Computing Invasive Species Population Based on a Generalized Random Walk Process: Application to Blue Crab (Callinectes sapidus)
Abstract
:1. Introduction
1.1. Animal Population Estimation Methods
1.2. Challenges in Estimating Population Abundance in Aquatic Environments
1.3. The Invasive Species Callinectes sapidus
1.4. Overview and Scope of the Study
2. Materials and Methods
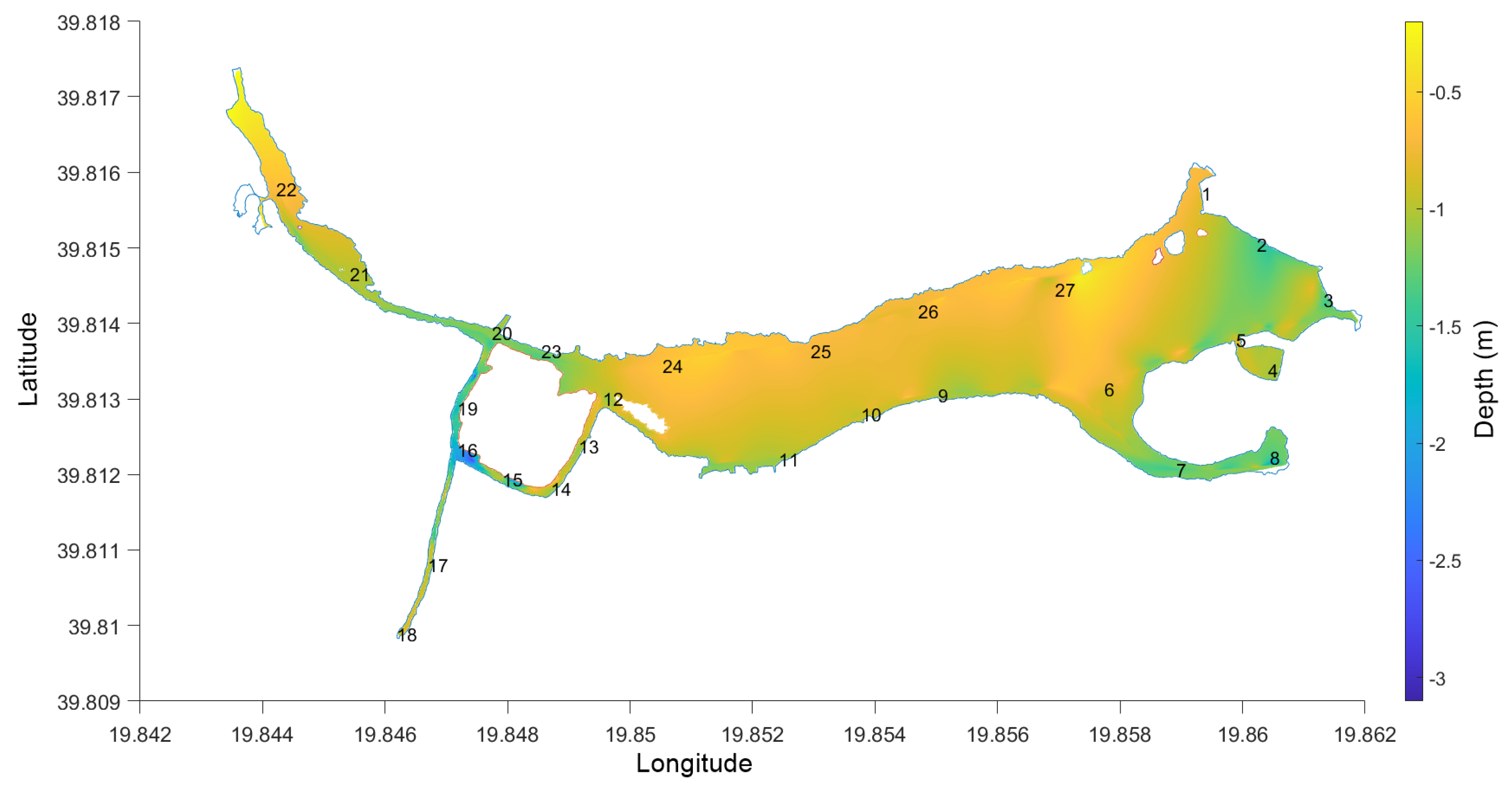
2.1. Study Site
2.2. Purpose
2.3. Entities, State Variables, and Scales
2.3.1. Field Measurements
2.3.2. Simulation Model
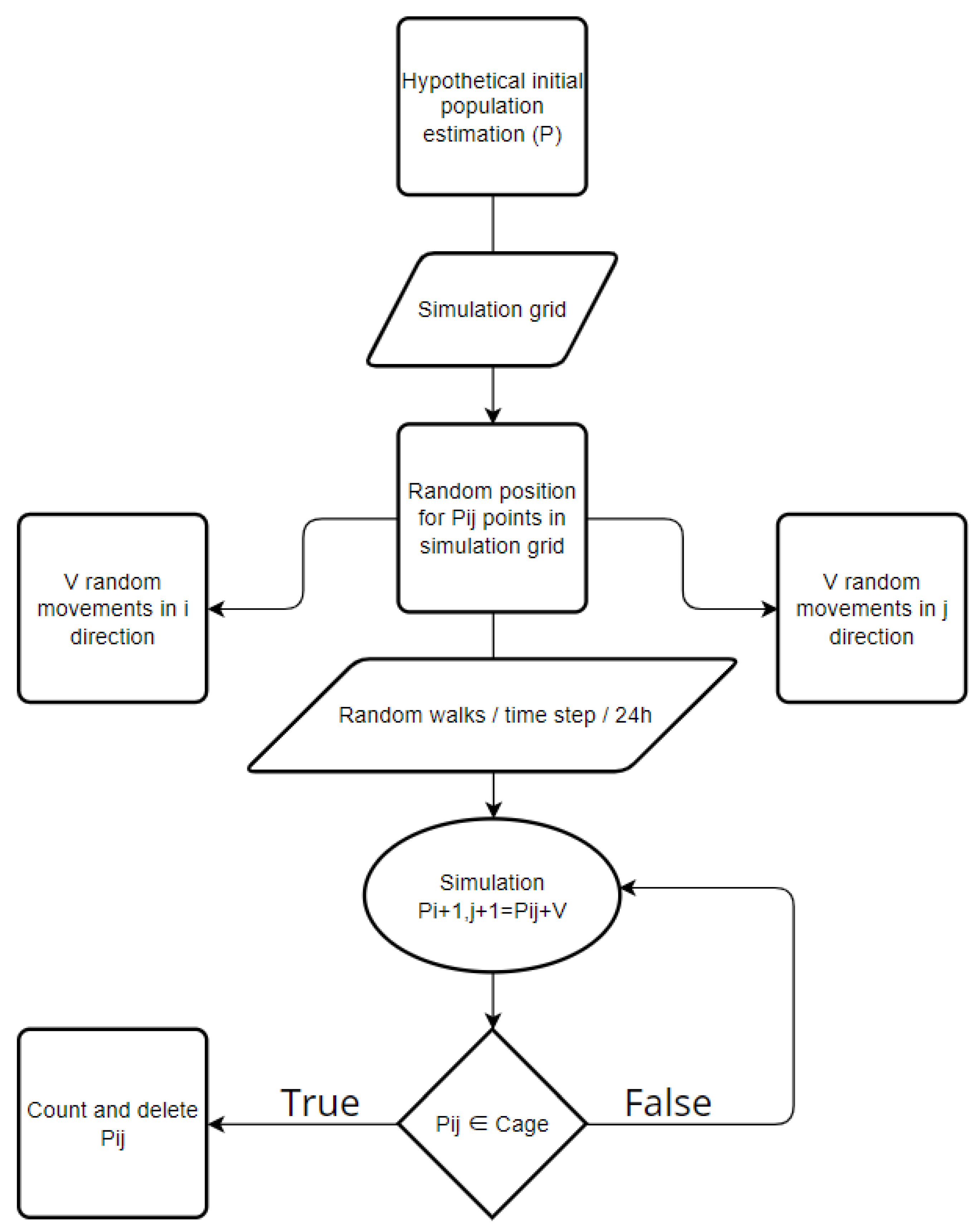
2.4. Process Overview and Scheduling
Governing Equations of Simulation Model
2.5. Design Concepts
2.5.1. Basic Principles
2.5.2. Emergence
2.5.3. Adaptation
2.6. Prediction
2.6.1. Sensing
2.6.2. Interaction
2.6.3. Stochasticity
2.6.4. Collectives
2.6.5. Observation
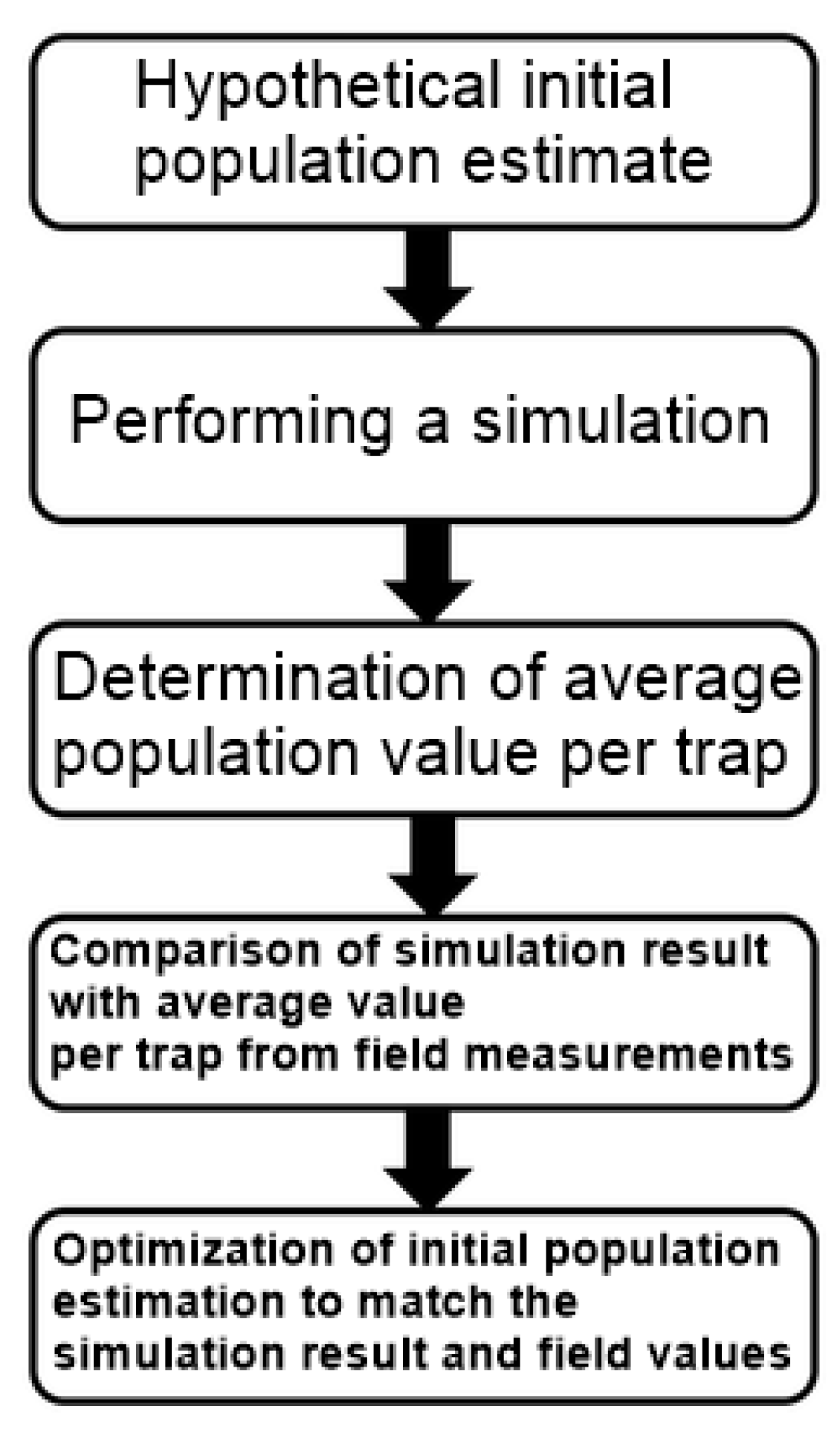
2.7. Initialization
Simulation Model
2.8. Input Data
2.9. Validation Method
Field Measurements—Juveniles
2.10. Assumptions/Limitations
3. Results
3.1. Field Measurements—Cages
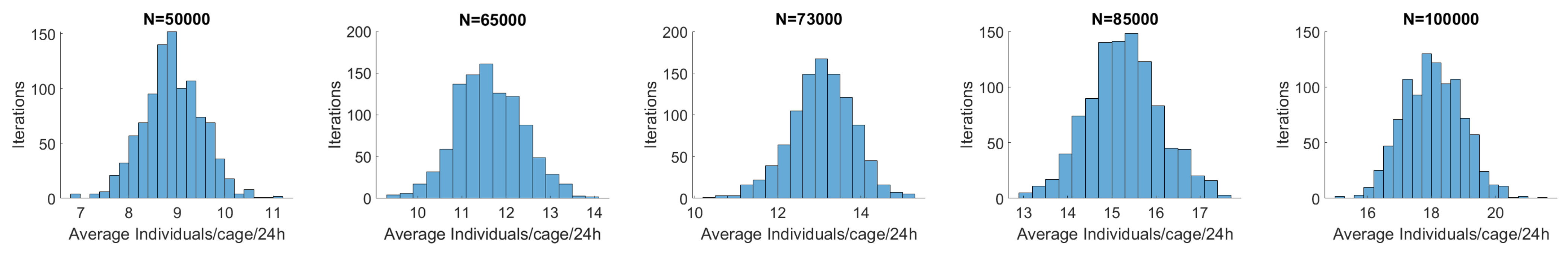
Simulation Model
3.2. Juveniles Samples
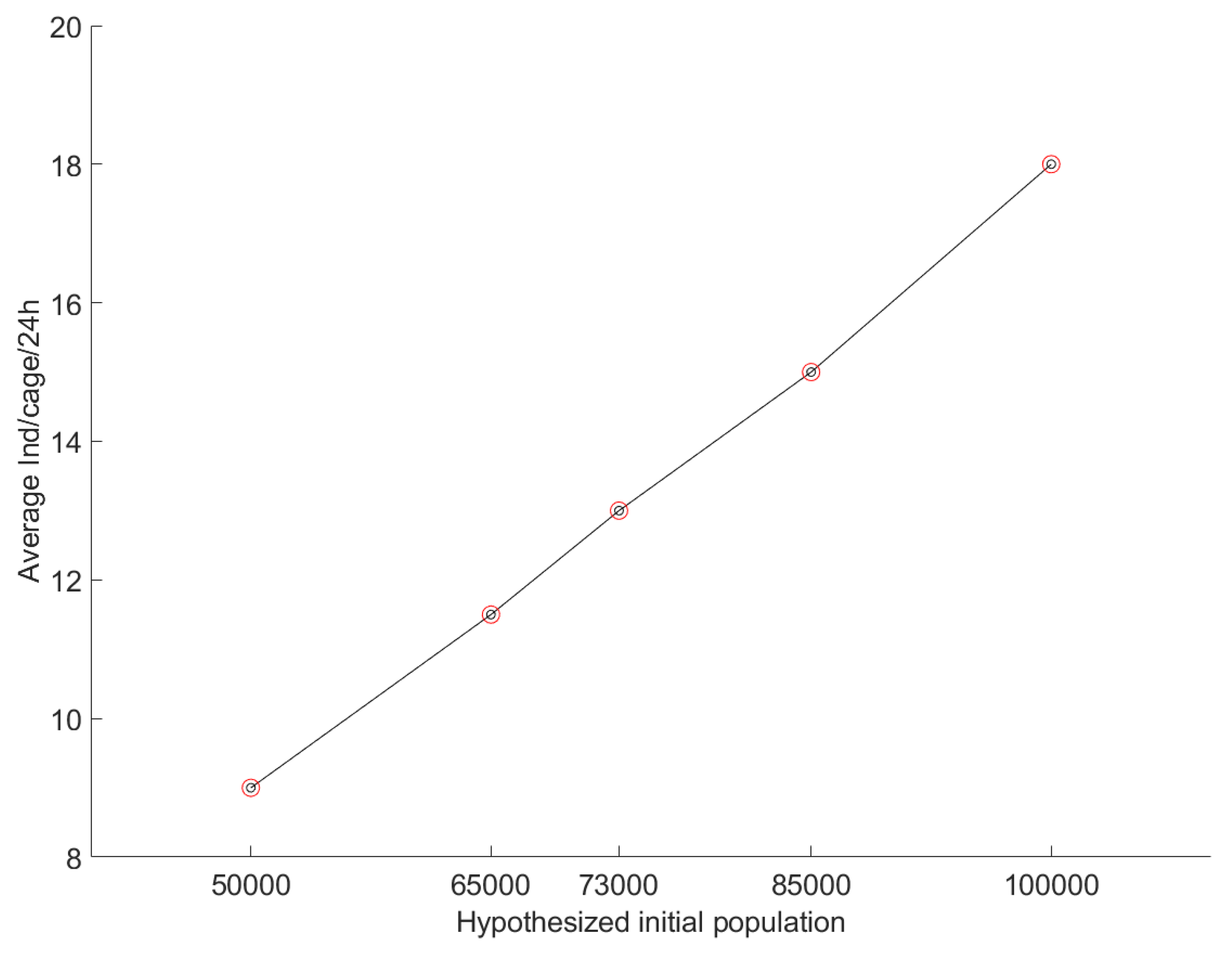
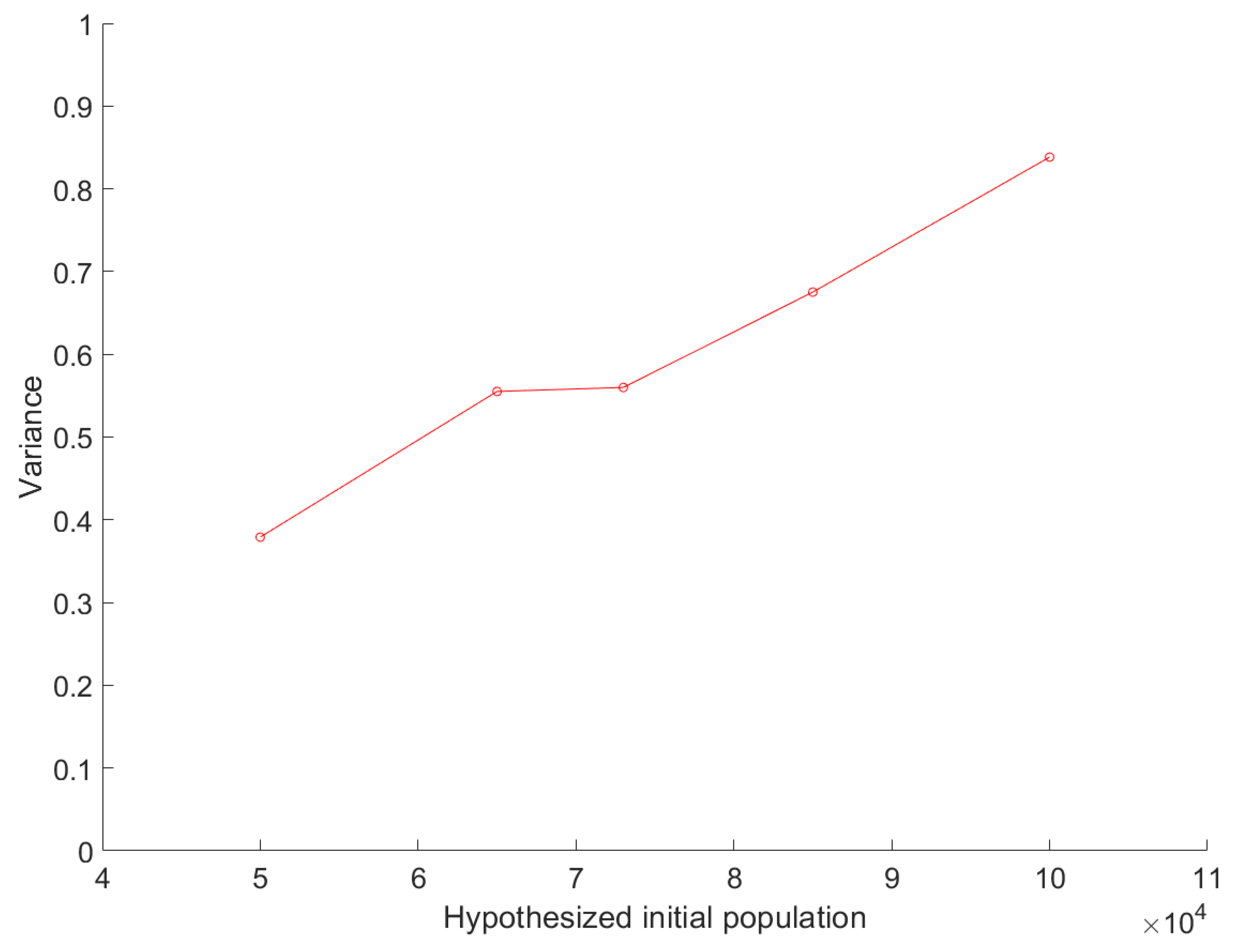
3.3. Sensitivity
4. Discussion
4.1. Applicability of Proposed Methodology in Confined Environments
4.2. Blue Crab Activity Periods in Antinioti lagoon
4.3. Factors Influencing the Validation Method
4.4. Limitations of the Juvenile Approach
4.5. Implications of Proposed Methodology
4.6. Adaptation to Large-Scale Areas
4.7. Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eberhardt, L. Appraising variability in population studies. J. Wildl. Manag. 1978, 42, 207–238. [Google Scholar] [CrossRef]
- Downing, R.L.; Moore, W.H.; Kight, J. Comparison of deer census techniques applied to a known population in a Georgia enclosure. Proc. Annu. Conf. Southeast. Assoc. Game Fish Comm. 1965, 19, 26–30. [Google Scholar]
- Schmidt, J.L. A comparison of census techniques of common duiker and bushbuck in timber plantations. S. Afr. For. J. 1983, 126, 15–19. [Google Scholar] [CrossRef]
- Koster, S.H.; Hart, J.A. Methods of estimating ungulate populations in tropical forests. Afr. J. Ecol. 1988, 26, 117–126. [Google Scholar] [CrossRef]
- Mayle, B.A.; Peace, A.J.; Gill, R. How Many Deer? Forestry Commission: Edinburgh, UK, 1999. [Google Scholar]
- Hirst, S. Road-strip census techniques for wild ungulates in African woodland. J. Wildl. Manag. 1969, 33, 40–48. [Google Scholar] [CrossRef]
- Dinerstein, E. An ecological survey of the Royal Karnali-Bardia Wildlife Reserve, Nepal: Part III: Ungulate populations. Biol. Conserv. 1980, 18, 5–37. [Google Scholar] [CrossRef]
- Underwood, R. On surveying ungulate groups. Afr. J. Ecol. 1982, 20, 105–111. [Google Scholar] [CrossRef]
- Peel, M.; Bothma, J.D.P. Comparison of the accuracy of four methods commonly used to count impala. S. Afr. J. Wildl. Res. 1995, 25, 41–43. [Google Scholar]
- Reilly, B. Precision of helicopter-based total-area counts of large ungulates in bushveld. Koedoe 2002, 45, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Bothma, J.P.; Peel, M.; Pettit, S.; Grossman, D. Evaluating the accuracy of some commonly used game-counting methods. S. Afr. J. Wildl. Res./S.-Afr. Tydskr. Natuurnavors. 1990, 20, 26–32. [Google Scholar]
- Bowland, A. Density estimate methods for blue duikers Philantomba monticola and red duikers Cephalophus natalensis in Natal, South Africa. J. Afr. Zool. 1994, 108, 505–519. [Google Scholar]
- Mandujano, S.; Gallina, S. Comparison of deer censusing methods in tropical dry forest. Wildl. Soc. Bull. 1995, 23, 180–186. [Google Scholar]
- Focardi, S.; Isotti, R.; Tinelli, A. Line transect estimates of ungulate populations in a Mediterranean forest. J. Wildl. Manag. 2002, 66, 48–58. [Google Scholar] [CrossRef]
- Lannoy, L.; Gaidet, N.; Chardonnet, P.; Fanguinoveny, M. Abundance estimates of duikers through direct counts in a rain forest, Gabon. Afr. J. Ecol. 2003, 41, 108–110. [Google Scholar] [CrossRef]
- Marsh, H.; Sinclair, D.F. Correcting for visibility bias in strip transect aerial surveys of aquatic fauna. J. Wildl. Manag. 1989, 53, 1017–1024. [Google Scholar] [CrossRef]
- Ellis, A.M. An Assessment of Density Estimation Methods for Forest Ungulates. Ph.D. Thesis, Rhodes University, Makhanda, South Africa, 2003. [Google Scholar]
- Stenson, G.; Myers, R. Accuracy of pup classifications and its effect on population estimates in the hooded seal (Cystophora cristata). Can. J. Fish. Aquat. Sci. 1988, 45, 715–719. [Google Scholar] [CrossRef]
- Bowen, W.; Myers, R.; Hay, K. Abundance estimation of a dispersed, dynamic population: Hooded seals (Cystophora cristata) in the Northwest Atlantic. Can. J. Fish. Aquat. Sci. 1987, 44, 282–295. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R.; Laake, J.L. Estimation of density from line transect sampling of biological populations. Wildl. Monogr. 1980, 72, 3–202. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. The need for distance data in transect counts. J. Wildl. Manag. 1984, 48, 1248–1254. [Google Scholar] [CrossRef]
- Bailey, R.; Putman, R. Estimation of fallow deer (Dama dama) populations from faecal accumulation. J. Appl. Ecol. 1981, 18, 697–702. [Google Scholar] [CrossRef]
- Mulligan, T.; Kieser, R. Comparison of acoustic population estimates of salmon in a lake with a weir count. Can. J. Fish. Aquat. Sci. 1986, 43, 1373–1385. [Google Scholar] [CrossRef]
- Kieser, R.; Mulligan, T.; Williamson, N.; Nelson, M. Intercalibration of two echo integration systems based on acoustic backscattering measurements. Can. J. Fish. Aquat. Sci. 1987, 44, 562–572. [Google Scholar] [CrossRef]
- Rudstam, L.G.; Clay, C.S.; Magnuson, J.J. Density and size estimates of cisco (Coregonus artedii) using analysis of echo peak PDF from a single-transducer sonar. Can. J. Fish. Aquat. Sci. 1987, 44, 811–821. [Google Scholar] [CrossRef]
- Do, M. Minimising errors in estimating fish population and biomass densities using the ‘acoustic volume backscattering strength’method. N. Z. J. Mar. Freshw. Res. 1987, 21, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Burczynski, J.J.; Johnson, R.L. Application of dual-beam acoustic survey techniques to limnetic populations of juvenile sockeye salmon (Oncorhynchus nerka). Can. J. Fish. Aquat. Sci. 1986, 43, 1776–1788. [Google Scholar] [CrossRef]
- Buerkle, U. Estimation of fish length from acoustic target strengths. Can. J. Fish. Aquat. Sci. 1987, 44, 1782–1785. [Google Scholar] [CrossRef]
- Thomas, G.; Jackson, D.R. Acoustic measurement of fish schools using array phase information. Can. J. Fish. Aquat. Sci. 1987, 44, 1544–1550. [Google Scholar] [CrossRef]
- Nero, R.; Magnuson, J. Characterization of patches along transects using high-resolution 70-kHz integrated acoustic data. Can. J. Fish. Aquat. Sci. 1989, 46, 2056–2064. [Google Scholar] [CrossRef]
- Marques, T.A.; Thomas, L.; Martin, S.W.; Mellinger, D.K.; Ward, J.A.; Moretti, D.J.; Harris, D.; Tyack, P.L. Estimating animal population density using passive acoustics. Biol. Rev. 2013, 88, 287–309. [Google Scholar] [CrossRef] [Green Version]
- Seber, G.A. A review of estimating animal abundance II. Int. Stat. Rev. Int. Stat. 1992, 60, 129–166. [Google Scholar] [CrossRef]
- Keiter, D.A.; Davis, A.J.; Rhodes, O.E.; Cunningham, F.L.; Kilgo, J.C.; Pepin, K.M.; Beasley, J.C. Effects of scale of movement, detection probability, and true population density on common methods of estimating population density. Sci. Rep. 2017, 7, 9446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laplace, P.S. Sur les naissances, les mariages et les mortsa Paris depuis 1771 jusqu’a 1784 et dans toute l’étendue de la France, pendant les années 1781 et 1782. Mem. Acad. Des Sci. Present. Var. Sch. 1786, 35–46. [Google Scholar]
- Lincoln, F.C. Calculating Waterfowl Abundance on the Basis of Banding Returns; Number 118; US Department of Agriculture: Washington, DC, USA, 1930. [Google Scholar]
- White, G.C. Capture-Recapture and Removal Methods for Sampling Closed Populations; Los Alamos National Laboratory: Los Alamos, NM, USA, 1982. [Google Scholar]
- Pollock, K.H. Review papers: Modeling capture, recapture, and removal statistics for estimation of demographic parameters for fish and wildlife populations: Past, present, and future. J. Am. Stat. Assoc. 1991, 86, 225–238. [Google Scholar] [CrossRef]
- Jolly, G.M. Explicit estimates from capture-recapture data with both death and immigration-stochastic model. Biometrika 1965, 52, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Seber, G.A. A note on the multiple-recapture census. Biometrika 1965, 52, 249–259. [Google Scholar] [CrossRef]
- Seber, G.A.F. The estimation of Animal Abundance and Related Parameters; Blackburn Press: Caldwell, NJ, USA, 1982. [Google Scholar]
- Parmenter, R.R.; Yates, T.L.; Anderson, D.R.; Burnham, K.P.; Dunnum, J.L.; Franklin, A.B.; Friggens, M.T.; Lubow, B.C.; Miller, M.; Olson, G.S.; et al. Small-mammal density estimation: A field comparison of grid-based vs. web-based density estimators. Ecol. Monogr. 2003, 73, 1–26. [Google Scholar] [CrossRef]
- Buckland, S. A mark-recapture survival analysis. J. Anim. Ecol. 1982, 51, 833–847. [Google Scholar] [CrossRef]
- Janečka, J.E.; Munkhtsog, B.; Jackson, R.M.; Naranbaatar, G.; Mallon, D.P.; Murphy, W.J. Comparison of noninvasive genetic and camera-trapping techniques for surveying snow leopards. J. Mammal. 2011, 92, 771–783. [Google Scholar] [CrossRef] [Green Version]
- Burgar, J.M.; Stewart, F.E.; Volpe, J.P.; Fisher, J.T.; Burton, A.C. Estimating density for species conservation: Comparing camera trap spatial count models to genetic spatial capture-recapture models. Glob. Ecol. Conserv. 2018, 15, e00411. [Google Scholar] [CrossRef]
- Davis, A.J.; Keiter, D.A.; Kierepka, E.M.; Slootmaker, C.; Piaggio, A.J.; Beasley, J.C.; Pepin, K.M. A comparison of cost and quality of three methods for estimating density for wild pig (Sus scrofa). Sci. Rep. 2020, 10, 2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCallum, J. Changing use of camera traps in mammalian field research: Habitats, taxa and study types. Mammal Rev. 2013, 43, 196–206. [Google Scholar] [CrossRef]
- Royle, J.A.; Young, K.V. A hierarchical model for spatial capture—Recapture data. Ecology 2008, 89, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Efford, M.G.; Dawson, D.K.; Borchers, D.L. Population density estimated from locations of individuals on a passive detector array. Ecology 2009, 90, 2676–2682. [Google Scholar] [CrossRef]
- Burton, A.C.; Neilson, E.; Moreira, D.; Ladle, A.; Steenweg, R.; Fisher, J.T.; Bayne, E.; Boutin, S. Wildlife camera trapping: A review and recommendations for linking surveys to ecological processes. J. Appl. Ecol. 2015, 52, 675–685. [Google Scholar] [CrossRef]
- Rowcliffe, J.M.; Field, J.; Turvey, S.T.; Carbone, C. Estimating animal density using camera traps without the need for individual recognition. J. Appl. Ecol. 2008, 45, 1228–1236. [Google Scholar] [CrossRef]
- Rowcliffe, J.M.; Kays, R.; Kranstauber, B.; Carbone, C.; Jansen, P.A. Quantifying levels of animal activity using camera trap data. Methods Ecol. Evol. 2014, 5, 1170–1179. [Google Scholar] [CrossRef] [Green Version]
- Anile, S.; Ragni, B.; Randi, E.; Mattucci, F.; Rovero, F. Wildcat population density on the E tna volcano, I taly: A comparison of density estimation methods. J. Zool. 2014, 293, 252–261. [Google Scholar] [CrossRef]
- Balestrieri, A.; Ruiz-González, A.; Vergara, M.; Capelli, E.; Tirozzi, P.; Alfino, S.; Minuti, G.; Prigioni, C.; Saino, N. Pine marten density in lowland riparian woods: A test of the Random Encounter Model based on genetic data. Mamm. Biol. 2016, 81, 439–446. [Google Scholar] [CrossRef]
- Caravaggi, A.; Zaccaroni, M.; Riga, F.; Schai-Braun, S.C.; Dick, J.T.; Montgomery, W.I.; Reid, N. An invasive-native mammalian species replacement process captured by camera trap survey random encounter models. Remote Sens. Ecol. Conserv. 2016, 2, 45–58. [Google Scholar] [CrossRef] [Green Version]
- Carbajal-Borges, J.P.; Godínez-Gómez, O.; Mendoza, E. Density, abundance and activity patterns of the endangered Tapirus bairdii in one of its last strongholds in southern Mexico. Trop. Conserv. Sci. 2014, 7, 100–114. [Google Scholar] [CrossRef] [Green Version]
- Manzo, E.; Bartolommei, P.; Rowcliffe, J.M.; Cozzolino, R. Estimation of population density of European pine marten in central Italy using camera trapping. Acta Theriol. 2012, 57, 165–172. [Google Scholar] [CrossRef]
- Cusack, J.J.; Swanson, A.; Coulson, T.; Packer, C.; Carbone, C.; Dickman, A.J.; Kosmala, M.; Lintott, C.; Rowcliffe, J.M. Applying a random encounter model to estimate lion density from camera traps in Serengeti National Park, Tanzania. J. Wildl. Manag. 2015, 79, 1014–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rademaker, M.; Meijaard, E.; Semiadi, G.; Blokland, S.; Neilson, E.W.; Rode-Margono, E.J. First ecological study of the Bawean warty pig (Sus blouchi), one of the rarest pigs on earth. PLoS ONE 2016, 11, e0151732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovero, F.; Marshall, A.R. Camera trapping photographic rate as an index of density in forest ungulates. J. Appl. Ecol. 2009, 46, 1011–1017. [Google Scholar] [CrossRef] [Green Version]
- Zero, V.H.; Sundaresan, S.R.; O’Brien, T.G.; Kinnaird, M.F. Monitoring an endangered savannah ungulate, Grevy’s zebra Equus grevyi: Choosing a method for estimating population densities. Oryx 2013, 47, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, Y.; Fukasawa, K.; Samejima, H. Estimating animal density without individual recognition using information derivable exclusively from camera traps. J. Appl. Ecol. 2018, 55, 735–744. [Google Scholar] [CrossRef]
- Nakashima, Y. Potentiality and limitations of N-mixture and Royle-Nichols models to estimate animal abundance based on noninstantaneous point surveys. Popul. Ecol. 2020, 62, 151–157. [Google Scholar] [CrossRef]
- consortium, E.; Grignolio, S.; Apollonio, M.; Brivio, F.; Vicente, J.; Acevedo, P.; Petrovic, K.; Keuling, O. Guidance on estimation of abundance and density data of wild ruminant population: Methods, challenges, possibilities. EFSA Support. Publ. 2020, 17, 1876E. [Google Scholar]
- Schwarz, C.J.; Seber, G.A. Estimating animal abundance: Review III. Stat. Sci. 1999, 14, 427–456. [Google Scholar] [CrossRef]
- Vargas Soto, J.S.; Castañeda, R.A.; Mandrak, N.E.; Molnár, P.K. Estimating animal density in three dimensions using capture-frequency data from remote detectors. Remote Sens. Ecol. Conserv. 2021, 7, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Bell, M.; Eaton, D.; Bannister, R.; Addison, J. A mark-recapture approach to estimating population density from continuous trapping data: Application to edible crabs, Cancer pagurus, on the east coast of England. Fish. Res. 2003, 65, 361–378. [Google Scholar] [CrossRef]
- Millikin, M.R. Synopsis of Biological Data on the Blue Crab, Callinectes sapidus Rathbun; Number 138; National Oceanic and Atmospheric Administration, National Marine Fisheries: Washington, DC, USA, 1984. [Google Scholar]
- Nehring, S. Invasion history and success of the American blue crab Callinectes sapidus in European and adjacent waters. In the Wrong Place-Alien Marine Crustaceans: Distribution, Biology and Impacts; Springer: Berlin/Heidelberg, Germany, 2011; pp. 607–624. [Google Scholar]
- Williams, A.B. The swimming crabs of the genus Callinectes (Decapoda: Portunidae). Fish. Bull. 1971, 72, 685. [Google Scholar]
- Hines, A.; Kennedy, V.; Cronin, L. The Blue Crab Callinectes sapidus; MD Sea Grant College Press: College Park, MD, USA, 2007. [Google Scholar]
- Perkins-Visser, E.; Wolcott, T.G.; Wolcott, D.L. Nursery role of seagrass beds: Enhanced growth of juvenile blue crabs (Callinectes sapidus Rathbun). J. Exp. Mar. Biol. Ecol. 1996, 198, 155–173. [Google Scholar] [CrossRef]
- Mancinelli, G.; Chainho, P.; Cilenti, L.; Falco, S.; Kapiris, K.; Katselis, G.; Ribeiro, F. The Atlantic blue crab Callinectes sapidus in southern European coastal waters: Distribution, impact and prospective invasion management strategies. Mar. Pollut. Bull. 2017, 119, 5–11. [Google Scholar] [CrossRef]
- BouvıER, E. Sur un Callinectes sapidus M. Rathbun trouvé à Rocheford. Bull. Mus. Hist. Naf, Paris 1901, 7, 66. [Google Scholar]
- Enzenro, R.; Enzenro, L.; Bİngel, F. Occurrence of blue crab, Callinectes sapidus (Rathbun, 1896) (Crustacea, Brachyura) on the Turkish Mediterranean and the adjacent Aegean coast and its size distribution in the bay of Iskenderun. Turk. J. Zool. 1997, 21, 113–122. [Google Scholar] [CrossRef]
- Labrune, C.; Amilhat, E.; Amouroux, J.M.; Coraline, J.; Alexandra, G.; Noël, P.Y. The arrival of the American blue crab, Callinectes sapidus Rathbun, 1896 (Decapoda: Brachyura: Portunidae), in the Gulf of lions (Mediterranean Sea). Bioinvasions Rec. 2019, 8, 876–881. [Google Scholar] [CrossRef]
- Massé, C.; Viard, F.; Humbert, S.; Antajan, E.; Auby, I.; Bachelet, G.; Bernard, G.; Bouchet, V.M.; Burel, T.; Dauvin, J.C.; et al. An overview of marine non-indigenous species found in three contrasting biogeographic metropolitan French regions: Insights on distribution, origins and pathways of introduction. Diversity 2023, 15, 161. [Google Scholar] [CrossRef]
- Kapiris, K.; Apostolidis, C.; Baldacconi, R.; Başusta, N.; Bilecenoglu, M.; Bitar, G.; Bobori, D.; Boyaci, Y.Ö.; Dimitriadis, C.; Djurović, M.; et al. New Mediterranean Biodiversity Records (April, 2014). Mediterr. Mar. Sci. 2014, 15, 198–212. [Google Scholar] [CrossRef] [Green Version]
- Bilecenoglu, M.; Alfaya, J.E.; Azzurro, E.; Baldacconi, R.; Boyaci, Y.; Circosta, V.; Compagno, L.; Coppola, F.; Deidun, A.; Durgham, H.; et al. New Mediterranean marine biodiversity records (December, 2013). Mediterr. Mar. Sci. 2013, 14, 463–480. [Google Scholar] [CrossRef] [Green Version]
- Perdikaris, C.; Konstantinidis, E.; Gouva, E.; Ergolavou, A.; Klaoudatos, D.; Nathanailides, C.; Paschos, I. Occurrence of the invasive crab species Callinectes sapidus Rathbun, 1896, in NW Greece. Walailak J. Sci. Technol. (WJST) 2016, 13, 503–510. [Google Scholar]
- Carrozzo, L.; Potenza, L.; Carlino, P.; Costantini, M.L.; Rossi, L.; Mancinelli, G. Seasonal abundance and trophic position of the Atlantic blue crab Callinectes sapidus Rathbun 1896 in a Mediterranean coastal habitat. Rend. Lincei 2014, 25, 201–208. [Google Scholar] [CrossRef]
- Codling, E.A.; Plank, M.J.; Benhamou, S. Random walk models in biology. J. R. Soc. Interface 2008, 5, 813–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimm, V.; Berger, U.; Bastiansen, F.; Eliassen, S.; Ginot, V.; Giske, J.; Goss-Custard, J.; Grand, T.; Heinz, S.K.; Huse, G.; et al. A standard protocol for describing individual-based and agent-based models. Ecol. Model. 2006, 198, 115–126. [Google Scholar] [CrossRef]
- Grimm, V.; Berger, U.; DeAngelis, D.L.; Polhill, J.G.; Giske, J.; Railsback, S.F. The ODD protocol: A review and first update. Ecol. Model. 2010, 221, 2760–2768. [Google Scholar] [CrossRef] [Green Version]
- Simantiris, N.; Theocharis, A.; Avlonitis, M. Environmental effects on zooplankton dynamics of a shallow Mediterranean coastal lagoon (October 2020–March 2021). Reg. Stud. Mar. Sci. 2021, 48, 102001. [Google Scholar] [CrossRef]
- Simantiris, N.; Avlonitis, M. Effects of future climate conditions on the zooplankton of a Mediterranean coastal lagoon. Estuarine, Coast. Shelf Sci. 2023, 282, 108231. [Google Scholar] [CrossRef]
- Hines, A.H.; Lipcius, R.N.; Haddon, A.M. Population dynamics and habitat partitioning by size, sex, and molt stage of blue crabs Callinectes sapidus in a subestuary of central Chesapeake Bay. Mar. Ecol. Prog. Ser. 1987, 36, 55–64. [Google Scholar] [CrossRef]
- Cai, A.Q.; Landman, K.A.; Hughes, B.D. Modelling directional guidance and motility regulation in cell migration. Bull. Math. Biol. 2006, 68, 25–52. [Google Scholar] [CrossRef]
- Vuilleumier, S.; Metzger, R. Animal dispersal modelling: Handling landscape features and related animal choices. Ecol. Model. 2006, 190, 159–170. [Google Scholar] [CrossRef] [Green Version]
- Kara, M.H.; Chaoui, L. Strong invasion of Mellah lagoon (South-Western Mediterranean) by the American blue crab Callinectes sapidus Rathbun, 1896. Mar. Pollut. Bull. 2021, 164, 112089. [Google Scholar] [CrossRef] [PubMed]
- Leffler, C. Some effects of temperature on the growth and metabolic rate of juvenile blue crabs, Callinectes sapidus, in the laboratory. Mar. Biol. 1972, 14, 104–110. [Google Scholar] [CrossRef]
- Bauer, L.J.; Miller, T.J. Temperature-, salinity-, and size-dependent winter mortality of juvenile blue crabs (Callinectes sapidus). Estuaries Coasts 2010, 33, 668–677. [Google Scholar] [CrossRef]
- Jakov, D.; Glamuzina, B. Six years from first record to population establishment: The case of the blue crab, Callinectes sapidus Rathbun, 1896 (Brachyura, Portunidae) in the Neretva River delta (South-eastern Adriatic Sea, Croatia). Crustaceana 2011, 84, 1211–1220. [Google Scholar]
- Couch, J.N. A new fungus on crab eggs. J. Elisha Mitchell Sci. Soc. 1942, 58, 158–162. [Google Scholar]
- Van Engel, W.A. The blue crab and its fishery in Chesapeake Bay. Part 1. Reproduction, early development, growth and migration. Commer. Fish. Rev. 1958, 20, 6. [Google Scholar]
- Lampert, W. The adaptive significance of diel vertical migration of zooplankton. Funct. Ecol. 1989, 3, 21–27. [Google Scholar] [CrossRef]
- Buchanan, B.A.; Stoner, A.W. Distributional patterns of blue crabs (Callinectes sp.) in a tropical estuarine lagoon. Estuaries 1988, 11, 231–239. [Google Scholar] [CrossRef]
- Hines, A.H.; Jivoff, P.R.; Bushmann, P.J.; van Montfrans, J.; Reed, S.A.; Wolcott, D.L.; Wolcott, T.G. Evidence for sperm limitation in the blue crab, Callinectes sapidus. Bull. Mar. Sci. 2003, 72, 287–310. [Google Scholar]
- Dittel, A.I.; Hines, A.H.; Ruiz, G.M.; Ruffin, K.K. Effects of shallow water refuge on behavior and density-dependent mortality of juvenile blue crabs in Chesapeake Bay. Bull. Mar. Sci. 1995, 57, 902–916. [Google Scholar]
- Orth, R.J.; VANMONTFRANS, J. Utilization of a seagrass meadow and tidal marsh creek by blue crabs Callinectes sapidus. I. Seasonal and annual variations in abundance with emphasis on post-settlement juveniles. Mar. Ecol. Prog. Ser. 1987, 41, 283. [Google Scholar] [CrossRef]
- Hovel, K.A.; Lipcius, R.N. Habitat fragmentation in a seagrass landscape: Patch size and complexity control blue crab survival. Ecology 2001, 82, 1814–1829. [Google Scholar] [CrossRef]
- Hovel, K.A.; Fonseca, M.S. Influence of seagrass landscape structure on the juvenile blue crab habitat-survival function. Mar. Ecol. Prog. Ser. 2005, 300, 179–191. [Google Scholar] [CrossRef]
- Beck, M.W.; Heck, K.L.; Able, K.W.; Childers, D.L.; Eggleston, D.B.; Gillanders, B.M.; Halpern, B.; Hays, C.G.; Hoshino, K.; Minello, T.J.; et al. The identification, conservation, and management of estuarine and marine nurseries for fish and invertebrates: A better understanding of the habitats that serve as nurseries for marine species and the factors that create site-specific variability in nursery quality will improve conservation and management of these areas. Bioscience 2001, 51, 633–641. [Google Scholar]
- Seitz, R.D.; Knick, K.E.; Westphal, M. Diet selectivity of juvenile blue crabs (Callinectes sapidus) in Chesapeake Bay. Integr. Comp. Biol. 2011, 51, 598–607. [Google Scholar] [CrossRef] [Green Version]
- Litvin, S.Y.; Weinstein, M.P.; Sheaves, M.; Nagelkerken, I. What makes nearshore habitats nurseries for nekton? An emerging view of the nursery role hypothesis. Estuaries Coasts 2018, 41, 1539–1550. [Google Scholar] [CrossRef]
- Epifanio, C.E.; Dittel, A.; Rodriguez, R.A.; Targett, T.E. The role of macroalgal beds as nursery habitat for juvenile blue crabs, Callinectes sapidus. J. Shellfish Res. 2003, 22, 881–886. [Google Scholar]
- Ruiz, G.M.; Hines, A.H.; Posey, M.H. Shallow water as a refuge habitat for fish and crustaceans in non-vegetated estuaries: An example from Chesapeake Bay. Mar. Ecol. Prog. Ser. 1993, 99, 1–16. [Google Scholar] [CrossRef]
- Hines, A.H.; Ruiz, G.M. Temporal variation in juvenile blue crab mortality: Nearshore shallows and cannibalism in Chesapeake Bay. Bull. Mar. Sci. 1995, 57, 884–901. [Google Scholar]
- Rogers-Talbert, R. The fungus Lagenidium callinectes Couch (1942) on eggs of the blue crab in Chesapeake Bay. Biol. Bull. 1948, 95, 214–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shields, J.D. Research priorities for diseases of the blue crab Callinectes sapidus. Bull. Mar. Sci. 2003, 72, 505. [Google Scholar]
- Pearson, W.H.; Olla, B.L. Chemoreception in the blue crab, Callinectes sapidus. Biol. Bull. 1977, 153, 346–354. [Google Scholar] [CrossRef]
- Devine, D.V.; Atema, J. Function of chemoreceptor organs in spatial orientation of the lobster, Homarus americanus: Differences and overlap. Biol. Bull. 1982, 163, 144–153. [Google Scholar] [CrossRef]
- Moore, P.A.; Scholz, N.; Atema, J. Chemical orientation of lobsters, Homarus americanus, in turbulent odor plumes. J. Chem. Ecol. 1991, 17, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Derby, C.D.; Atema, J. The function of chemo-and mechanoreceptors in lobster (Homarus americanus) feeding behaviour. J. Exp. Biol. 1982, 98, 317–327. [Google Scholar] [CrossRef]
- Hayden, D.; Jennings, A.; Müller, C.; Pascoe, D.; Bublitz, R.; Webb, H.; Breithaupt, T.; Watkins, L.; Hardege, J. Sex-specific mediation of foraging in the shore crab, Carcinus maenas. Horm. Behav. 2007, 52, 162–168. [Google Scholar] [CrossRef]
- Hardege, J.; Bartels-Hardege, H.; Fletcher, N.; Terschak, J.; Harley, M.; Smith, M.; Davidson, L.; Hayden, D.; Müller, C.T.; Lorch, M.; et al. Identification of a female sex pheromone in Carcinus maenas. Mar. Ecol. Prog. Ser. 2011, 436, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Draper, A.M.; Weissburg, M.J. Impacts of global warming and elevated CO2 on sensory behavior in predator-prey interactions: A review and synthesis. Front. Ecol. Evol. 2019, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Richardson, B.; Martin, H.; Bartels-Hardege, H.; Fletcher, N.; Hardege, J.D. The role of changing pH on olfactory success of predator–prey interactions in green shore crabs, Carcinus maenas. Aquat. Ecol. 2022, 56, 409–418. [Google Scholar] [CrossRef]
- Page, J.L.; Dickman, B.D.; Webster, D.R.; Weissburg, M.J. Getting ahead: Context-dependent responses to odorant filaments drive along-stream progress during odor tracking in blue crabs. J. Exp. Biol. 2011, 214, 1498–1512. [Google Scholar] [CrossRef] [Green Version]
- Aggio, J.F.; Tieu, R.; Wei, A.; Derby, C.D. Oesophageal chemoreceptors of blue crabs, Callinectes sapidus, sense chemical deterrents and can block ingestion of food. J. Exp. Biol. 2012, 215, 1700–1710. [Google Scholar] [CrossRef] [Green Version]
- Aggio, J.F.; Derby, C.D. Hydrogen peroxide and other components in the ink of sea hares are chemical defenses against predatory spiny lobsters acting through non-antennular chemoreceptors. J. Exp. Mar. Biol. Ecol. 2008, 363, 28–34. [Google Scholar] [CrossRef]
- Kamio, M.; Grimes, T.V.; Hutchins, M.H.; van Dam, R.; Derby, C.D. The purple pigment aplysioviolin in sea hare ink deters predatory blue crabs through their chemical senses. Anim. Behav. 2010, 80, 89–100. [Google Scholar] [CrossRef]
- Ferner, M.C.; Smee, D.L.; Chang, Y.P. Cannibalistic crabs respond to the scent of injured conspecifics: Danger or dinner? Mar. Ecol. Prog. Ser. 2005, 300, 193–200. [Google Scholar] [CrossRef] [Green Version]
- Moir, F.; Weissburg, M. Cautious cannibals: Behavioral responses of juvenile and adult blue crabs to the odor of injured conspecifics. J. Exp. Mar. Biol. Ecol. 2009, 369, 87–92. [Google Scholar] [CrossRef]
- Grosholz, E.; Ashton, G.; Bradley, M.; Brown, C.; Ceballos-Osuna, L.; Chang, A.; de Rivera, C.; Gonzalez, J.; Heineke, M.; Marraffini, M.; et al. Stage-specific overcompensation, the hydra effect, and the failure to eradicate an invasive predator. Proc. Natl. Acad. Sci. USA 2021, 118, e2003955118. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, S.; Fu, C.; Gong, X.; Huang, L.; Song, X.; Zhao, Y. Molecular genetic structure and evolution in native and colonized populations of the Chinese mitten crab, Eriocheir sinensis. Biol. Invasions 2009, 11, 389–399. [Google Scholar] [CrossRef]
- Simantiris, N.; Vardaki, M.Z.; Koralli, P.; Chochos, C.L.; Gregoriou, V.G.; Kourkoumelis, N.; Avlonitis, M. Seasonal evaluation of floating microplastics in a shallow Mediterranean coastal lagoon: Abundance, distribution, chemical composition, and influence of environmental factors. Estuarine, Coast. Shelf Sci. 2022, 272, 107859. [Google Scholar] [CrossRef]
- Simantiris, N.; Avlonitis, M.; Theocharis, A. Simulation of the transport of marine microplastic particles in the Ionian Archipelago (NE Ionian Sea) using a Lagrangian model and the control mechanisms affecting their transport. J. Hazard. Mater. 2022, 437, 129349. [Google Scholar] [CrossRef]
- Simantiris, N.; Vardaki, M.Z.; Kourkoumelis, N.; Avlonitis, M.; Theocharis, A. Microplastics in the Mediterranean and Elsewhere in Coastal Seas; Elsevier: Amsterdam, The Netherlands, 2023; in press. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simantiris, N.; Violaris, I.G.; Avlonitis, M. Computing Invasive Species Population Based on a Generalized Random Walk Process: Application to Blue Crab (Callinectes sapidus). J. Mar. Sci. Eng. 2023, 11, 1282. https://doi.org/10.3390/jmse11071282
Simantiris N, Violaris IG, Avlonitis M. Computing Invasive Species Population Based on a Generalized Random Walk Process: Application to Blue Crab (Callinectes sapidus). Journal of Marine Science and Engineering. 2023; 11(7):1282. https://doi.org/10.3390/jmse11071282
Chicago/Turabian StyleSimantiris, Nikolaos, Ioannis G. Violaris, and Markos Avlonitis. 2023. "Computing Invasive Species Population Based on a Generalized Random Walk Process: Application to Blue Crab (Callinectes sapidus)" Journal of Marine Science and Engineering 11, no. 7: 1282. https://doi.org/10.3390/jmse11071282




