Algal Community Membership of Estuarine Mudflats from the Savannah River, United States
Abstract
:1. Introduction
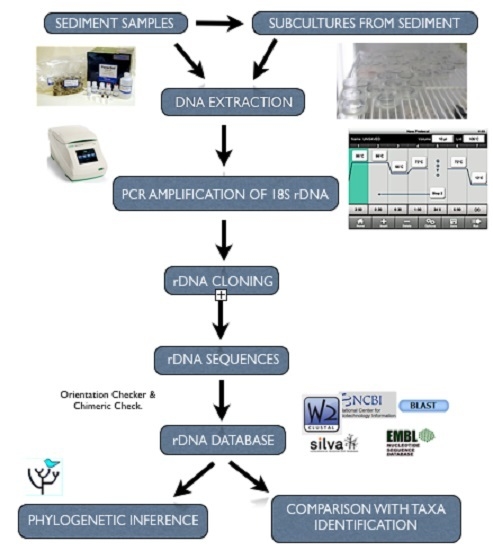
2. Experimental Section
2.1. Study Sites and Sampling
2.2. Morphological Assessment
2.3. Culturing of Isolates from Environmental Samples
2.4. Genomic DNA Isolation
2.5. PCR Amplification Using 18S rDNA Primers
2.6. 18S rDNA Cloning and Plasmid DNA Isolation
2.7. Restriction Fragment Length Polymorphism (RFLP) Analyses of 18S rDNA
2.8. DNA Sequencing and Analyses
3. Results and Discussion
3.1. Morphological Analyses
3.2. Molecular Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pritchard, D. What is an estuary: Physical viewpoint. In Estuaries; Lauff, G., Ed.; American Association for the Advancement of Science: Washington, DC, USA, 1967. [Google Scholar]
- NRC (National Research Council). Clean Coastal Waters: Understanding and Reducing the Effects of Nutrient Pollution; National Academy Press: Washington, DC, USA, 2000. [Google Scholar]
- Rabalais, N.N.; Turner, R.E.; Wiseman, W.J.; Boesch, D.F. A brief summary of hypoxia on the northern Gulf of Mexico continental shelf: 1985–1988. In Geological Society; Tyson, R.V., Pearson, T.H., Eds.; The Geological Society: London, UK, 1991. [Google Scholar]
- Vos, P.C.; de Wolf, H. Methodological aspects of palaeoecological diatom research in coastal areas of the Netherlands. Geol. Mijnb. 1988, 67, 31–40. [Google Scholar]
- Morrisey, D.J. Differences in effects of grazing by deposit-feeders Hydrobia ulvae (Pennant) (Gastropoda: Prosobranchia) and Corophium arenarium Crawford (Amphipoda) on sediment microalgal populations. II. Quantitative effects. J. Exp. Mar. Biol. Ecol. 1988, 118, 43–53. [Google Scholar] [CrossRef]
- Underwood, G.J.C.; Paterson, D.M. Recovery of intertidal benthic diatoms after biocide treatment and associated sediment dynamics. J. Mar. Biol. Assoc. UK 1993, 73, 25–45. [Google Scholar] [CrossRef]
- Underwood, G.J.C.; Paterson, D.M. Seasonal changes in diatom biomass, sediment stability and biogenic stabilization in the Severn Estuary. J. Exp. Mar. Biol. Ecol. 1993, 73, 871–887. [Google Scholar] [CrossRef]
- Admiraal, W.; Peletier, H.; Brouwer, T. The seasonal succession patterns of diatom species on an intertidal mudflat: An experimental analysis. Oikos 1984, 42, 30–40. [Google Scholar] [CrossRef]
- Underwood, G.J.C. Seasonal and Spatial variation in epipelic diatom assemblages in the Severn estuary. Diatom Res. 1994, 9, 451–472. [Google Scholar] [CrossRef]
- Graham, J.E.; Graham, L.E.; Wilcox, L.W. Algae, 2nd ed.; Benjamin Cummings: San Francisco, CA, USA, 2009; p. 616. [Google Scholar]
- Andersen, R.A. Diversity of eukaryotic algae. Biodivers. Conserv. 1992, 1, 267–292. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Beolchini, F.; Dell’Anno, A. Damage and degradation rates of extracellular DNA in marine sediments: Implications for the preservation of gene sequences. Mol. Ecol. 2008, 17, 3939–3951. [Google Scholar] [CrossRef] [PubMed]
- Keck, F.; Rimet, F.; Franc, A.; Bouchez, A. Phylogenetic signal in diatom ecology: Perspectives for aquatic ecosystems biomonitoring. Ecol. Appl. 2015. [Google Scholar] [CrossRef]
- Chessman, B.C.; Bate, N.; Gell, P.A.; Newall, P. A diatom species index for bioassessment of Australian rivers. Mar. Freshw. Res. 2007, 58, 542–557. [Google Scholar] [CrossRef]
- Stevenson, R.J.; Pan, Y.; Manoylov, K.M.; Parker, C.A.; Larsen, D.P.; Herlihy, A.T. Development of diatom indicators of ecological conditions for streams of the western US. J. N. Am. Benthol. Soc. 2008, 27, 1000–1016. [Google Scholar] [CrossRef]
- Manoylov, K.M.; Dominy, J.N., Jr. Changes in epipelic diatom diversity from the Savannah River Estuary. J. Environ. Prot. 2013, 4, 172–179. [Google Scholar] [CrossRef]
- Manoylov, K.M. Taxonomic identifications of algae (morphological and molecular), species concepts, methodologies, and their implications for ecological bioassessment. J. Phycol. 2014, 50, 409–424. [Google Scholar] [CrossRef]
- Savin, M.C.; Martin, J.L.; LeGresley, M.; Giewat, M.; Rooney-Varga, J. Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microb. Ecol. 2004, 48, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.K. Application of molecular techniques for genetic differentiation of microalgae. In Out of the Past. Collected Reviews to Celebrate the Jubilee of the British Phycological Society; Northon, T.A., Ed.; Dataplus Print & Design (Dunmurry): Belfast, UK, 2003; pp. 31–48. [Google Scholar]
- Moon-van der Staay, S.Y.; van der Staay, G.W.M.; Guillou, L.; Vaulot, D.; Claustre, H.; Medlin, L.K. Abundance and diversity of prymnesiophytes in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences. Limnol. Oceanogr. 2000, 45, 98–109. [Google Scholar] [CrossRef]
- Pace, N.R.; Stahl, D.A.; Lane, D.J.; Olsen, G.J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 1986, 9, 1–55. [Google Scholar]
- López-García, P.; Rodríguez-Valera, F.; Pedrós-Alió, C.; Moreira, D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 2001, 409, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Moon-van der Staay, S.Y.; de Wachter, R.; Vaulot, D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 2001, 409, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Viprey, M.; Guillou, L.; Ferreol, M.; Vaulot, D. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ. Microbiol. 2008, 10, 1804–1822. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA). Standard Methods for Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1998. [Google Scholar]
- Dominy, J.D., Jr. Algal Population Dynamics within the Savannah River Estuary. Master’s Thesis, Georgia College & State University, Milledgeville, GA, USA, 2012. [Google Scholar]
- Palmer, C.M.; Maloney, T.E. A New Counting Slide for Nanoplankton; American Society of Limnology and Oceanography Special Publication: Waco, TX, USA, 1954; p. 6. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 1. Teil: Naviculaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Jena, Germany, 1986; pp. 1–876. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 2. Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Jena, Germany, 1988; pp. 1–596. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 3. Teil: Centrales, Fragilariaceae, Eunotiaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Jena, Germany, 1991; pp. 1–576. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 4. Teil: Achnanthaceae. Kritische Ergänzungen zu Navicula (Lineolatae) und Gomphonema. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Jena, Germany, 1991; pp. 1–437. [Google Scholar]
- Witkowski, A.; Lange-Bertalot, H.; Metzeltin, D. Diatom Flora of Marine Coasts I. In Iconographia Diatomologica Annotated Diatom Micrograph; Lange-Bertalot, H., Ed.; A.R.G. Gantner Verlag K. G.: Ruggell, Germany, 2008. [Google Scholar]
- Hofmann, G.; Werum, M.; Lange-Bertalot, H. Diatomeen im Süßwasser Benthos von Mitteleuropa. Bestimmungsflora Kieselalgen für die ökologische Praxis. Über 700 der häufigsten Arten und ihre Ökologie; Koeltz Scientific Books: Oberreifenberg, Germany, 2013. [Google Scholar]
- Manoylov, K.M.; Marsh, T.; Stevenson, R.J. Testing molecular tools for assessment of taxonomic composition of a benthic algal community. Nova Hedwig. Beih. 2009, 135, 121–136. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Ashelford, K.E.; Chuzhanova, N.A.; Fry, J.C.; Jones, A.J.; Weightman, A.J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 2006, 72, 5734–5741. [Google Scholar] [CrossRef] [PubMed]
- Ashelford, K.E.; Chuzhanova, N.A.; Fry, J.C.; Jones, A.J.; Weightman, A.J. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 2005, 71, 7724–7736. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Higgins, D.G. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Riley-Buckley, M.S. Microbial Communities in Pristine and Tetrachloroethylene-Contaminated Aquifer Sediment. Ph.D. Thesis, Michigan State University, East Lansing, MI, USA, 2001. [Google Scholar]
- Nguyen, T.N.; Berzano, M.; Gualerzi, C.O.; Spurio, R. Development of molecular tools for the detection of freshwater diatoms. J. Microbiol. Method 2011, 84, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Geletu, A. Assessment of Molecular Approaches in Studying Algal Diversity. Master’s Thesis, Georgia College & State University, Milledgeville, GA, USA, 2012. [Google Scholar]
- Kermarrec, L.; Franc, A.; Rimet, F.; Chaumeil, P.; Frigerio, J.M.; Humbert, J.F.; Bouchez, A. A next-generation sequencing approach to river biomonitoring using benthic diatoms. Freshw. Sci. 2014, 33, 349–364. [Google Scholar] [CrossRef]
- Lefebvre, K.E.; Hamilton, P.E. Morphology and molecular studies on large Neidium species (Bacillariophyta) of North America, including an examination of Ehrenberg’s types. Phytotaxa 2015, 220, 201–223. [Google Scholar] [CrossRef]
- Quaiser, A.; Zivanovic, Y.; Moreira, D.; Lopez-Garcia, P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2011, 5, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Flynn, D.F.B.; Mirotchnick, N.; Jain, M.; Palmer, M.I.; Naeem, S. Functional and phylogenetic diversity as predictors of biodiversity-ecosystem-function relationships. Ecology 2011, 92, 1573–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, J.; Glöckner, G.; Jahn, R.; Enke, N.; Gemeinholzer, B. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol. Ecol. Res. 2015, 15, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Li, C.L.; Ashworth, M.P.; Witkowski, A.; Dąbek, P.; Medlin, L.K.; Kooistra, W.H.C.F.; Sato, S.; Zgłobicka, I.; Kurzydłowski, K.J.; Theriot, E.C.; et al. New insights into Plagiogrammaceae (Bacillariophyta) based on multigene phylogenies and morphological characteristics with the description of a new genus and three new species. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Capo, E.; Debroas, D.; Domaizon, I. Is planktonic diversity well recorded in sedimentary DNA? Toward the Reconstruction of Past Protistan Diversity. Microbiol. Ecol. 2015, 70, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, F. Marine Littoral Diatoms of Beaufort, North Carolina; Duke University Press: Durham, NC, USA, 1955; Volume 6, p. 167. [Google Scholar]
- Kermarrec, L.; Franc, A.; Rimet, F.; Chaumeil, P.; Humbert, J.F.; Bouchez, A. Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: A test for freshwater diatoms. Mol. Ecol. Res. 2013, 13, 607–619. [Google Scholar] [CrossRef] [PubMed]




| Taxon | Savannah River DNA Source | Similarity | Group | Level Identification | Genbank # |
|---|---|---|---|---|---|
| Odontella sinensis | A | 89 | Bacillariophyta | Species | Y10570 |
| Biddulphia sp. CCMP 14718Sr RNA gene | B | 95 | Bacillariophyta | Genus | EF585585 |
| Surirella sp. | B | 97 | Bacillariophyta | Genus | FR865515 |
| Pleurosira cf. laevis clone p203 | B | 99 | Bacillariophyta | Species | AJ535188 |
| Cerataulus smithii | B | 94 | Bacillariophyta | Species | HQ912666 |
| Paraphysomonas vestita | B | 99 | Chrysophyta | Species | Z28335 |
| Cymbella aspera | B | 98 | Bacillariophyta | Species | AM502016 |
| Hydrosera sp. CYTX025 | C | 98 | Bacillariophyta | Genus | HQ912683 |
| Goniomonas truncata clone | C | 99 | Chrysophyta | Species | DQ980479 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manoylov, K.M.; France, Y.E.; Geletu, A.; Dominy, J.N. Algal Community Membership of Estuarine Mudflats from the Savannah River, United States. J. Mar. Sci. Eng. 2016, 4, 11. https://doi.org/10.3390/jmse4010011
Manoylov KM, France YE, Geletu A, Dominy JN. Algal Community Membership of Estuarine Mudflats from the Savannah River, United States. Journal of Marine Science and Engineering. 2016; 4(1):11. https://doi.org/10.3390/jmse4010011
Chicago/Turabian StyleManoylov, Kalina M., Yenkang Ellen France, Abeselom Geletu, and Joseph N. Dominy. 2016. "Algal Community Membership of Estuarine Mudflats from the Savannah River, United States" Journal of Marine Science and Engineering 4, no. 1: 11. https://doi.org/10.3390/jmse4010011
APA StyleManoylov, K. M., France, Y. E., Geletu, A., & Dominy, J. N. (2016). Algal Community Membership of Estuarine Mudflats from the Savannah River, United States. Journal of Marine Science and Engineering, 4(1), 11. https://doi.org/10.3390/jmse4010011