A.; Hermsmeier, M.C.; Rogowski, P.A.; Terrill, E.; Burton, R.S. Monitoring spawning activity in a southern California marine protected area using molecular identification of fish eggs. PLoS ONE 2015, 10, e0134647. [Google Scholar] [CrossRef]
- Choi, H.Y.; Chin, B.S.; Park, G.S.; Kim, S. Evidence of intrusion of a rare species, Peristedion liorhynchus, into Korean waters based on high-throughput sequencing of the mixed fish eggs. Korean J. Ichthyol. 2022, 34, 8–15. [Google Scholar] [CrossRef]
- Vihtakari, M. GgOceanMaps: Plot Data on Oceanographic Maps Using ‘ggplot2’. R Package Version 1.3.7. Available online: https://mikkovihtakari.github.io/ggOceanMaps/ (accessed on 6 December 2022).
- Illumina, I. 16S Metagenomic Sequencing Library Preparation. Preparing 16S ribosomal RNA gene amplicons for the illumina MiSeq system. Microb. Genom. 2013, 1, 28. [Google Scholar]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Smith, M.M. , Heemstra, P.C. Smiths’ Sea Fishes; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J.; Wangensteen, O.S.; Chapman, D.D.; Boussarie, G.; Buddo, D.; Guttridge, T.L.; Hertler, H.; Mouillot, D.; Vigliola, L.; Mariani, S. Environmental DNA reveals tropical shark diversity in contrasting levels of anthropogenic impact. Sci. Rep. 2017, 7, 16886. [Google Scholar] [CrossRef] [PubMed]
- Boussarie, G.; Bakker, J.; Wangensteen, O.S.; Mariani, S.; Bonnin, L.; Juhel, J.-B.; Kiszka, J.J.; Kulbicki, M.; Manel, S.; Robbins, W.D.; et al. Environmental DNA illuminates the dark diversity of sharks. Sci. Adv. 2018, 4, eaap9661. [Google Scholar] [CrossRef]
- Kim, G.; Song, Y. Identification of freshwater fish species in Korea using environmental DNA technique from the experiment at the freshwater fish ecological learning center in Yangpyeong, Gyeonggi Do. J. Environ. Impact Assess. 2021, 30, 1–12. [Google Scholar]
- Checkley, D.M., Jr.; Ortner, P.B.; Settle, L.R.; Cummings, S.R. A continuous, underway fish egg sampler. Fish. Oceanogr. 1997, 6, 58–73. [Google Scholar] [CrossRef]
- Lelièvre, S.; Verrez-Bagnis, V.; Jérôme, M.; Vaz, S. PCR-RFLP analyses of formalin-fixed fish eggs for the mapping of spawning areas in the Eastern Channel and Southern North Sea. J. Plankton Res. 2010, 32, 1527–1539. [Google Scholar] [CrossRef]
- Fox, C.J.; Taylor, M.; Dickey-Collas, M.; Fossum, P.; Kraus, G.; Rohlf, N.; Munk, P.; van Damme, C.J.G.; Bolle, L.J.; Maxwell, D.L.; et al. Mapping the spawning grounds of North Sea cod (Gadus morhua) by direct and indirect means. Proc. R. Soc. B: Biol. Sci. 2008, 275, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, K.; Chow, S.; Otake, T.; Kurogi, H.; Mochioka, N.; Miller, M.J.; Aoyama, J.; Kimura, S.; Watanabe, S.; Yoshinaga, T.; et al. Oceanic spawning ecology of freshwater eels in the western North Pacific. Nat. Commun. 2011, 2, 179. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y.; Choi, H.C.; Kim, S.; Oh, H.J.; Youn, S.H. Discovery of pelagic eggs of two species from the rare mesopelagic fish genus Trachipterus (Lampriformes: Trachipteridae). J. Mar. Sci. Eng. 2022, 10, 637. [Google Scholar] [CrossRef]
- Kim, J.Y. Relationship Between Anchovy, Engraulis japonica, egg and larval density and environmental factors in the eastern waters of Korea. Korean J. Fish. Aquat. Sci. 1992, 25, 495–500. [Google Scholar]
- Kim, S.; Yoo, J.M. Distribution of Eggs and Larvae of Maurolicus muelleri in the Thermal Front of the Korea Strait. Korean J. Ichthyol. 1999, 11, 62–71. [Google Scholar]
- Song, C.-U.; Choi, H.; Jeon, M.-S.; Kim, E.-J.; Jeong, H.G.; Kim, S.; Kim, C.-G.; Hwang, H.; Purnaningtyas, D.W.; Lee, S.; et al. Zooplankton diversity monitoring strategy for the urban coastal region using metabarcoding analysis. Sci. Rep. 2021, 11, 24339. [Google Scholar] [CrossRef]
- Kimmerling, N.; Zuqert, O.; Amitai, G.; Gurevich, T.; Armoza-Zvuloni, R.; Kolesnikov, I.; Berenshtein, I.; Melamed, S.; Gilad, S.; Benjamin, S.; et al. Quantitative species-level ecology of reef fish larvae via metabarcoding. Nat. Ecol. Evol. 2018, 2, 306–316. [Google Scholar] [CrossRef]
- Duke, E.M.; Burton, R.S. Efficacy of metabarcoding for identification of fish eggs evaluated with mock communities. Ecol. Evol. 2020, 10, 3463–3476. [Google Scholar] [CrossRef]
- Chase, J.W.; Dawid, I.B. Biogenesis of mitochondria during Xenopus laevis development. Dev. Biol. 1972, 27, 504–518. [Google Scholar] [CrossRef]
- Pikó, L.; Taylor, K.D. Amounts of mitochondrial DNA and 132 abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. 1987, 123, 364–374. [Google Scholar] [CrossRef]
- Artuso, L.; Romano, A.; Verri, T.; Domenichini, A.; Argenton, F.; Santorelli, F.M.; Petruzzella, V. Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim. Biophys. Acta BBA Bioenerg. 2012, 1817, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y. Diagnosis and Change Prediction of Spawning Areas along the Coasts of the Korean Peninsula Using Pelagic Fish Eggs. Ph.D. Thesis, University of Science and Technology, Daejeon, Republic of Korea, 2021; p. 415. [Google Scholar]
- FishBase. World Wide Web Electronic Publication. Froese, R., Pauly D., Eds. Available online: http://www.fishbase.org (accessed on 6 December 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).