Seasonal Dynamics of Bathyarchaeota-Dominated Benthic Archaeal Communities Associated with Seagrass (Zostera japonica) Meadows
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. DNA Extraction and ILLUMINA Sequencing
2.3. Sequence Processing, Diversity Analysis, and Predicted Functional Profiles
2.4. Phylogenetic Analysis of Bathyarchaeota Sequences
2.5. Statistical Analysis
3. Results
3.1. Seasonal Variations in Physicochemical Parameters
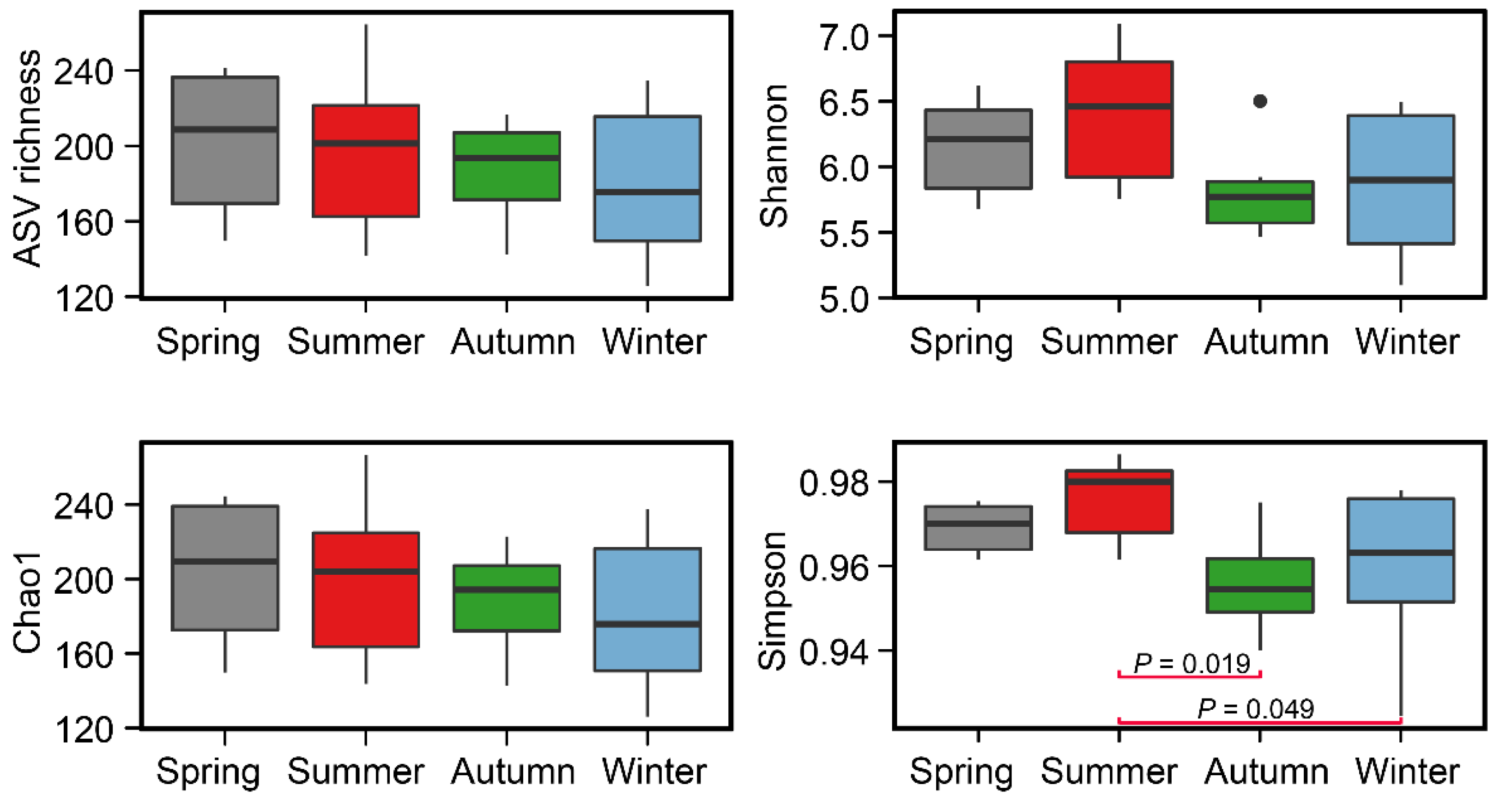
3.2. Variations in Alpha Diversity of Benthic Archaea Community
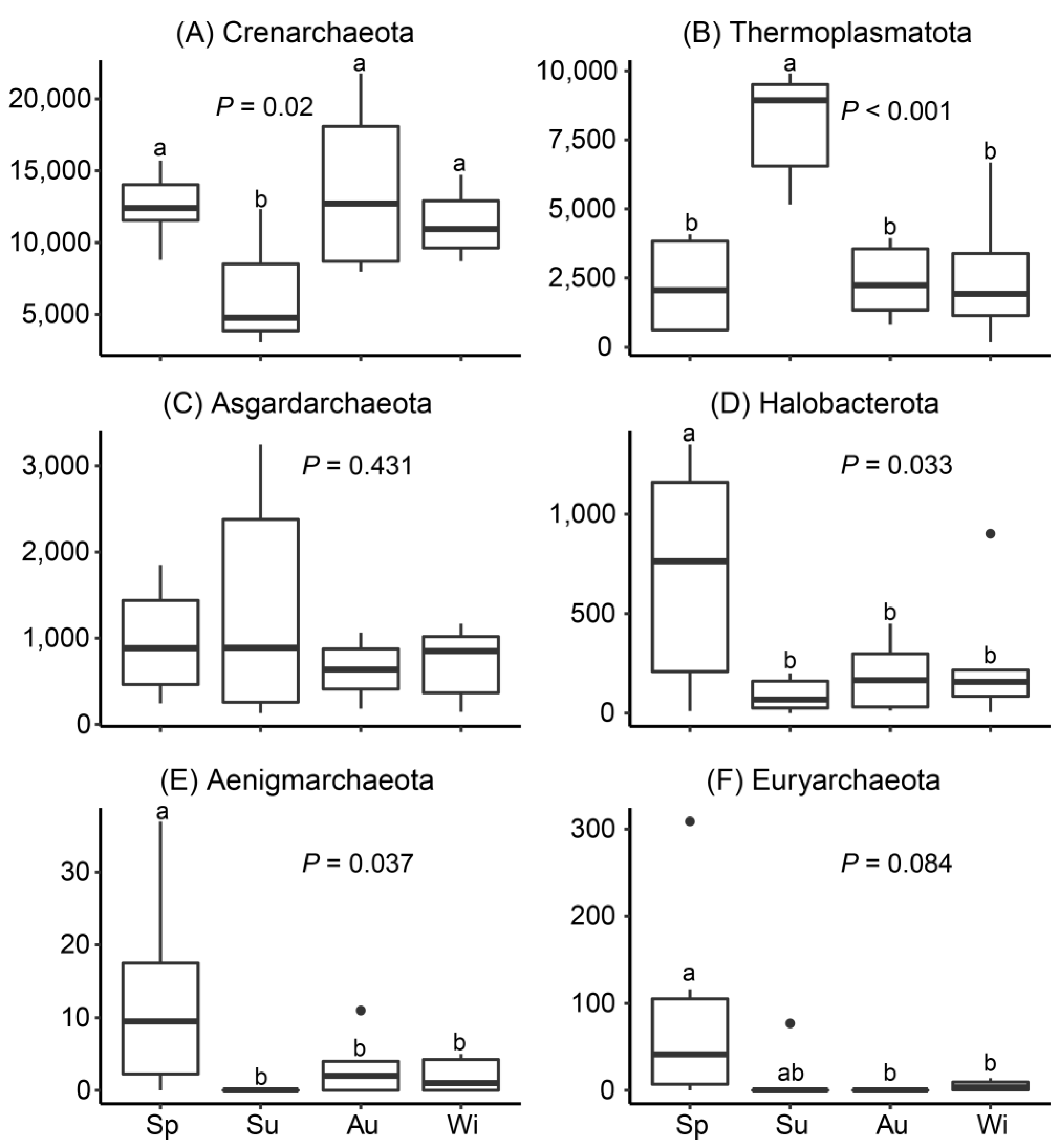
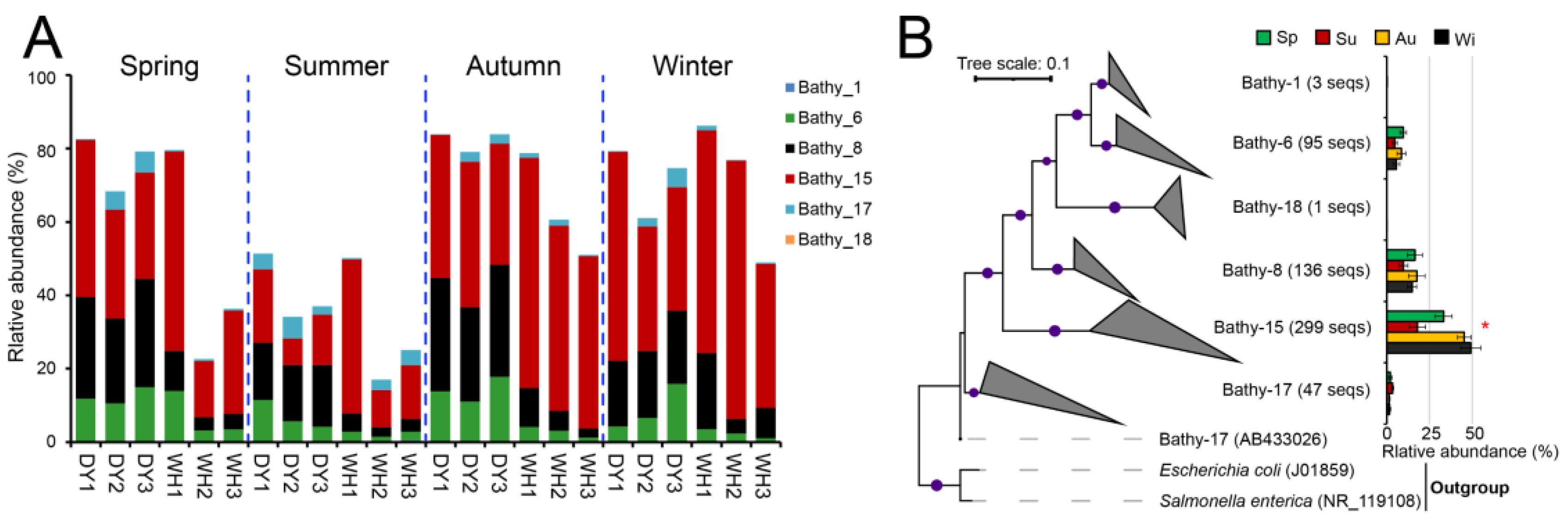
3.3. Archaeal Community Composition and Bathyarchaeota Populations
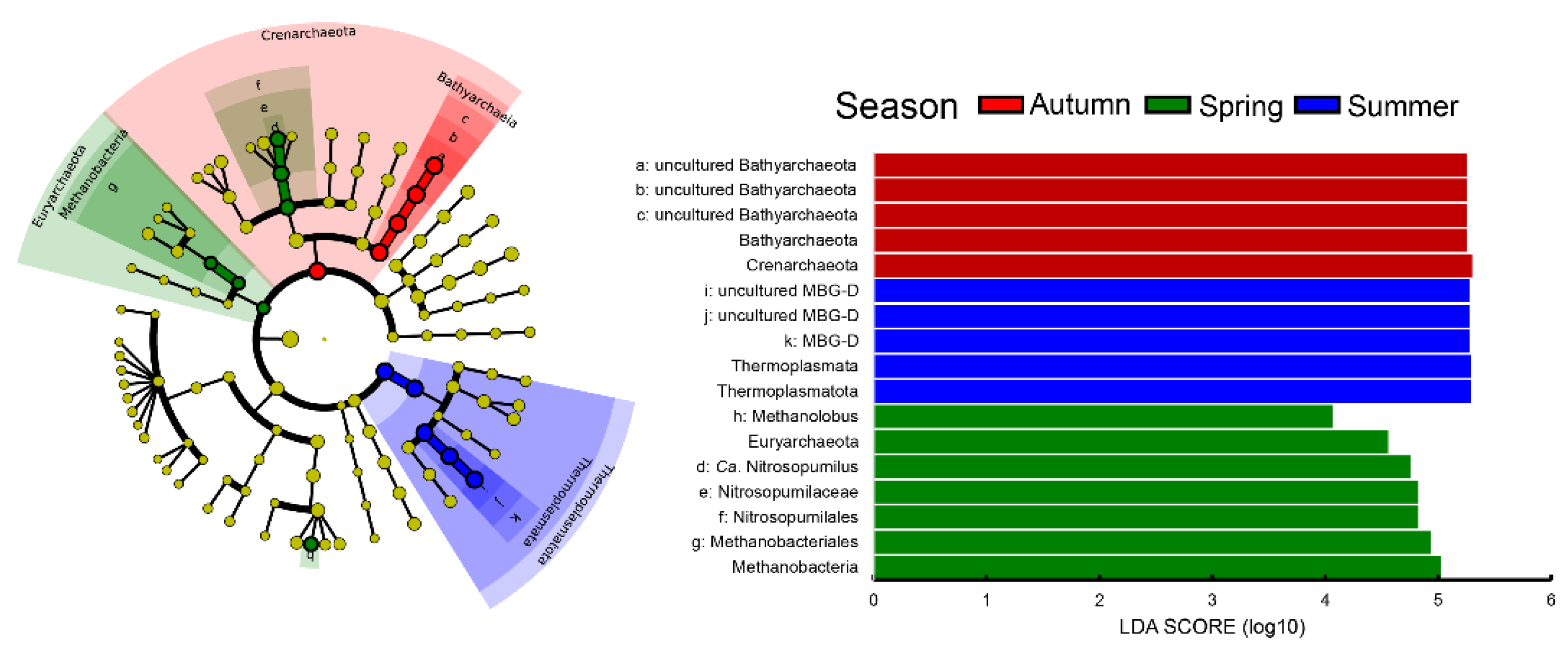
3.4. Seasonal Variability of Benthic Archaeal Communities in Z. japonica Meadows
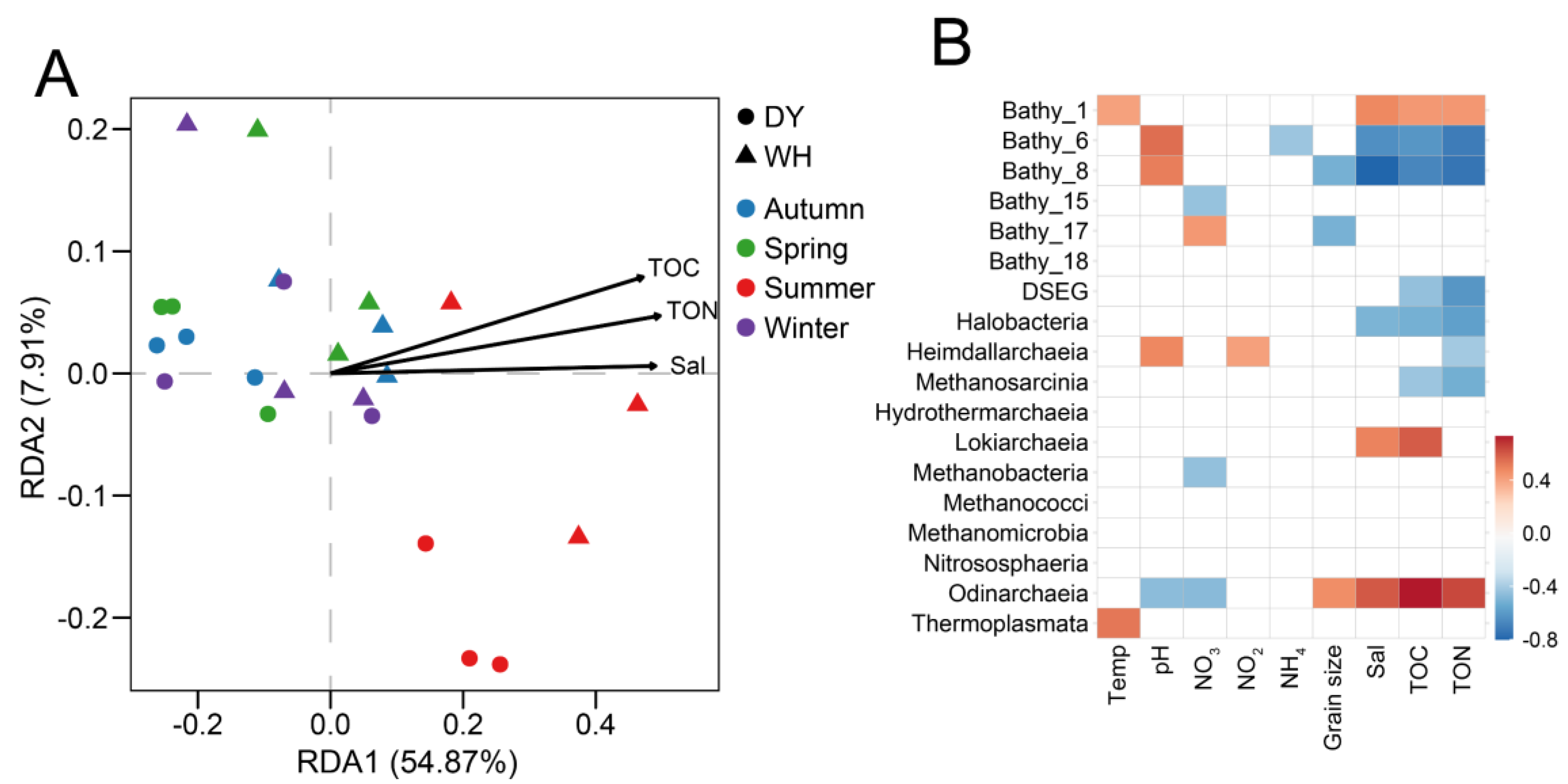
3.5. Environmental Drivers of Variation in Archaeal Community Structure
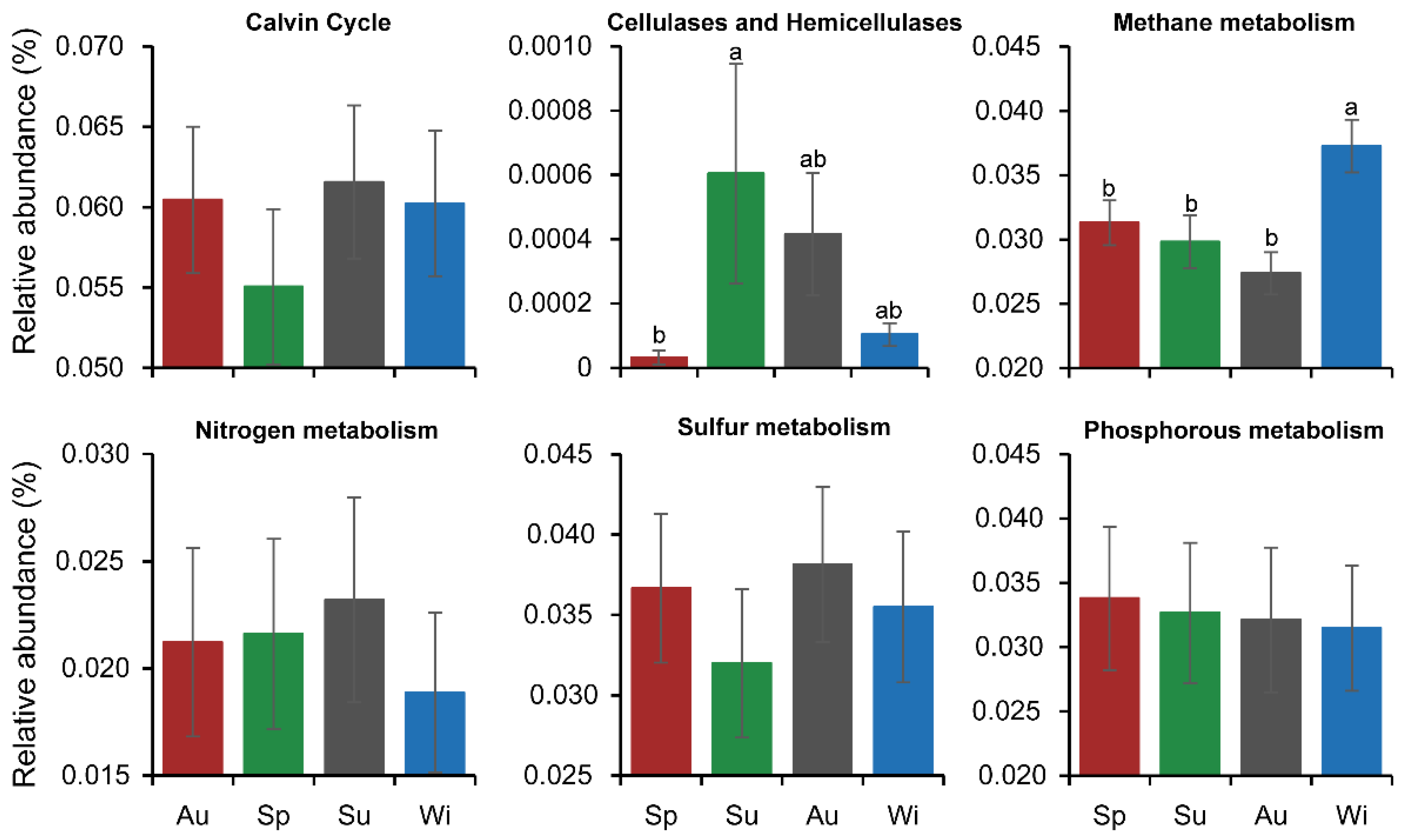
3.6. Seasonal Variation in the Functional Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orth, R.J.; Carruthers, T.J.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A Global Crisis for Seagrass Ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Ugarelli, K.; Chakrabarti, S.; Laas, P.; Stingl, U. The Seagrass Holobiont and Its Microbiome. Microorganisms 2017, 5, 81. [Google Scholar] [CrossRef]
- Mazarrasa, I.; Marbà, N.; Lovelock, C.E.; Serrano, O.; Lavery, P.S.; Fourqurean, J.W.; Kennedy, H.; Mateo, M.A.; Krause-Jensen, D.; Steven, A.D.L.; et al. Seagrass meadows as a globally significant carbonate reservoir. Biogeosciences 2015, 12, 4993–5003. [Google Scholar] [CrossRef]
- Guannel, G.; Arkema, K.; Ruggiero, P.; Verutes, G. The Power of Three: Coral Reefs, Seagrasses and Mangroves Protect Coastal Regions and Increase Their Resilience. PLoS ONE 2016, 11, e0158094. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.M.; Unsworth, R.K. Protecting the hand that feeds us: Seagrass (Zostera marina) serves as commercial juvenile fish habitat. Mar. Pollut. Bull. 2014, 83, 425–429. [Google Scholar] [CrossRef]
- Lilley, R.J.; Unsworth, R.K. Atlantic Cod (Gadus morhua) benefits from the availability of seagrass (Zostera marina) nursery habitat. Glob. Ecol. Conserv. 2014, 2, 367–377. [Google Scholar] [CrossRef]
- Reynolds, P.L.; Duffy, E.; Knowlton, N. Seagrass and Seagrass Beds. Available online: https://ocean.si.edu/ocean-leaf/plants-algae/seagrass-and-seagrass-beds (accessed on 22 March 2018).
- Unsworth, R.K.; Nordlund, L.M.; Cullen-Unsworth, L.C. Seagrass meadows support global fisheries production. Conserv. Lett. 2018, 12, e12566. [Google Scholar] [CrossRef]
- Fraser, M.W.; Statton, J.; Hovey, R.K.; Laverock, B.; Kendrick, G.A. Seagrass derived organic matter influences biogeochemistry, microbial communities, and seedling biomass partitioning in seagrass sediments. Plant Soil 2015, 400, 133–146. [Google Scholar] [CrossRef]
- Kannan, R.R.R.; Arumugam, R.; Anantharaman, P.; Rengasamy, K.R. Antibacterial potential of three seagrasses against human pathogens. Asian Pac. J. Trop. Med. 2010, 3, 890–893. [Google Scholar] [CrossRef]
- Lamb, J.B.; van de Water, J.A.J.M.; Bourne, D.G.; Altier, C.; Hein, M.Y.; Fiorenza, E.A.; Abu, N.; Jompa, J.; Harvell, C.D. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 2017, 355, 731–733. [Google Scholar] [CrossRef]
- Fahimipour, A.K.; Kardish, M.R.; Lang, J.M.; Green, J.L.; Eisen, J.A.; Stachowicz, J.J. Global-Scale Structure of the Eelgrass Microbiome. Appl. Environ. Microbiol. 2017, 83, e03391-16. [Google Scholar] [CrossRef]
- Könneke, M.; Bernhard, A.E.; José, R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, K.G.; Schreiber, L.; Petersen, D.G.; Kjeldsen, K.U.; Lever, M.A.; Steen, A.; Stepanauskas, R.; Richter, M.; Kleindienst, S.; Lenk, S.; et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 2013, 496, 215–218. [Google Scholar] [CrossRef]
- Orphan, V.; House, C.H.; Hinrichs, K.-U.; McKeegan, K.D.; DeLong, E.F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 2002, 99, 7663–7668. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, S.; Liu, X.; Yao, P.; Ge, T.; Zhang, X.-H. Spatiotemporal dynamics of the archaeal community in coastal sediments: Assembly process and co-occurrence relationship. ISME J. 2020, 14, 1463–1478. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Voerman, S.E.; Lang, J.M.; Stachowicz, J.J.; Eisen, J.A. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ 2017, 5, e3246. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wang, C.; Zhang, X.; Gong, J. Community Structure and Abundance of Archaea in a Zostera marina Meadow: A Comparison between Seagrass-Colonized and Bare Sediment Sites. Archaea 2019, 2019, 5108012. [Google Scholar] [CrossRef]
- Zheng, S.; Wang, B.; Xu, G.; Liu, F. Effects of Organic Phosphorus on Methylotrophic Methanogenesis in Coastal Lagoon Sediments With Seagrass (Zostera marina) Colonization. Front. Microbiol. 2020, 11, 1770. [Google Scholar] [CrossRef]
- Kalanetra, K.M.; Bano, N.; Hollibaugh, J.T. Ammonia-oxidizing Archaeain the Arctic Ocean and Antarctic coastal waters. Environ. Microbiol. 2009, 11, 2434–2445. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Majumder, N.S.; Basak, P.; Mukherji, S.; Roy, D.; Nag, S.; Haldar, A.; Chattopadhyay, D.; Mitra, S.; Bhattacharyya, M.; et al. Diversity and Distribution of Archaea in the Mangrove Sediment of Sundarbans. Archaea 2015, 2015, 968582. [Google Scholar] [CrossRef]
- Zhou, Z.; Meng, H.; Liu, Y.; Gu, J.-D.; Li, M. Stratified Bacterial and Archaeal Community in Mangrove and Intertidal Wetland Mudflats Revealed by High Throughput 16S rRNA Gene Sequencing. Front. Microbiol. 2017, 8, 2148. [Google Scholar] [CrossRef]
- Herfort, L.; Schouten, S.; Abbas, B.; Veldhuis, M.J.W.; Coolen, M.J.L.; Wuchter, C.; Boon, J.P.; Herndl, G.J.; Sinninghe Damsté, J.S. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. FEMS Microbiol. Ecol. 2007, 62, 242–257. [Google Scholar] [CrossRef][Green Version]
- Wong, A.H.L.; Visscher, P.; III, R.A.W.; Smith, D.-L.; Patterson, M.M.; Burns, B.P. Dynamics of archaea at fine spatial scales in Shark Bay mat microbiomes. Sci. Rep. 2017, 7, 46160. [Google Scholar] [CrossRef]
- Smith, A.C.; Kostka, J.E.; Devereux, R.; Yates, D.F. Seasonal composition and activity of sulfate-reducing prokaryotic communities in seagrass bed sediments. Aquat. Microb. Ecol. 2004, 37, 183–195. [Google Scholar] [CrossRef]
- Segovia, B.T.; Sanders-Smith, R.; Adamczyk, E.M.; Forbes, C.; Hessing-Lewis, M.; O’Connor, M.I.; Parfrey, L.W. Microeukaryotic Communities Associated With the Seagrass Zostera marina Are Spatially Structured. J. Eukaryot. Microbiol. 2021, 68, e12827. [Google Scholar] [CrossRef]
- Seyler, L.; McGuinness, L.M.; Kerkhof, L.J. Crenarchaeal heterotrophy in salt marsh sediments. ISME J. 2014, 8, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Teske, A.; Sørensen, K.B. Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2008, 2, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.; Parks, D.H.; Chadwick, G.; Robbins, S.J.; Orphan, V.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef]
- He, Y.; Li, M.; Perumal, V.; Feng, X.; Fang, J.; Xie, J.; Sievert, S.; Wang, F. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 2016, 1, 16035. [Google Scholar] [CrossRef]
- Lazar, C.S.; Baker, B.; Seitz, K.; Hyde, A.S.; Dick, G.J.; Hinrichs, K.-U.; Teske, A.P. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 2016, 18, 1200–1211. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, X.; Zhang, Q.; Liu, F.; Zhang, J.; Gong, J. Seagrass (Zostera marina) Colonization Promotes the Accumulation of Diazotrophic Bacteria and Alters the Relative Abundances of Specific Bacterial Lineages Involved in Benthic Carbon and Sulfur Cycling. Appl. Environ. Microbiol. 2015, 81, 6901–6914. [Google Scholar] [CrossRef] [PubMed]
- Pires, A.; Cleary, D.; Almeida, A.; Cunha, A.; Dealtry, S.; Mendonça-Hagler, L.C.S.; Smalla, K.; Gomes, N.C.M. Denaturing Gradient Gel Electrophoresis and Barcoded Pyrosequencing Reveal Unprecedented Archaeal Diversity in Mangrove Sediment and Rhizosphere Samples. Appl. Environ. Microbiol. 2012, 78, 5520–5528. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A language and environment for statistical computing. In Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2018; Volume 3.6.3. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Murali, A.; Bhargava, A.; Wright, E.S. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 2018, 6, 140. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 1–12. [Google Scholar] [CrossRef]
- Zhou, Z.; Pan, J.; Wang, F.; Gu, J.-D.; Li, M. Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 2018, 42, 639–655. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Cifuentes, A.; Antón, J.; Benlloch, S.; Donnelly, A.; Herbert, R.A.; Rodríguez-Valera, F. Prokaryotic Diversity in Zostera noltii Colonized Marine Sediments. Appl. Environ. Microbiol. 2000, 66, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Song, Z.; Zhang, H.; Liu, P.; Hu, X. Seagrass vegetation affect the vertical organization of microbial communities in sediment. Mar. Environ. Res. 2020, 162, 105174. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.S.; Biddle, J.; Meador, T.B.; Blair, N.; Hinrichs, K.-U.; Teske, A.P. Environmental controls on intragroup diversity of the uncultured benthic archaea of the miscellaneous Crenarchaeotal group lineage naturally enriched in anoxic sediments of the White Oak River estuary (North Carolina, USA). Environ. Microbiol. 2015, 17, 2228–2238. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Pan, J.; Liu, Z.; Zhang, C.; Liu, H.; Li, M. The Distribution of Bathyarchaeota in Surface Sediments of the Pearl River Estuary Along Salinity Gradient. Front. Microbiol. 2020, 11, 285. [Google Scholar] [CrossRef]
- Yu, T.; Wu, W.; Liang, W.; Lever, M.A.; Hinrichs, K.-U.; Wang, F. Growth of sedimentaryBathyarchaeotaon lignin as an energy source. Proc. Natl. Acad. Sci. USA 2018, 115, 6022–6027. [Google Scholar] [CrossRef]
- Davies, P.; Morvan, C.; Sire, O.; Baley, C. Structure and properties of fibres from sea-grass (Zostera marina). J. Mater. Sci. 2007, 42, 4850–4857. [Google Scholar] [CrossRef]
- Kaal, J.; Serrano, O.; Nierop, K.G.; Schellekens, J.; Cortizas, A.M.; Mateo, M.-Á. Molecular composition of plant parts and sediment organic matter in a Mediterranean seagrass (Posidonia oceanica) mat. Aquat. Bot. 2016, 133, 50–61. [Google Scholar] [CrossRef]
- Klap, V.; Hemminga, M.; Boon, J. Retention of lignin in seagrasses: Angiosperms that returned to the sea. Mar. Ecol. Prog. Ser. 2000, 194, 1–11. [Google Scholar] [CrossRef]
- Meng, J.; Xu, J.; Qin, D.; He, Y.; Xiao, X.; Wang, F. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 2014, 8, 650–659. [Google Scholar] [CrossRef]
- Pan, J.; Chen, Y.; Wang, Y.; Zhou, Z.; Li, M. Vertical Distribution of Bathyarchaeotal Communities in Mangrove Wetlands Suggests Distinct Niche Preference of Bathyarchaeota Subgroup 6. Microb. Ecol. 2019, 77, 417–428. [Google Scholar] [CrossRef]
- Virta, L.; Soininen, J.; Norkko, A. Stable Seasonal and Annual Alpha Diversity of Benthic Diatom Communities Despite Changing Community Composition. Front. Mar. Sci. 2020, 7, 88. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Wang, Y.; Shi, T.; Zhang, X.; Huang, G.; Gong, J. Intertidal zonation affects diversity and functional potentials of bacteria in surface sediments: A case study of the Golden Bay mangrove, China. Appl. Soil Ecol. 2018, 130, 159–168. [Google Scholar] [CrossRef]
- Kawashima, T.; Amano, N.; Koike, H.; Makino, S.-I.; Higuchi, S.; Kawashima-Ohya, Y.; Watanabe, K.; Yamazaki, M.; Kanehori, K.; Kawamoto, T.; et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 2000, 97, 14257–14262. [Google Scholar] [CrossRef] [PubMed]
- Walsh, D.A.; Papke, R.; Doolittle, W.F. Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 2005, 7, 1655–1666. [Google Scholar] [CrossRef]
- Valenzuela, E.I.; Prieto-Davó, A.; López-Lozano, N.E.; Eligio, A.H.; Vega-Alvarado, L.; Juárez, K.; García-González, A.S.; López, M.G.; Cervantes, F.J. Anaerobic Methane Oxidation Driven by Microbial Reduction of Natural Organic Matter in a Tropical Wetland. Appl. Environ. Microbiol. 2017, 83, e00645-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Jiang, N.; Liu, X.; Dong, X. Methanogenesis from Methanol at Low Temperatures by a Novel Psychrophilic Methanogen, “Methanolobus psychrophilus” sp. nov., Prevalent in Zoige Wetland of the Tibetan Plateau. Appl. Environ. Microbiol. 2008, 74, 6114–6120. [Google Scholar] [CrossRef] [PubMed]
- Dukunde, A.; Schneider, D.; Schmidt, M.; Veldkamp, E.; Daniel, R. Tree Species Shape Soil Bacterial Community Structure and Function in Temperate Deciduous Forests. Front. Microbiol. 2019, 10, 1519. [Google Scholar] [CrossRef] [PubMed]
- Fillol, M.; Sànchez-Melsió, A.; Gich, F.; Borrego, C.M. Diversity of Miscellaneous Crenarchaeotic Group archaea in freshwater karstic lakes and their segregation between planktonic and sediment habitats. FEMS Microbiol. Ecol. 2015, 91, fiv020. [Google Scholar] [CrossRef]
- Wemheuer, F.; Von Hoyningen-Huene, A.J.E.; Pohlner, M.; Degenhardt, J.; Engelen, B.; Daniel, R.; Wemheuer, B. Primary Production in the Water Column as Major Structuring Element of the Biogeographical Distribution and Function of Archaea in Deep-Sea Sediments of the Central Pacific Ocean. Archaea 2019, 2019, 3717239. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Song, Y.; Li, D.; Liu, R.; Niu, Q. The auto fluorescence characteristics, specific activity, and microbial community structure in batch tests of mono-chicken manure digestion. Waste Manag. 2019, 83, 57–67. [Google Scholar] [CrossRef]

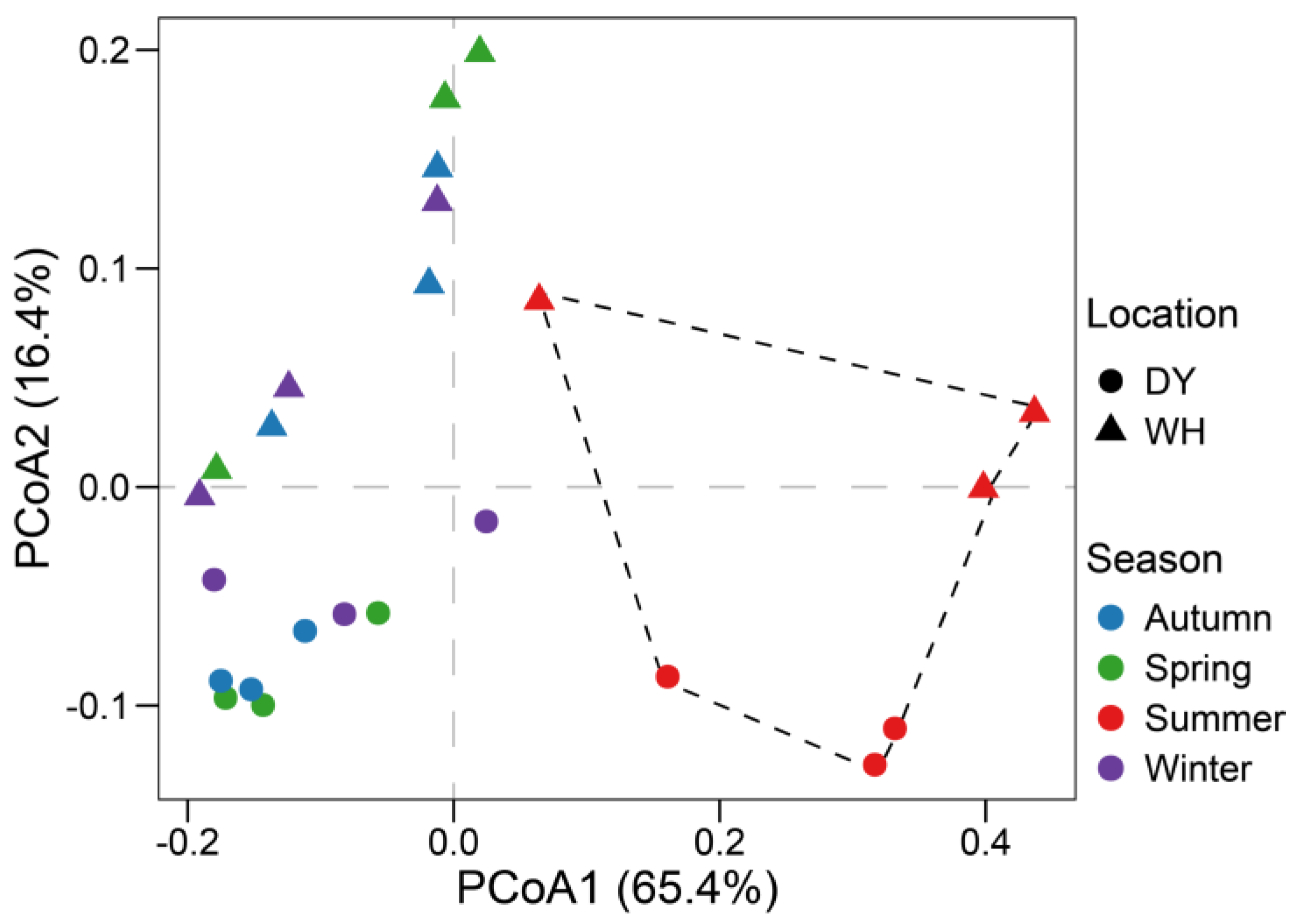
| Grouping | All Samples (n = 6) | Within WH (n = 3) | Within DY (n = 3) | |||
|---|---|---|---|---|---|---|
| R | p | R | p | R | p | |
| Season (global test) | 0.337 | 0.001 | 0.869 | 0.001 | 0.426 | 0.013 |
| Spring vs. Summer | 0.761 | 0.002 | 0.761 | 0.002 | 0.593 | 0.100 |
| Spring vs. Autumn | −0.02 | 0.452 | −0.020 | 0.452 | 0.111 | 0.500 |
| Spring vs. Winter | −0.013 | 0.432 | −0.013 | 0.432 | 0.037 | 0.300 |
| Summer vs. Autumn | 0.787 | 0.002 | 0.815 | 0.002 | 0.630 | 0.100 |
| Summer vs. Winter | 0.759 | 0.002 | 0.833 | 0.002 | 0.667 | 0.100 |
| Autumn vs. Winter | −0.169 | 0.959 | 0.792 | 0.100 | 0.111 | 0.500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhang, H.; Song, Z.; Huang, Y.; Hu, X. Seasonal Dynamics of Bathyarchaeota-Dominated Benthic Archaeal Communities Associated with Seagrass (Zostera japonica) Meadows. J. Mar. Sci. Eng. 2021, 9, 1304. https://doi.org/10.3390/jmse9111304
Liu P, Zhang H, Song Z, Huang Y, Hu X. Seasonal Dynamics of Bathyarchaeota-Dominated Benthic Archaeal Communities Associated with Seagrass (Zostera japonica) Meadows. Journal of Marine Science and Engineering. 2021; 9(11):1304. https://doi.org/10.3390/jmse9111304
Chicago/Turabian StyleLiu, Pengyuan, Haikun Zhang, Zenglei Song, Yanyan Huang, and Xiaoke Hu. 2021. "Seasonal Dynamics of Bathyarchaeota-Dominated Benthic Archaeal Communities Associated with Seagrass (Zostera japonica) Meadows" Journal of Marine Science and Engineering 9, no. 11: 1304. https://doi.org/10.3390/jmse9111304
APA StyleLiu, P., Zhang, H., Song, Z., Huang, Y., & Hu, X. (2021). Seasonal Dynamics of Bathyarchaeota-Dominated Benthic Archaeal Communities Associated with Seagrass (Zostera japonica) Meadows. Journal of Marine Science and Engineering, 9(11), 1304. https://doi.org/10.3390/jmse9111304





