1. Introduction
Analysis, visualization, and distribution of coastal ocean model data is challenging due to the sheer size of the data involved, with regional simulations commonly in the 10GB–1TB range. The traditional workflow is to download data to local workstations or file servers from which the data needed can be extracted via services such as OPeNDAP [
1]. Analysis and visualization take place with environments like MATLAB® and Python running on local computers. Not only are these datasets becoming too large to effectively download and analyze locally, but this approach requires acquiring and maintaining local hardware, software, and personnel to ensure reliable and efficient processing. Archiving is an additional challenge for many centers. Effective sharing with collaborators is often limited by unreliable services that cannot scale with demand. In some cases, a subset of analysis and visualization tools are made available through custom web portals (e.g., [
2,
3]). These portals can satisfy the needs of data dissemination to the public but don’t have the suite of scientific analysis tools needed for collaborative research use. In addition, the development and maintenance of these portals require dedicated web software developers and is out of the reach of most scientists. The traditional method of data access and use is becoming time and cost inefficient.
The Cloud and recent advances in technology provide new opportunities for analysis, visualization, and distribution of model data, overcoming these problems [
4]. Data can be stored in the Cloud efficiently in object storage which allows performant access by providers or end users alike. Analysis and visualization can take place in the Cloud, close to the data, allowing efficient and cost-effective access, as the only data that needs to leave the Cloud are graphics and text returned to the browser. As these tools have matured, they have lowered the barrier of entry and are poised to transform the ability of regular scientists and engineers to collaborate on difficult research problems without being constrained by their local resources.
The Pangeo project [
5,
6,
7] was created to take advantage of these advances for the scientific community. The specific goals of Pangeo are to: “(1) Foster collaboration around the open source scientific Python ecosystem for ocean/atmosphere/land/climate science. (2) Support the development with domain-specific geoscience packages. (3) Improve scalability of these tools to handle petabyte-scale datasets on HPC and Cloud platforms.” It makes progress toward these goals by building on open-source packages already widely used in the Python ecosystem, and supporting a flexible and modular framework for interactive, scalable, data-proximate computing on large gridded datasets. Here we first describe the essential components of this framework, then demonstrate two coastal ocean modeling use cases: (1) Calculating the maximum water level at each grid cell from a 53 GB, 720 time step, 9 million node triangular mesh ADCIRC [
8] simulation of Hurricane Ike; (2) creating a dashboard for visualizing data from the curvilinear orthogonal COAWST/ROMS [
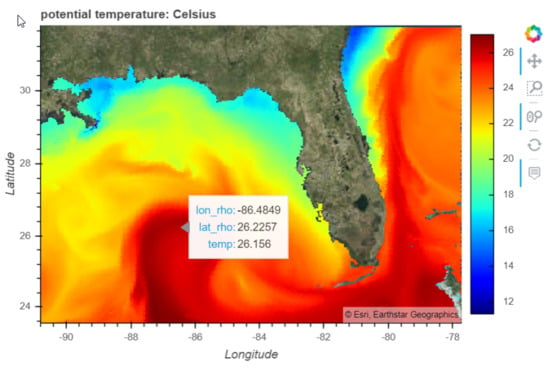
9] forecast model.
2. Framework Description
Pangeo is a flexible framework which can be deployed in different types of platforms with different components, so here we describe the specific framework we used for this work, consisting of:
Zarr [
10] format files with model output for cloud-friendly access
Dask [
11] for parallel scheduling and execution
Xarray [
12] for working effectively with model output using the NetCDF/CF [
13,
14] Data Model
PyViz [
15] for interactive visualization of the output
Jupyter [
16] to allow user interaction via their web browser (
Figure 1).
We will briefly describe these and several other important components in more detail.
2.1. Zarr
Zarr is a Cloud-friendly data format. The Cloud uses object storage. Access to NetCDF files (the most commonly used format for model data) in object storage is poor, due to the latency of object requests and the numerous small requests involved with accessing data from a NetCDF file. Therefore, we converted the model output from NetCDF format to Zarr format which was developed specifically to allow Cloud-friendly access to n-dimensional array data. With Zarr, the metadata is stored in JSON format, and the data chunks are stored as separate storage objects, typically with chunk sizes of 10–100 mb, which enables concurrent reads by multiple processors. The major features of the HDF5 and NetCDF4 data models are supported: Self-describing datasets with variables, dimensions and attribute, supporting groups, chunking, and compression. It is being developed in an open community fashion on GitHub, with contributions from multiple research organizations. Currently, only a Python interface exists, but it has a well-documented specification and other language bindings are being developed. The Unidata NetCDF team is working on the adoption of Zarr as a back-end to the NetCDF C library. Data can be converted from NetCDF, HDF5 or other n-dimensional array formats to Zarr using the Xarray library (described below).
2.2. Dask
Dask is a component that facilitates out-of-memory and parallel computations. Dask arrays allow handling very large array operations using many small arrays known as “chunks”. Dask workers perform operations in parallel, and dask worker clusters can be created on local machines with multiple CPUs, on HPC with job submission, and on the Cloud via Kubernetes [
17] orchestration of Docker [
18] containers.
2.3. Xarray
Xarray is a component which implements the NetCDF Data model, with the concept of a dataset that contains named shared dimensions, global attributes, and a collection of variables that have identified dimensions and variable attributes. It can read from a variety of sources, including NetCDF, HDF, OPeNDAP, Zarr and many raster data formats. Xarray automatically uses Dask for parallelization when the data are stored in a format that uses chunks, or when chunking is explicitly specified by the user.
2.4. PyViz
PyViz is a coordinated effort to make data visualization in Python easier to use, easier to learn, and more powerful. It is a collection of visualization packages built on top of a foundation of mature, widely used data structures and packages in the scientific Python ecosystem. The functions of these packages are described separately below along with the associated EarthSim project that is instrumental in advancing and extending PyViz capabilities.
2.5. EarthSim
EarthSim [
19,
20] is a project that acts as a testing ground for PyViz workflows specifying, launching, visualizing and analyzing environmental simulations such as hydrologic, oceanographic, weather and climate modeling. It contains both experimental tools and example workflows. Approaches and tools developed in this project often are incorporated into the other PyViz packages upon maturity. Specifically, key improvements in the ability to represent large curvilinear mesh and triangular mesh grids made the PyViz tools practical for use by modelers, e.g., TriMesh and QuadMesh.
2.6. PyViz: Datashader
Datashader [
21] renders visualizations of large data into rasters, allowing accurate, dynamic representation of datasets that would otherwise be impossible to display in the browser.
2.7. PyViz: HoloViews
HoloViews [
22] is a package that allows the visualization of data objects through annotation of the objects. It supports different back-end plotting packages, including Matplotlib, Plotly, and Bokeh. The Matplotlib backend provides static plots, while Bokeh generates visualizations in JavaScript that are rendered in the browser and allow user interaction such as zooming, panning, and selection. We used Bokeh here, and the visualizations work both in Jupyter Notebooks and deployed as web pages running with Python backends. A key aspect of using HoloViews for large data is that it can dynamically rasterize the plot to screen resolution using Datashader.
2.8. PyViz: GeoViews
GeoViews [
23] is a package that layers geographic mapping on top of HoloViews, using the Cartopy [
24] package for map projections and plotting. It also allows a consistent interface to many different map elements, including Web Map Tile Services, vector-based geometry formats such as Shapefiles and GeoJSON, raster data and QuadMesh and TriMesh objects useful for representing model grids.
2.9. PyViz HvPlot
HvPlot [
25] is a high-level package that makes it easy to create HoloViews/GeoViews objects by allowing users to replace their normal object .plot() commands with .hvplot(). Sophisticated visualizations can therefore be created with one plot call, and then if needed supplemented with additional lower-level HoloViews information for finer-grained control.
2.10. PyViz: Panel
Panel [
26] is a package that provides a framework for creating dashboards that contain multiple visualizations, control widgets and explanatory text. It works within Jupyter and the dashboards can also be deployed as web applications that work dynamically with Dask-powered Python backends.
2.11. JupyterHub
JupyterHub [
27] is a component that allows multi-user login, with each user getting their own Jupyter server and persisted disk space. The Jupyter server runs on the host system, and users interact with the server via the Jupyter client, which runs in any modern web browser. Users type code into cells in a Jupyter Notebook, which get processed on the server and the output (e.g., figures and results of calculations) return as cell output directly below the code. The notebooks themselves are simple text files that may be shared and reused by others.
2.12. Kubernetes
Kubernetes [
17] is a component that orchestrates containers like Docker, automating deployment, scaling, and operations of containers across clusters of hosts. Although developed by Google, the project is open-source and Cloud agnostic. It allows JupyterHub to scale with the number of users, and individual tasks to scale with the number of requested Dask workers.
2.13. Conda: Reproducible Software Environment
Utilizing both Pangeo and PyViz components, the system contains 300+ packages. With these many packages, we need an approach that minimizes the possibility of conflicts. We use Conda [
28], “an open source, cross-platform, language agnostic package manager and environment management system”. Conda allows installation of pre-built binary packages, and providers can deliver packages via channels at anaconda.org. To provide a consistent and reliable build environment, the community has created conda-forge [
29], a build infrastructure that relies on continuous integration to create packages for Windows, macOS, and Linux. We specify the conda-forge channel only when we create our environment, and use specific packages from other channels only when absolutely necessary. For example, currently, over 90% of the 300+ packages we use to build the Pangeo Docker containers are from conda-forge.
2.14. Community
The Pangeo collaborator community [
30] plays a critical role in making this framework deployable and usable by domain scientists like ocean modelers. The community discusses technical and usage challenges on GitHub issues [
31], during weekly check-in meetings, and in a blog [
32].
4. Discussion
The Pangeo framework demonstrated here works not only on the Cloud, but can run on HPC or even on a local desktop. On the local desktop, however, data needs to be downloaded for analysis by each user, and parallel computations are limited to the locally available CPUs. On HPC, there may be access to more CPUs, but the data still needs to be downloaded to the HPC center. On the Cloud, however, anyone can access the data without it having to be moved and have virtually unlimited processing power available to them. On the Cloud, Pangeo allows similar functionality to Google Earth Engine [
37], allowing computation at a scale close to the data, but can be run on any Cloud, and with any type of data. Let us review the advantages of the Cloud in more detail:
Data access: Data in object storage like S3, can be accessed directly from a URL without the need of a special data service like OPeNDAP. This prevents the data service from being a bottleneck on operations, or data access failing because the data service has failed. It also means that data storage on the Cloud is immediately available for use by your collaborators or users. While data services like OPeNDAP can become overwhelmed by too many concurrent requests, this doesn’t happen with access from object storage. Object storage is also extremely reliable, 99.999999999% with default storage on Amazon, which means if 10,000 objects are stored, you may lose one every 10 million years. Finally, data in object storage are not just available for researchers to analyze, but are also available for Cloud-enabled web applications to use. This includes applications that have been developed by scientists as PyViz dashboards, then published using Panel as dynamic web applications with one additional line of code.
Computing on demand: On the Cloud, costs accrue per hour for each machine type in use. It costs the same to run 60 CPUs for 1 min as it does to run 1 CPU for 60 min, and because nearly instantaneous access is available, with virtually unlimited numbers of CPUs, big data analysis tasks can be conducted interactively instead of being limited to batch operations. The Pangeo instance automatically spins up and down Cloud instances based on computational demands.
Freedom from local infrastructure: Because the data, analysis, and visualization are on the Cloud, buying or maintaining local computer centers, high power computer systems, or even fast internet connections is not necessary. Researchers and their colleagues can analyze and visualize data from anywhere with a simple laptop computer and the WiFi from a cell phone hotspot.
We have demonstrated the Pangeo framework for coastal ocean modeling here, but the framework is flexible and is being used increasingly by a wide variety of research projects, including climate scale modeling [
38] and remote sensing [
39]. While the framework clearly benefits the analysis and visualization of large datasets, it is useful for other applications as well. For example, the AWS Pangeo instance we deployed was used by the USGS for two multi-day machine learning workshops that each had 40 students from various institutions with a diversity of computer configurations, operating systems, and versions. The students were able to do the coursework on the Cloud using their web browsers, avoiding the challenges encountered when the course computing environment needs to be installed on a number of heterogeneous personal computers.
While there are numerous benefits to this framework, there are also some remaining challenges [
40,
41]. One important challenge is cost. The Cloud often appears expensive to researchers because much of the true cost of computing is covered by local overhead (e.g., the physical structure, electricity, internet costs, support staff). Gradual adoption, training, and subsidies for Cloud computing are some of the approaches that can help researchers and institutions make the transition to the Cloud more effectively. Another challenge is cultural: Scientists are accustomed to having their data local, and some do not trust storage on the Cloud, despite the reliability. Security issues are also a perceived concern with non-local data. Finally, converting the large collection of datasets designed for file systems to datasets that work well on object storage is a non-trivial task even with the tools discussed above.
Once these challenges are overcome, we can look forward to a day when all model data and analysis takes place on the Cloud, with all data directly accessible and connected by high-speed networks (e.g., Internet 2) and common computing environments can be shared easily. This will lead to unprecedented levels of performance, reliability, and reproducibility for the scientific community, leading to more efficient and effective science.
Several agencies have played key roles in the development of these open-source tools that support the entire community: DARPA (U.S. Defense Advanced Research Projects Agency) provided significant funding for Dask, and ERDC (U.S. Army Engineer Research and Development Center) has provided significant funding through their EarthSim project for developing modeling-related functionality in the PyViz package. We hope that more agencies will participate in this type of open source development, accelerating our progress on expanding this framework to more use cases and more communities.