A Package of Script Codes, POSIBIOM for Vegetation Acoustics: POSIdonia BIOMass
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algorithm of ‘Lost’ Bottom (AbdezdeR1)
2.1.1. Real Bottom Recovery
- (i)
- “Chirping” relates to the Sv of the bottom echo having priority over the next two bottom correction methods. This method worked well in cases where there was a clear bottom echo in the underestimated bottom portion of the data.
- (ii)
- The “running average” would be automatically called on for by the processor when there was no clear (weak) bottom echo. This method worked by correcting the lost bottom between the previous and next corrected bottom depths and angles.
- (iii)
- The “fill gap” would then be used to correct the bottom. In some cases, when there was no bottom echo (a gap) due to the occurrence of surface or volume reverberations or intense fish shoals, the bottom echo would disappear. Before filling the gap, the method would check the depths and angles.
- (i)
- The data loaded from file to file were converted into an extended echogram of Sv on depth (Y-axis) vs. ping (X-axis);
- (ii)
- The bottom line estimated with the Visual Analyzer program was then plotted on the echogram;
- (iii)
- The bottom angle in degrees was calculated between pings (optional to use the horizontal distance depending on the ping rate, since the GPS transmitted the position every second), taking into account the bottom depth differences;
- (iv)
- The depth and angle differences in successive pings were plotted as a basic bottom estimation algorithm;
- (v)
- Taking into account the differences (default bottom angle > 80 degrees with tolerance together) in the algorithm, the misestimated bottom depth (green line) was plotted on the echogram;
- (vi)
- After all the settings and conditions (Table S1) were checked and entered in the menu and the run was started, the algorithm would estimate the real bottom depth within the horizontal differences in the single or block area, which was recognized as the “lost” bottom from the first ping;
- (vii)
- To find the real bottom, the three methods above were first ordered in “chirping”, “running average” and “fill gap”;
- (viii)
- The chirping had a range from −46 dB to −5 dB of the Sv by the depth per single ping or blocked (segregated) ping estimated as the “lost” bottom, taking into account the real bottom depth around the pings of the “lost” bottom by ranging the pings back and forth one by one;
- (ix)
- If there was no bottom echo, the program proceeded to the next method (running average); similar tracking by chirping to the correct bottom would be applied to this method by increasing the factor of the running average until the bottom was fixed for the lost bottom. If the range was too long and the difference in the estimated real bottom was highly variable for the bottom within the range, the method would pass to the last method (filling gap);
- (x)
- Similar to the previous lost bottom search methods, the filling gap was highly dependent on the range of the lost bottom and the range was kept very short. Overall, the priority, preference and frequency of the use of the methods decreased from the first to the last;
- (xi)
- There were auxiliary submethods and options controlled by the user to recover the real bottom among the “lost” bottoms (Table S1);
- (xii)
- After the real bottom was recovered for a ping, the algorithm was assured to process the next pings regardless of the previous pings corrected. In the menu, all settings reset by the user were interactive;
- (xiii)
- All files for the process were finished, followed by the estimation of the dead zone, and then the algorithm would call for the next data file.
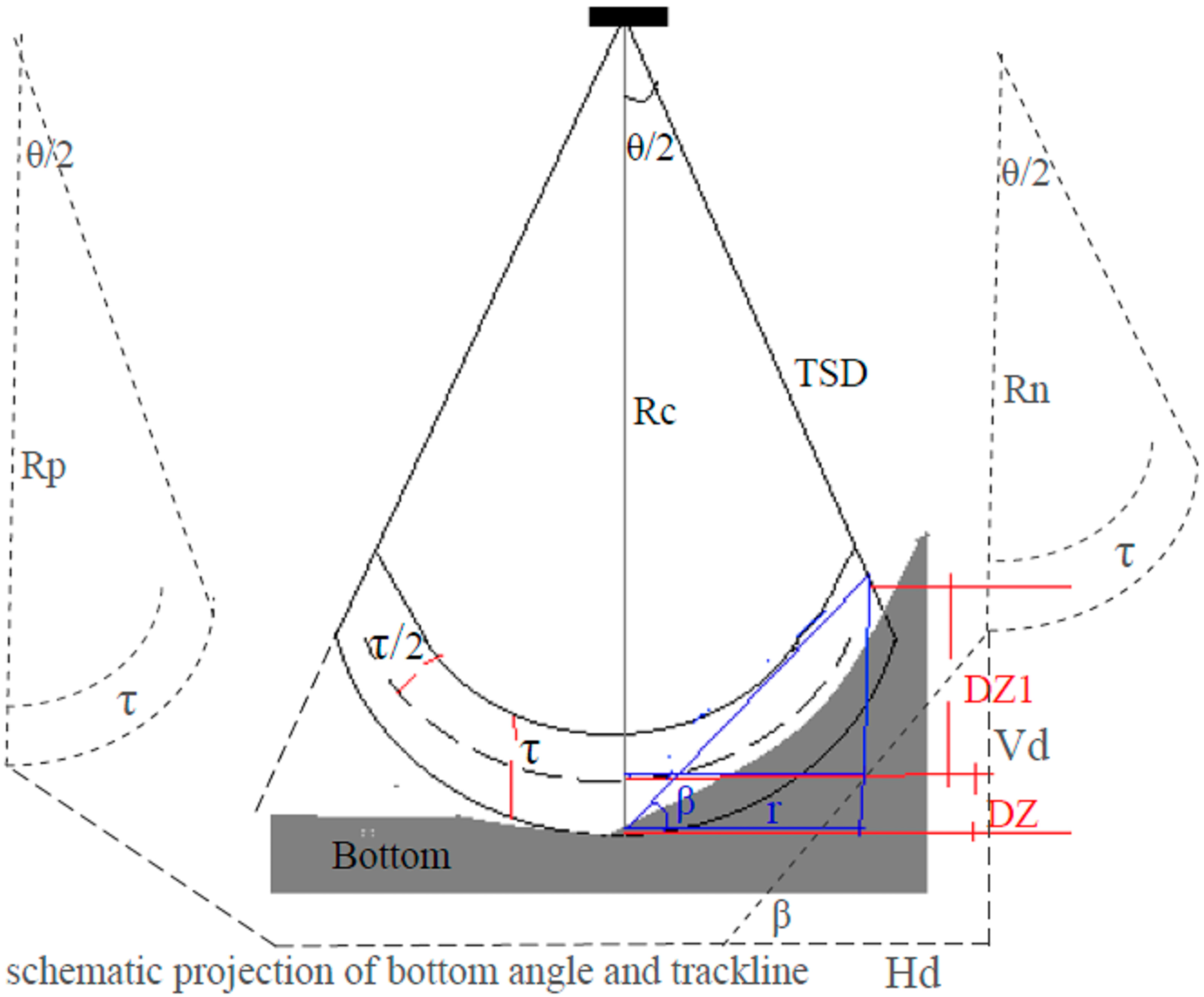
2.1.2. Dead Zone Estimates
2.2. Algorithm of Noise and Reverberation (AbemsiR1)
- (i)
- If MSNRb > ESNRb, discard the data;
- (ii)
- If MSNRir > ESNRir, remove the data;
- (iii)
- If MSNRir/MSNRb > an automatically estimated criterion value between the ratios for each acoustic data file, remove the data;
- (i)
- Considering the inherent occurrence of natural and artificial noises (noise, reverberation and interference), the solution of the described method was modified by referring to the measured noises as a base of the background threshold for each season;
- (ii)
- Both the measured noise and the study acoustic data were subjected to the described SNR method to estimate the unwanted noise through the water column;
- (iii)
- Noise was removed or replaced using the three cases above;
- (iv)
- Case iii would estimate a criterion threshold that was automatically calculated specifically for each acoustic data file containing different reverberations and interferences in terms of their distribution.
2.3. Algorithm of “SheathFinder” and Leaf Length and Biomass (Sheathfinder1 and 2)
- (i)
- The algorithm used the output file of “Noise Removal” as the input file;
- (ii)
- The algorithm setting to first remove weak and strong scatterers and then to fix the sheath + vertical rhizome (all data based on Sv) above the dead zone was visually calibrated by referring to a file containing a clear P. oceanica section;
- (iii)
- According to each setting above, the program first removed weak and then strong scatterers, and then scanned ping-to-ping data of the water column to the bottom to check the availability of sheaths + vertical rhizomes above the dead zone;
- (iv)
- After completing the scanning, the algorithm fixed real P. oceanica to estimate the leaf of each ping;
- (v)
- The leaf was estimated by checking the decreasing order of the Sv of the count-to-count (pixel) data from the sheaths or vertical rhizomes to the leaf tip, where the next few Sv increased (representing a school of fish) or were zero (free of a strong scatter);
- (vi)
- The pixels of the fixed ping of the leaf were checked for strong scatterers that broke a rule of a decreasing order of Sv along the leaf height;
- (vii)
- The strong scatterers were removed from the leaf;
- (viii)
- Sv was estimated by averaging all Sv of the leaf;
- (ix)
- Sa was calculated by summing the height-wise Sv of the leaf;
- (x)
- The leaf or canopy height was the distance from the top to the bottom of the leaf to the sheaths;
- (xi)
- The Sv and Sa of each ping were converted to absolute biomass (g/m2) using the seasonal (annual data) acoustic–biomass relationships estimated by Mutlu and Olguner ([60]).
3. Results
3.1. Flowchart of POSIBIOM
3.2. Lost Bottom and Dead Zone
3.3. Noise, Reverberation and Interference
3.4. Leaf and Biomass Estimation
3.5. Advantages of the Package
- (i)
- The package was simple and easy to use and was not a destructive method for protected seagrasses;
- (ii)
- Users with little knowledge of acoustics could use the package;
- (iii)
- The package solved one of the commonly important postprocessing problems (bottom depth correction) by correcting misestimated bottoms;
- (iv)
- The package identified spurious scatterers through the water column and bottom other than the targeted seagrass;
- (v)
- Removed unwanted noise, reverberations and interference using subcalls of the modified SNR;
- (vi)
- Removal of strong and weak scatterers among the seagrass;
- (vii)
- Most of the algorithms ran autonomously without user intervention;
- (viii)
- The package could distinguish P. oceanica without the need to sea-truth the scatterers;
- (ix)
- No further destructive sampling was required;
- (x)
- The package could separate the sheaths–vertical rhizomes from the leaves;
- (xi)
- The package worked well for the extraction of P. oceanica found on different substrata;
- (xii)
- The relative biomass was converted to the absolute biomass of the leaves, regarding the seasonal–EDSU–biomass relation by using different equations of the in/ex situ experiments;
- (xiii)
- As for many programs, the package plotted the distribution of the results (leaf biomass and canopy).
3.6. Disadvantages (Troubleshooting/Negatives) of the Package
- (i)
- The package was only associated with acoustic data collected with the BioSonics echosounder;
- (ii)
- The package could not read line acoustic data and required a postprocessor belonging to BioSonics;
- (iii)
- Bottom recovery was not possible in the case of strong volume reverberations, especially produced by SCUBA divers;
- (iv)
- A cyclic solution that took time to complete bottom correction could occur during a ping solution of an incorrectly estimated bottom;
- (v)
- The deletion of the first pings may be necessary if the first pings had no bottom echo or the first echo was incorrectly estimated with the postprocessor;
- (vi)
- Bottom recovery was achieved at 90–95%, which biased seagrass detection;
- (vii)
- The accuracy and precision of the leaf biomass and canopy height depended on the vertical resolution of the data obtained with the postprocessor, which took time to convert raw data to the CSV format;
- (viii)
- The package did not allow for measurements of the true leaf length, but only canopy height;
- (ix)
- Leaf biomass estimation depended only on the conversion equations performed for a bottom depth of 15 m, where the biometry of P. oceanica was highly variable compared to other depths, regardless of shallower or deeper depths;
- (x)
- Almost 7–8 equations [60] for the conversions were used to display the results in the “Results Table”, but three of them were saved in the output file;
- (xi)
- The package results were limited with estimates of biomass, height and canopy cover. Vertical rhizome + sheath length could be measured. Other biometrics (shoot density, number of leaves per shoot, leaf type, internode distance, leaf width, etc.) could not be estimated;
- (xii)
- False seagrass could be detected using the package, but was then eliminated due to the package;
- (xiii)
- Some large macrophytes (algae; Caulerpa prolifera) could be insignificantly misclassified as P. oceanica due to the occurrence of negligible cases when the bottom echo was not perfectly removed;
- (xiv)
- Effect of significant epiphytes on leaves could not be removed from the leaf echo.
3.7. Preliminary Demonstration of Results from Each Algorithm
4. Discussion
- (i)
- The algorithm should read acoustic data from echosounders produced by other companies in addition to BioSonics;
- (ii)
- A way could be developed to read the acoustic line data directly without the need to postprocess it first;
- (iii)
- The debugs of the package could be corrected as the feedback is performed by the users who would use the package in the C computer language;
- (iv)
- Adapt the algorithm to operate with multiple frequencies when the data are collected, and then the algorithm could be modified and configured accordingly;
- (v)
- The experiments should be repeated to establish the relationships between the EDSUs and the biometrics at different bottom depths other than 15 m;
- (vi)
- Leaf types should also be considered seasonally in the relationship equations;
- (vii)
- Mapping of the results should be considered to be integrated into different types of coastal maps;
- (viii)
- An algorithm of the bottom classifier (hard and soft substrates) could be added to the package through a method ratioing the first bottom echo to the second bottom echo using a proper statistical solution;
- (ix)
- Finally, the software of the package could be hardware connected to the echosounder, which currently requires too much work associated with the electronic integration.
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



| Info Variables | Description |
|---|---|
| Alpha | Absorption coefficient |
| Ping rate | Pulse rate per second |
| PW | Pulse width |
| C | Sound speed |
| Threshold | Minimum data collection threshold |
| Tot ping no * | Total number of pings |
| Strata * | Number of stratum (vertical resolution) |
| Report No * | Number of reporting data (horizontal resolution) |
| Beam width | Angle of main lobe of beam |


References
- Colantoni, P.; Gallignani, P.; Fresi, E.; Cinelli, F. Patterns of Posidonia oceanica (L.) DELILE Beds around the Island of Ischia (Gulf of Naples) and in adjacent waters. Pszni Mar. Ecol. 1982, 3, 53–74. [Google Scholar] [CrossRef]
- Brown, C.J.; Smith, S.J.; Lawton, P.; Anderson, J.T. Benthic habitat mapping: A review of progress towards improved understanding of the spatial ecology of the seafloor using acoustic techniques. Estuar. Coast. Shelf Sci. 2011, 92, 502–520. [Google Scholar] [CrossRef]
- Pal, D.; Hogland, W. An overview and assessment of the existing technological options for management and resource recovery from beach wrack and dredged sediments: An environmental and economic perspective. J. Environ. Manag. 2022, 302, 113971. [Google Scholar] [CrossRef]
- Boudouresque, C.F.; Meinesz, A. Découverte de l’herbier de Posidonies. Cahier Parc. Nation. Port-Cros. 1982, 4, 1–79. [Google Scholar]
- Mutlu, E.; Olguner, C.; Gökoğlu, M.; Özvarol, Y. Seasonal growth dynamics of Posidonia oceanica in a pristine Mediterranean gulf. Ocean. Sci. J. 2022, 57, 381–397. [Google Scholar] [CrossRef]
- Vacchi, M.; De Falco, G.; Simeone, S.; Montefalcone, M.; Morri, C.; Ferrar, M.; Bianchi, C.N. Biogeomorphology of the Mediterranean Posidonia oceanica seagrass meadows. Earth Surf. Proc. Land. 2017, 42, 42–54. [Google Scholar] [CrossRef]
- Spalding, M.; Taylor, M.; Ravilious, C.; Short, F.; Green, E. The distribution and status of seagrasses. In World Atlas of Seagrasses; Green, E.P., Short, F., Eds.; University California Press: Berkeley, CA, USA, 2003; pp. 5–26. [Google Scholar]
- Aires, T.; Marbà, N.; Cunha, R.L.; Kendrick, G.A.; Walker, D.I.; Serrão, E.A.; Duarte, C.M.; Arnaud-Haond, S. Evolutionary history of the seagrass genus Posidonia. Mar. Ecol. Prog. Ser. 2011, 421, 117–130. [Google Scholar] [CrossRef]
- Bianchi, C.N.; Morri, C.; Chiantore, M.; Montefalcone, M.; Parravicini, V.; Rovere, A. Mediterranean Sea biodiversity between the legacy from the past and a future of change. In Life in the Mediterranean Sea: A Look at Habitat Changes; Science, N., Ed.; Stambler N. Publishers: New York, NY, USA, 2012; pp. 1–55. [Google Scholar]
- Planton, S.; Lionello, P.; Artale, V.; Aznar, R.; Carrillo, A.; Colin, J.; Congedi, L.; Dubois, C.; Elizalde, A.; Gualdi, S.; et al. The climate of the Mediterranean region in future climate projections. In The Climate of the Mediterranean Region, from the Past to the Future; Lionello, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 449–502. [Google Scholar]
- Pergent, G.; Bazairi, H.; Bianchi, C.N.; Boudouresque, C.F.; Buia, M.C.; Calvo, S.; Clabaut, P.; Harmelin-Vivien, M.; Mateo, M.A.; Montefalcone, M.; et al. Climate change and Mediterranean seagrass meadows: A synopsis for environmental managers. Mediterr. Mar. Sci. 2014, 15, 462–473. [Google Scholar] [CrossRef]
- Bernier, P.; Guidi, J.-B.; Biittcher, M.E. Coastal progradation and very early diagenesis of ultramafic sands as a result of rubble discharge from asbestos excavations (northern Corsica, western Mediterranean). Mar. Geol. 1997, 144, 163–175. [Google Scholar] [CrossRef]
- Peirano, A.; Damasso, V.; Montefalcone, M.; Morri, C.; Bianchi, C.N. Effects of climate, invasive species and anthropogenic impacts on the growth of the seagrass Posidonia oceanica (L.) Delile in Liguria (NW Mediterranean Sea). Mar. Pollut. Bull. 2005, 50, 817–822. [Google Scholar] [CrossRef]
- de Mendoza, F.P.; Fontolan, G.; Mancini, E.; Scanu, E.; Scanu, S.; Bonamano, S.; Marcelli, M. Sediment dynamics and resuspension processes in a shallow-water Posidonia oceanica meadow. Mar. Geol. 2018, 404, 174–186. [Google Scholar] [CrossRef]
- Bonamano, S.; Piazzolla, D.; Scanu, S.; Mancini, E.; Madonia, A.; Piermattei, V.; Marcell, M. Modelling approach for the evaluation of burial and erosion processes on Posidonia oceanica meadows. Estuar. Coast. Shelf Sci. 2021, 254, 107321. [Google Scholar] [CrossRef]
- Mateo-Ramírez, Á.; Marina, P.; Martín-Arjona, A.; Bañares-España, E.; García Raso, J.E.; Rueda, J.L.; Urra, J. Posidonia oceanica (L.) Delile at its westernmost biogeographical limit (northwestern Alboran Sea): Meadow features and plant phenology. Oceans 2023, 4, 27–48. [Google Scholar] [CrossRef]
- Mutlu, E.; Olguner, C.; Özvarol, Y.; Gökoğlu, M. Spatiotemporalbiometrics of Cymodocea nodosa in a western Turkish Mediterranean coast. Biologia 2022, 77, 649–670. [Google Scholar] [CrossRef]
- Mutlu, E.; Duman, G.S.; Karaca, D.; Özvarol, Y.; Şahin, A. Biometrical Variation of Posidonia oceanica with different bottom types along the entire Turkish Mediterranean coast. Ocean Sci. J. 2023, 58, 9. [Google Scholar] [CrossRef]
- Lawton, J.H. What do species do in ecosystems? Oikos 1994, 71, 367–374. [Google Scholar] [CrossRef]
- Ingrosso, G.; Abbiati, M.; Badalamenti, F.; Bavestrello, G.; Belmonte, G.; Cannas, R.; Benedetti-Cecchi, L.; Bertolino, M.; Bevilacqua, S.; Bianchi, C.N.; et al. Mediterranean Bioconstructions Along the Italian Coast. Adv. Mar. Biol. 2018, 79, 61–136. [Google Scholar]
- Edgar, G.J.; Shaw, C. The production and trophic ecology of shallow-water fish assemblages in southern Australia. 3. General relationships between sediments, seagrasses, invertebrates and fishes. J. Exp. Mar. Biol. Ecol. 1995, 194, 107–131. [Google Scholar] [CrossRef]
- Buia, M.C.; Gambi, M.C.; Zupo, V. Structure and functioning of Mediterranean seagrass ecosystems: An overview. Biol. Mar. Mediterr. 2000, 7, 167–190. [Google Scholar]
- Mazzella, L.; Scipione, M.B.; Buia, M.C. Spatio-temporal distribution of algal and animal communities in a Posidonia oceanica (L.) Delile meadow. Mar. Ecol. 1989, 10, 107–129. [Google Scholar] [CrossRef]
- Francour, P. Fish assemblages of Posidonia oceanica beds at Port-Cros (France, NW Mediterranean): Assessment of composition and long-term fluctuations by visual census. Mar. Ecol. 1997, 18, 157–173. [Google Scholar] [CrossRef]
- Dauby, P.; Bale, A.J.; Bloomer, N.; Canon, C.; Ling, R.D.; Norro, A.; Robertson, J.E.; Simon, A.; Théate, J.M.; Watson, A.J.; et al. Particle fluxes over a Mediterranean seagrass bed: A one-year sediment trap experiment. Mar. Ecol. Prog. Ser. 1995, 126, 233–246. [Google Scholar] [CrossRef]
- Mateo, M.A.; Sanchez-Lizaso, J.L.; Romero, J. Posidonia oceanica “banquettes”: A preliminary assessment for an ecosystem carbon and nutrient budget. Mar. Ecol. Prog. Ser. 2003, 151, 43–45. [Google Scholar] [CrossRef]
- Guala, I.; Simeone, S.; Buia, M.C.; Flagella, S.; Baroli, M.; De Falco, G. Posidonia oceanica ‘banquette’ removal: Environmental impact and management implications. Biol. Mar. Mediterr. 2006, 13, 149–153. [Google Scholar]
- Fonseca, M.S.; Koehl, M.A.R.; Kopp, B.S. Biomechanical factors contributing to self-organization in seagrass landscapes. J. Exp. Mar. Biol. Ecol. 2007, 340, 227–246. [Google Scholar] [CrossRef]
- Den Hartog, C. Structure, function, and classification in seagrass communities. In Seagrass Ecosystems: A Scientific Perspective; McRoy, C.P., Helfferich, C., Eds.; Marcel Dekker: New York, NY, USA, 1977; pp. 89–121. [Google Scholar]
- Catucci, E.; Scardi, M. Modeling Posidonia oceanica shoot density and rhizome primary production. Sci. Rep. 2020, 10, 16978. [Google Scholar] [CrossRef] [PubMed]
- Marba, N.; Duarte, C.M.; Holmer, M.; Martinez, R.; Basterretxea, G.; Orfila, A.; Jordi, A.; Tintore, J. Effectiveness of protection of seagrass (Posidonia oceanica) populations in Cabrera national park (Spain). Environ. Conserv. 2002, 29, 509–518. [Google Scholar] [CrossRef]
- Orth, R.; Carruthers, T.; Dennison, W.; Duarte, C.; Fourqurean, J.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Gobert, S.; Sartoretto, S.; Rico-Raimondino, V.; Andral, B.; Chery, A.; Lejeune, P.; Boissery, P. Assessment of the ecological status of Mediterranean French coastal waters as required by the water framework directive using the Posidonia oceanica rapid easy index: PREI. Mar. Pollut. Bull. 2009, 58, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Pergent, G.; Pergent-Martini, C.; Boudouresque, C.F. Utilisation de l’herbier a Posidonia oceanica comme indicateur biologique de la qualite du milieu littoral en Mediterranee: Etat des connaissances. Mesogee 1995, 54, 3–27. [Google Scholar]
- Mutlu, E.; Karaca, D.; Duman, G.S.; Şahin, A.; Özvarol, Y.; Olguner, C. Seasonality and phenology of an epiphytic calcareous red alga, Hydrolithon boreale, on the leaves of Posidonia oceanica (L) Delile in the Turkish water. Environ. Sci. Pollut. Res. 2023, 30, 17193–17213. [Google Scholar] [CrossRef]
- Gobert, S.; Lefebvre, L.; Boissery, P.; Richir, J. A non-destructive method to assess the status of Posidonia oceanica meadows. Ecol. Indicat. 2020, 119, 106838. [Google Scholar] [CrossRef]
- McGonigle, C.; Grabowski, J.H.; Brown, C.J.; Weber, T.C.; Quinn, R. Detection of deep water benthic macroalgae using image-based classification techniques on multibeam backscatter at Cashes Ledge, Gulf of Maine, USA. Estuar. Coast. Shelf Sci. 2011, 91, 87–101. [Google Scholar] [CrossRef]
- Jaubert, J.M.; Chisholm, J.R.M.; Minghelli-Roman, A.; Marchioretti, M.; Morrow, J.H.; Ripley, H.T. Re-evaluation of the extent of Caulerpa taxifolia development in the northern Mediterranean using airborne spectrographic sensing. Mar. Ecol. Prog. Ser. 2003, 263, 75–82. [Google Scholar] [CrossRef]
- Robinson, K.A.; Ramsay, K.; Lindenbaum, C.; Frost, N.; Moore, J.; Wright, A.P.; Petrey, D. Predicting the distribution of seabed biotopes in the southern Irish Sea. Continent. Shelf Res. 2011, 31, 120–131. [Google Scholar] [CrossRef]
- Mielck, F.; Bartsch, I.; Hass, H.C.; Wölfl, A.-C.; Bürk, D.; Betzler, C. Predicting spatial kelp abundance in shallow coastal waters using the acoustic ground discrimination system RoxAnn. Estuar. Coast. Shelf Sci. 2014, 143, 1–11. [Google Scholar] [CrossRef]
- Noiraksar, T.; Sawayama, S.; Phauk, S.; Komatsu, T. Mapping Sargassum beds off the coast of Chon Buri Province, Thailand, using ALOS AVNIR-2 satellite imagery. Bot. Mar. 2014, 57, 367–377. [Google Scholar] [CrossRef]
- Randall, J.; Hermand, J.-P.; Ernould, M.-E.; Ross, J.; Johnson, C. Measurement of acoustic material properties of macroalgae (Ecklonia radiata). J. Exp. Mar. Biol. Ecol. 2014, 461, 430–440. [Google Scholar] [CrossRef]
- Randall, J.; Johnson, C.R.; Ross, J.; Hermand, J.-P. Acoustic investigation of the primary production of an Australian temperate macroalgal (Ecklonia radiata) system. J. Exp. Mar. Biol. Ecol. 2020, 524, 151309. [Google Scholar] [CrossRef]
- Ware, S.; Anna-Leena, D. Challenges of habitat mapping to inform marine protected area (MPA) designation and monitoring: An operational perspective. Mar. Pol. 2020, 111, 103717. [Google Scholar] [CrossRef]
- Vis, C.; Hudon, C.; Carignan, R. An evaluation of approaches used to determine the distribution and biomass of emergent and submerged aquatic macrophytes over large spatial scales. Aquat. Bot. 2003, 77, 187–201. [Google Scholar] [CrossRef]
- Hossain, M.S.; Mazlan, H. Potential of Earth Observation (EO) technologies for seagrass ecosystem service assessments. Inter. J. Appl. Earth Observ. Geoinform. 2019, 77, 15–29. [Google Scholar] [CrossRef]
- McCarthy, E.; Sabol, B. Acoustic characterization of submerged aquatic vegetation: Military and environmental monitoring applications. In Proceedings of the Oceans 2000 MTS/IEEE Conference and Exhibition, Providence, RI, USA, 11–14 September 2000; pp. 1957–1961. [Google Scholar]
- Fakiris, E.; Zoura, D.; Ramfos, A.; Spinos, E.; Georgiou, N.; Ferentinos, G.; Papatheodorou, G. Object-based classification of sub-bottom profiling data for benthic habitat mapping. Comparison with sidescan and RoxAnn in a Greek shallow-water habitat. Estuar. Coast. Shelf Sci. 2018, 208, 219–234. [Google Scholar] [CrossRef]
- Dimas, X.; Fakiris, E.; Christodoulou, D.; Georgiou, N.; Geraga, M.; Papathanasiou, V.; Orfanidis, S.; Kotomatas, S.; Papatheodorou, G. Marine priority habitat mapping in a Mediterranean conservation area (Gyaros, South Aegean) through multi-platform marine remote sensing techniques. Front. Mar. Sci. 2022, 9, 953462. [Google Scholar] [CrossRef]
- van Rein, H.; Brown, C.J.; Quinn, R.; Breen, J.; Schoeman, D. An evaluation of acoustic seabed classification techniques for marine biotope monitoring over broad-scales (>1 km2) and meso-scales (10 m2 − 1 km2). Estuar. Coast. Shelf Sci. 2011, 93, 336–349. [Google Scholar] [CrossRef]
- Urick, R.J. Principles of Underwater Sound, 3rd ed.; Peninsula: Newport Beach, CA, USA, 2013. [Google Scholar]
- Simmonds, J.; Maclennan, D. Fisheries Acoustics: Theory and Practice, 2nd ed.; Blackwell Publishing: Hoboken, NJ, USA, 2005; p. 456. [Google Scholar]
- Depew, D.C.; Stevens, A.W.; Smith, R.E.H.; Hecky, R.E. Detection and characterization of benthic filamentous algal stands (Cladophora sp.) on rocky substrata using a high-frequency echosounder. Limnol. Oceanogr. Methods 2009, 7, 693–705. [Google Scholar] [CrossRef]
- Monpert, C.; Legris, M.; Noel, C.; Zerr, B.; Caillec, J.M.L. Studying and modeling of submerged aquatic vegetation environments seen by a single beam echosounder. In Proceedings of the Meetings on Acoustics: Acoustical Society of America, Edinburgh, UK, 2–6 July 2012; Volume 17, p. 070044. [Google Scholar] [CrossRef]
- Llorens-Escrich, S.; Tamarit, E.; Hernandis, S.; Sánchez-Carnero, N.; Rodilla, M.; Pérez-Arjona, I.; Moszynski, M.; Puig-Pons, V.; Tena-Medialdea, J.; Espinosa, V. Vertical configuration of a side scan sonar for the monitoring of Posidonia oceanica meadows. J. Mar. Sci. Engineer. 2021, 9, 1332. [Google Scholar] [CrossRef]
- Shao, H.; Minami, K.; Shirakawa, H.; Kawauchi, Y.; Matsukura, R.; Tomiyasu, M.; Miyashita, K. Target strength of a common kelp species, Saccharina japonica, measured using a quantitative echosounder in an indoor seawater tank. Fish. Res. 2019, 214, 110–116. [Google Scholar] [CrossRef]
- Minami, K.; Kita, C.; Shirakawa, H.; Kawauchi, Y.; Shao, H.; Tomiyasu, M.; Iwahara, Y.; Takahara, H.; Kitagawa, T.; Miyashita, K. Acoustic characteristics of a potentially important macroalgae, Sargassum horneri, for coastal fisheries. Fish. Res. 2021, 240, 105955. [Google Scholar] [CrossRef]
- Mutlu, E.; Balaban, C. New algorithms for the acoustic biomass estimation of Posidonia oceanica: A study in the Antalya gulf (Turkey). Fresen. Environ. Bull. 2018, 27, 2555–2561. [Google Scholar]
- Olguner, C.; Mutlu, E. Acoustic estimates of leaf height and biomass of Posidonia oceanica meadow in Gulf of Antalya, the eastern Mediterranean. COMU. J. Mar. Sci. Fish. 2020, 3, 79–94. [Google Scholar] [CrossRef]
- Mutlu, E.; Olguner, C. Acoustic scattering properties of seagrass: In/ex situ measurements of Posidonia oceanica. Medit. Mar. Sci. 2023, 24, 272–291. [Google Scholar] [CrossRef]
- Mutlu, E.; Olguner, C. Acoustic scattering properties of a seagrass, Cymodocea nodosa: In-situ measurements. Bot. Mar. 2023. under revision. [Google Scholar]
- Mutlu, E.; Olguner, C. Density-depended acoustical identification of two common seaweeds (Posidonia oceanica and Cymodocea nodosa) in the Mediterranean Sea. Thalassas 2023. [Google Scholar] [CrossRef]
- Bakiera, D.; Stepnowski, A. Method of the sea bottom classification with a division of the first echo signal. In Proceedings of the XIIIth Symposium on Hydroacoustics, Gdynia-Jurata, Poland, 12–16 May 1996; pp. 55–60. [Google Scholar]
- Bezdek, J.C. Pattern Recognition with Fuzzy Function Algorithms; Plenum Press: New York, NY, USA, 1981; pp. 43–93. [Google Scholar]
- Burczynski, J. Bottom Classification; BioSonics Inc.: Seattle, WA, USA, 1999; 14p. [Google Scholar]
- Stepnowski, A.; Moszynski, M.; Komendarczyk, R.; Burczynski, J. Visual real-time Bottom Typing System (VBTS) and neural networks experiment for seabed classification. In Proceedings of the 3rd European Conference on Underwater Acoustics Heraklion, Crete, Greece, 24–28 July 1996; pp. 685–690. [Google Scholar]
- Chivers, R.C. New acoustic processing for underway surveying. Hydrogr. J. 1990, 56, 9–17. [Google Scholar]
- Orlowski, A. Application of multiple echoes energy measurements for evaluation of sea bottom type. Oceanologia 1984, 19, 61–78. [Google Scholar]
- Pouliquen, E.; Lurton, X. Sea-bed identification using echo-sounder signals. In Proceedings of the European Conference on Underwater Acoustics, Luxembourg, 14–18 September 1992; Elsevier Applied Science: London, UK; New York, NY, USA, 1992; pp. 535–539. [Google Scholar]
- Tegowski, J. Acoustical classification of the bottom sediments in the southern Baltic Sea. Quat. Int. 2005, 130, 153–161. [Google Scholar] [CrossRef]
- Kruss, A.; Blondel, P.; Tegowski, J. Acoustic properties of macrophytes: Comparison of single-beam and multibeam imaging with modeling results. In Proceedings of the 11th European Conference on Underwater Acoustics, Edinburgh, UK, 2–6 July 2012; pp. 168–175. [Google Scholar]
- Maceina, M.J.; Shireman, J.V. The use of a recording fathometer for determination of distribution and biomass of hydrilla. J. Aquat. Plant Manag. 1980, 18, 34–39. [Google Scholar]
- Winfield, I.; Onoufriou, C.; O’Connel, M.; Godlewska, M.; Ward, R.; Brown, A.; Yallop, M. Assessment in two shallow lakes of a hydroacoustic system for surveying aquatic macrophytes. Hydrobiologia 2007, 584, 111–119. [Google Scholar] [CrossRef]
- Mutlu, E.; Balaban, C.; Gokoglu, M.; Ozvarol, Y.; Olguner, M.T. Acoustical Density-Dependent Calibration of the Dominant Sea Meadows and Seagrasses and Monitoring of Their Distribution; TUBITAK, Ankara, TUBITAK Project Final Report 110Y232; The Scientific and Technological Research Council of Turkey (TÜBİTAK): Ankara, Turkey, 2014; p. 368. [Google Scholar]
- Mutlu, E.; Özvarol, Y.; Şahin, A.; Duman, G.S.; Karaca, D. Acoustical Determination of Biomass Quantities and Monitoring of Distribution of Posidonia oceanica Meadows on the Turkish Entire Coasts in the Eastern Mediterranean; TUBITAK, Ankara, TUBITAK Project Final Report, 117Y133; The Scientific and Technological Research Council of Turkey (TÜBİTAK): Ankara, Turkey, 2020; p. 190. [Google Scholar]
- Marine Region. Available online: https://marineregions.org/ (accessed on 8 March 2023).
- EchoView. Available online: https://support.echoview.com/WebHelp/RH8_Popups/Phenomena/Acoustic_beam_dead_zone_referenced.htm#:~:text=The%20deadzone%20is%20an%20area,areas%20with%20steep%20bottom%20topography (accessed on 28 December 2022).
- Mello, L.G.S.; Rose, G.A. The acoustic dead zone: Theoretical vs. empirical estimates, and its effect on density measurements of semi-demersal fish. ICES J. Mar. Sci. 2009, 66, 1364–1369. [Google Scholar] [CrossRef]
- De Robertis, A.; Higginbottom, I. A post-processing technique to estimate the signal-to-noise ratio and remove echosounder background noise. ICES J. Mar. Sci. 2007, 64, 1282–1291. [Google Scholar] [CrossRef]
- Lee, W.S.; Lin, C.Y. Mapping of tropical marine benthic habitat: Hydroacoustic classification of coral reefs environment using single-beam (RoxAnn™) system. Continen. Shelf Res. 2018, 170, 1–10. [Google Scholar] [CrossRef]
- Lurton, X. An Introduction to Underwater Acoustics; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Quintino, V.; Freitas, R.; Mamede, R.; Ricardo, F.; Rodrigues, A.M.; Mota, J.; Pe’rez-Ruzafa, A.; Marcos, C. Remote sensing of underwater vegetation using single-beam acoustics. ICES J. Mar. Sci. 2010, 67, 594–605. [Google Scholar] [CrossRef]
- Canals, M.; Ballesteros, E. Production of carbonate particles by phytobenthic communities on the Mallorca-Menorca shelf, northwestern Mediterranean Sea. Deep-Sea Res. Part II 1997, 44, 611–629. [Google Scholar] [CrossRef]
- Terrados, J.; Medina-Pons, F.J. Inter-annual variation of shoot density and biomass nitrogen and phosphorus content of the leaves and epiphyte load of the seagrass Posidonia oceanica (L.) Delile off Mallorca Western Mediterranean. Sci. Mar. 2011, 75, 61–70. [Google Scholar] [CrossRef]
- Sghaier, Y.R.; Zakhama-Sraieb, R.Y.M.; Charfi-Cheikhrouha, F. Patterns of shallow seagrass (Posidonia oceanica) growth and flowering along the Tunisian coast. Aquat. Bot. 2013, 104, 185–192. [Google Scholar] [CrossRef]
- Mavko, G.; Mukerji, T.; Dvorkin, J. The Rock Physics Handbook; Cambridge University Press: Cambridge, UK, 1998. [Google Scholar]
- Merriam, C.O. Depositional History of Lower Permian (Wolfcampian-Leonardian) Carbonate Buildups, Midland Basin, Upton County, Texas. Master’s Thesis, Texas A&M University, College Station, TX, USA, 1999. [Google Scholar]
- Aleman, P.B. Acoustic Impedance Inversion of Lower Permian Carbonate Buildups in the Permian Basin, Texas. Master’s Thesis, The Office of Graduate Studies of Texas A&M University, College Station, TX, USA, 2004. [Google Scholar]
- Mateo, M.A.; Romero, J.; Perez, M.; Littler, M.M.; Littler, D.S. Dynamics of millenary organic deposits resulting from the growth of the Mediterranean seagrass Posidonia oceanica. Estuar. Coast. Shelf Sci. 1997, 44, 103–110. [Google Scholar] [CrossRef]
- Enriquez, S.; Schubert, N. Direct contribution of the seagrass Thalassia testudinum to lime mud production. Nat. Commun. 2004, 5, 3835. [Google Scholar] [CrossRef]

















Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutlu, E. A Package of Script Codes, POSIBIOM for Vegetation Acoustics: POSIdonia BIOMass. J. Mar. Sci. Eng. 2023, 11, 1790. https://doi.org/10.3390/jmse11091790
Mutlu E. A Package of Script Codes, POSIBIOM for Vegetation Acoustics: POSIdonia BIOMass. Journal of Marine Science and Engineering. 2023; 11(9):1790. https://doi.org/10.3390/jmse11091790
Chicago/Turabian StyleMutlu, Erhan. 2023. "A Package of Script Codes, POSIBIOM for Vegetation Acoustics: POSIdonia BIOMass" Journal of Marine Science and Engineering 11, no. 9: 1790. https://doi.org/10.3390/jmse11091790





