TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Tissue Collection
2.2. Cloning of cDNA Encoding Putative Kawakawa slc24a5
2.3. RT-PCR Analysis of Kawakawa slc24a5 Transcripts in Adult Tissues and Early Stages of Embryos
2.4. Quantitative Real-Time PCR (qPCR)
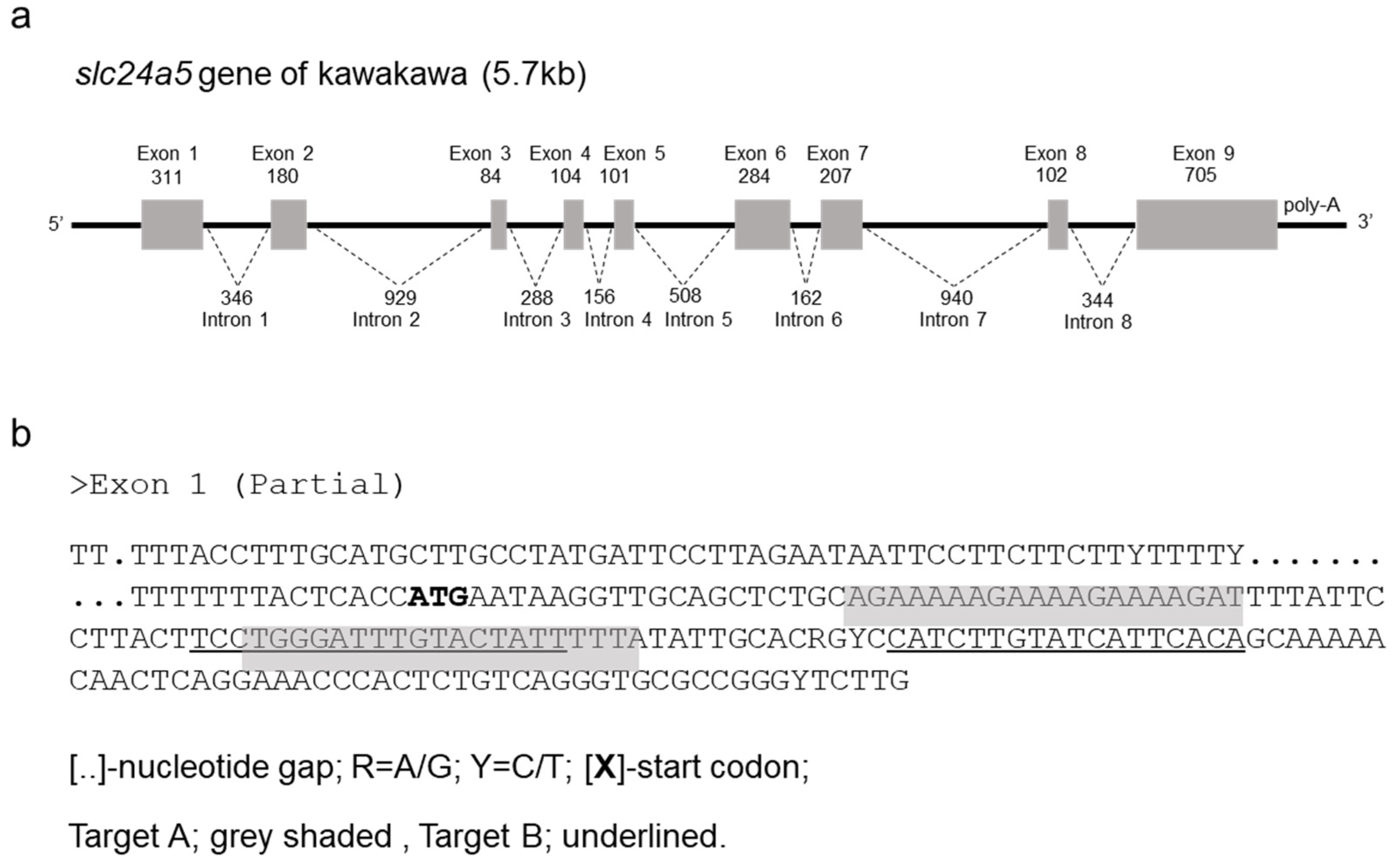
2.5. Genome Sequence and TALEN Design for Kawakawa slc24a5
2.6. Microinjection of ka-slc24a5 TALENs
2.7. Genotyping TALEN-Induced Mutations
2.8. Histology
2.9. Statistical Analysis
3. Results
3.1. Cloning and Characterization of Kawakawa slc24a5
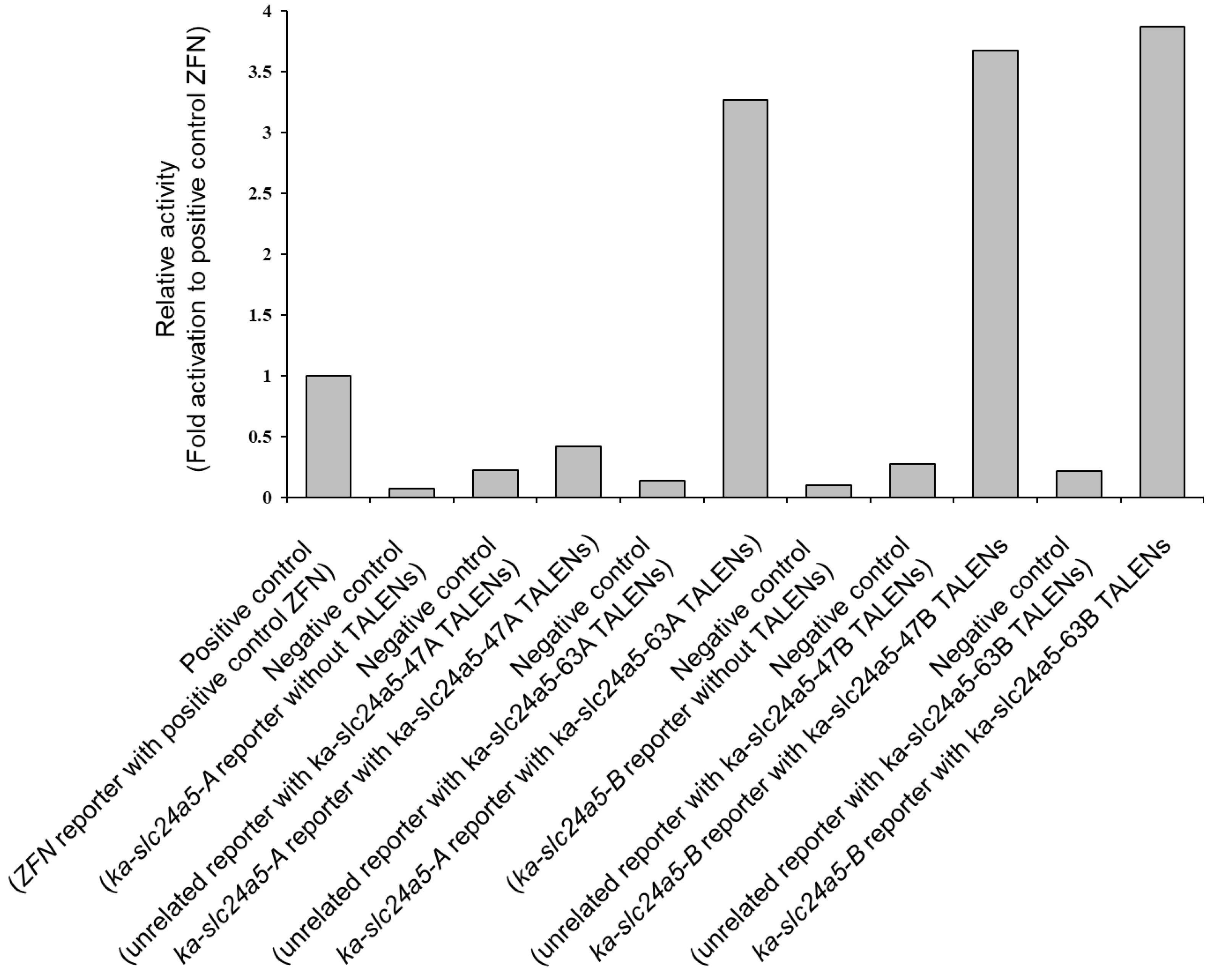
3.2. In Vitro Activity of Each TALEN by SSA Assay
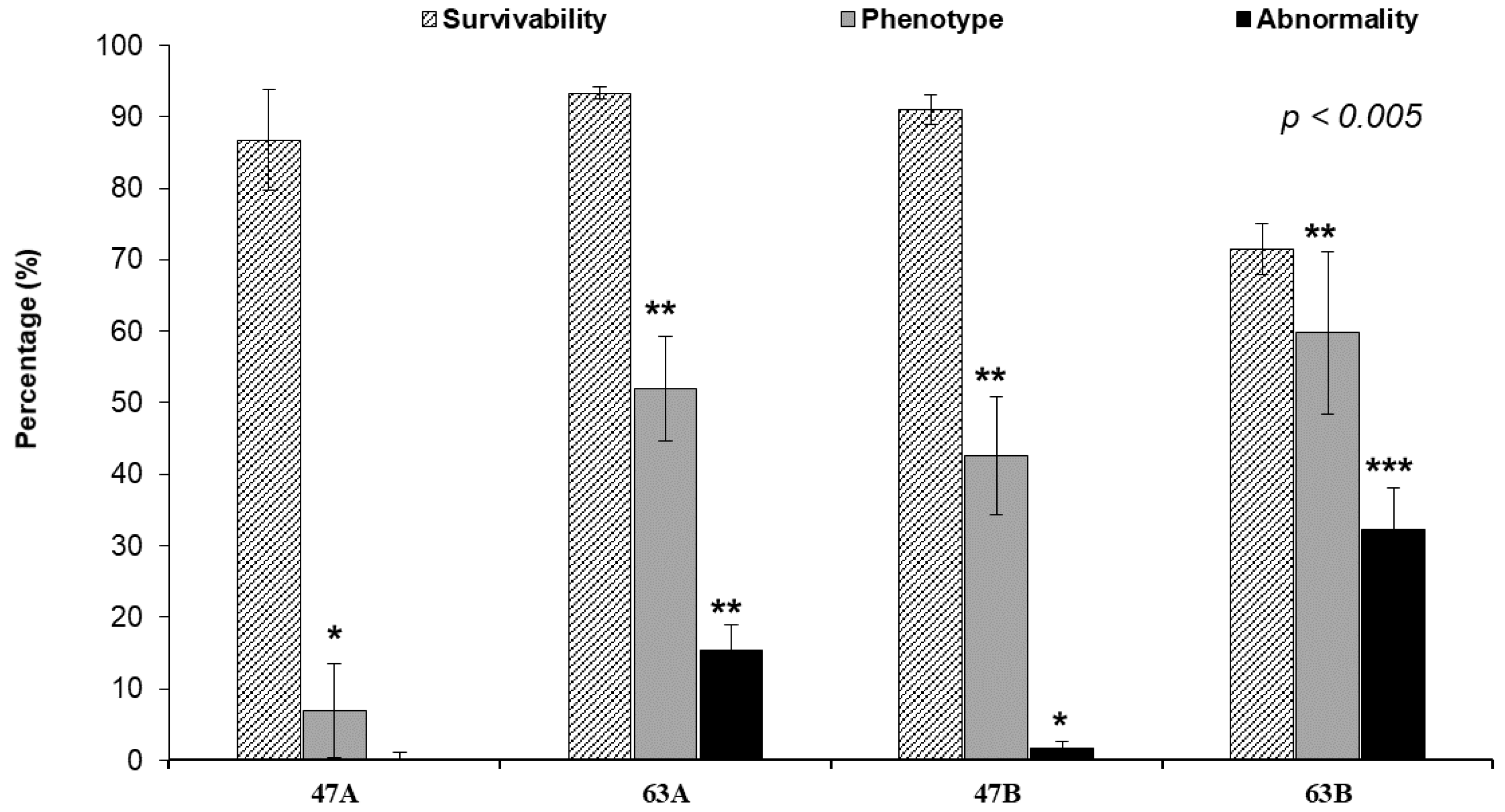
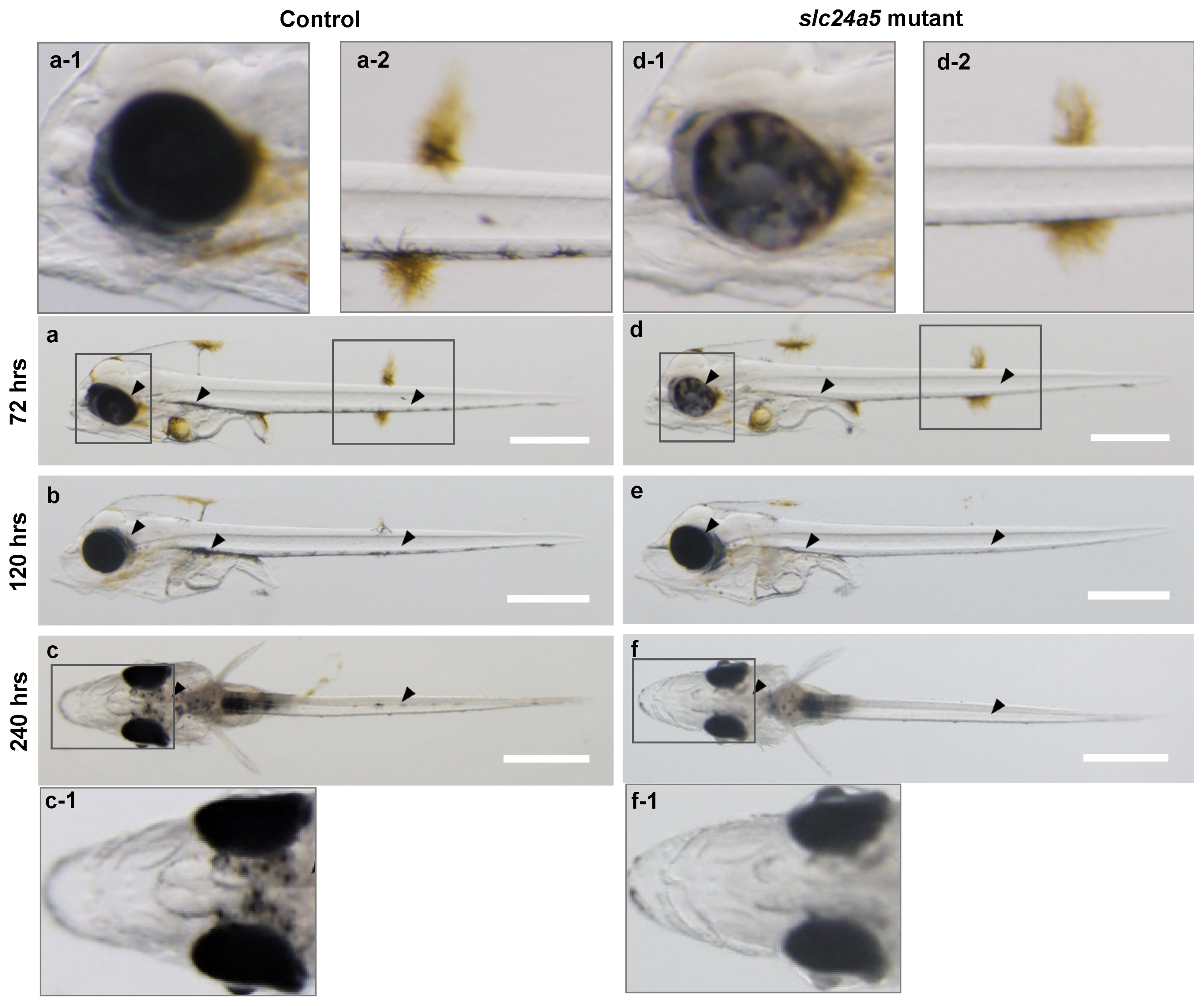
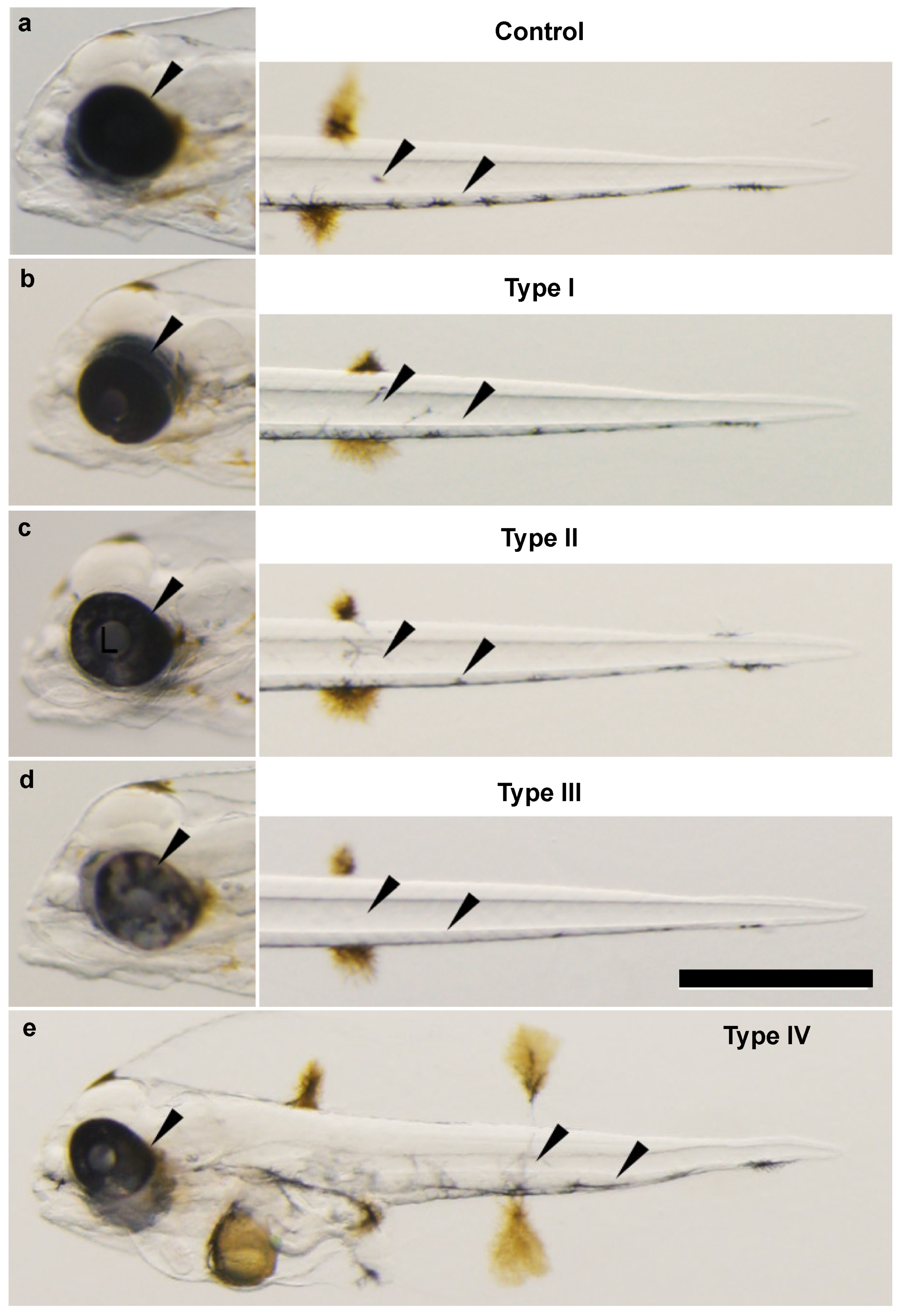
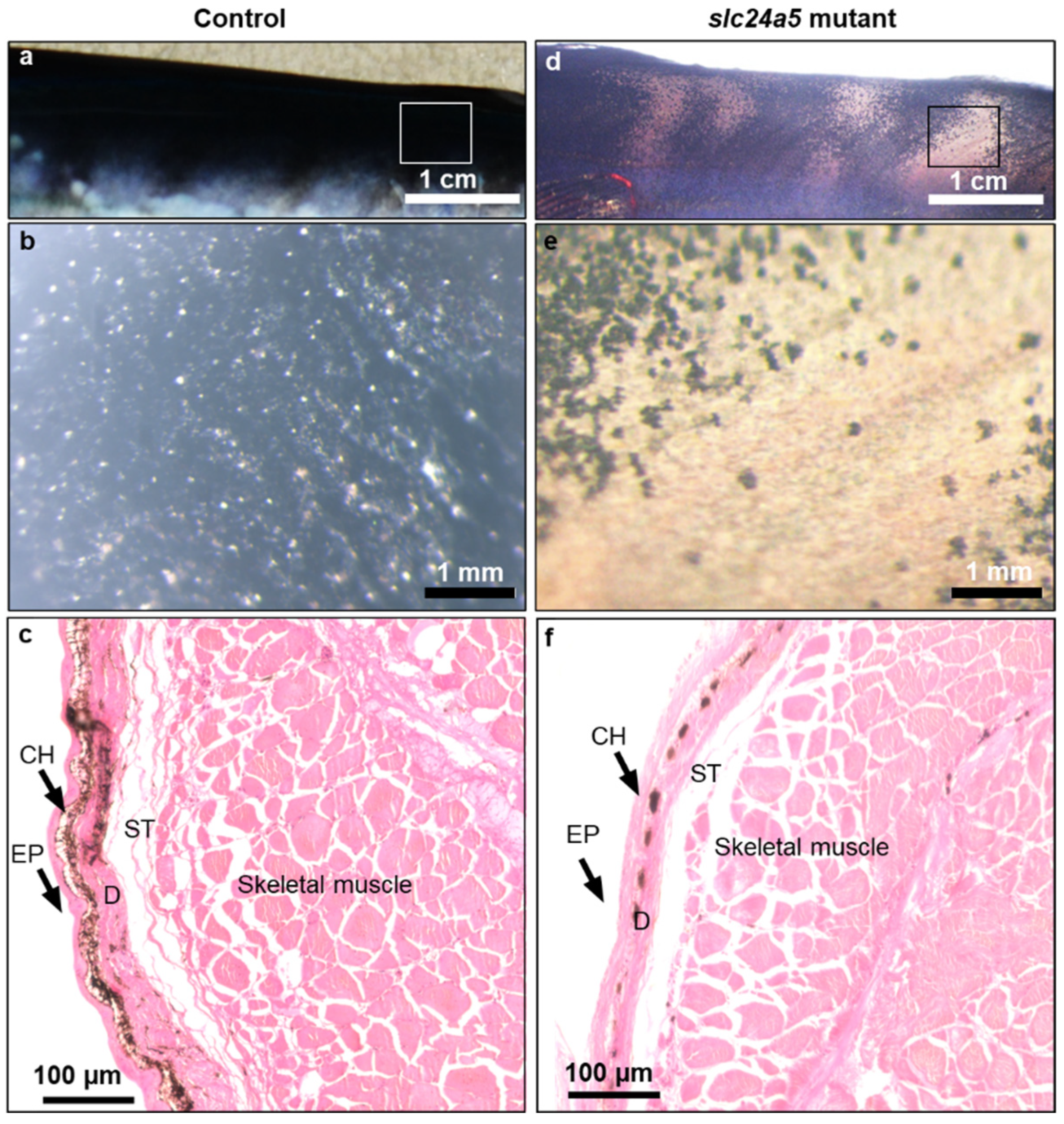
3.3. Phenotypic Change, Survival, and Abnormal Rate after Injecting Each TALENs RNA
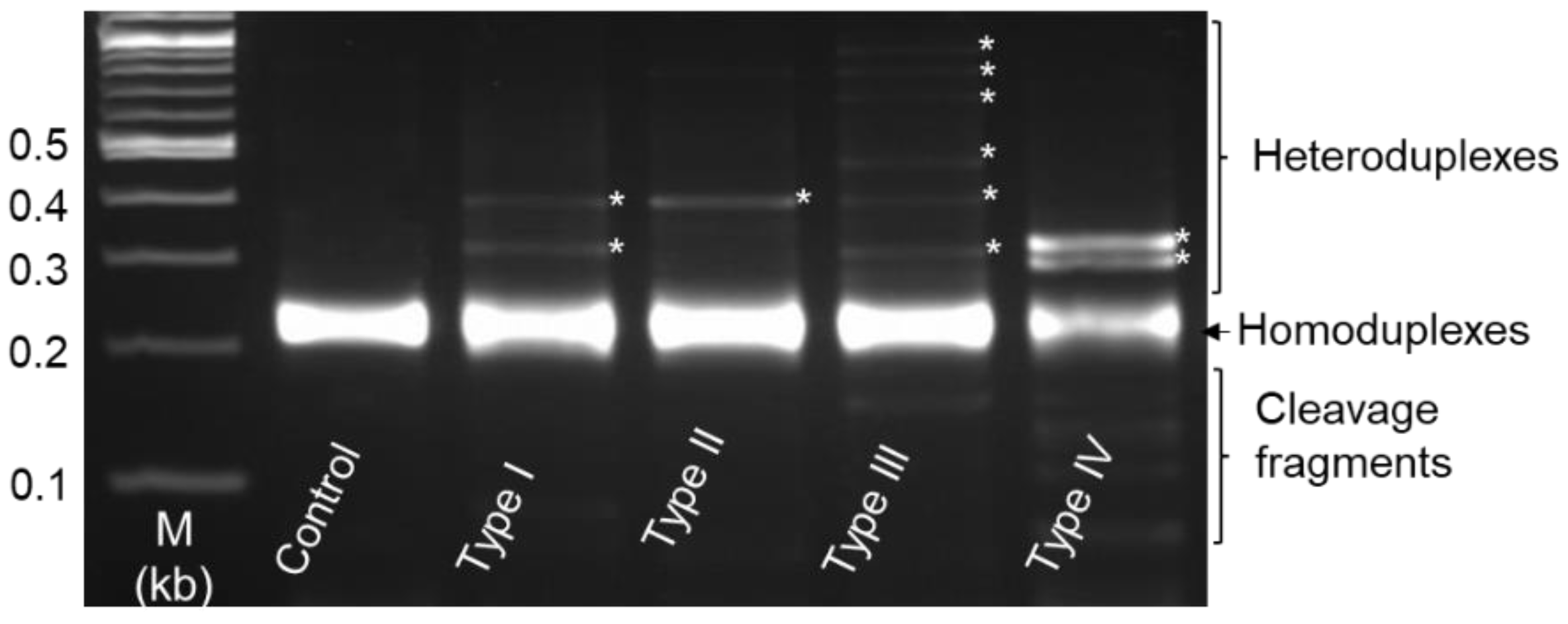
3.4. Heteroduplex Mobility Assay (HMA) and Genotype
3.5. Analysis of slc24a5 Transcripts by RT-PCR and Quantitative Real-Time PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collette, B.B.; Nauen, C.E. FAO Species Catalogue. An Annotated and Illustrated Catalogue of Tunas, Mackerels, Bonitos and Related Species Known to Date; Synopsis 125, Volume 2 Scombrids of the World; Food and Agriculture Organization of The United Nations: Rome, Italy, 1983; p. 137. [Google Scholar]
- Yesaki, M. A review of the biology and fisheries for Kawakawa (Euthynnus affinis) in the Indo-Pacific Region. Interact. Pac. Tuna Fish. 1994, 2, 3–11. [Google Scholar]
- Nissar, K.K.S.; Rashid, F.; Phadke, G.G.; Desai, A.Y. Reproductive biology of little tuna (Euthynnus affinis) in the Arabian sea. Ecol. Environ. Cons. 2015, 21, 115–118. [Google Scholar]
- Zhu, B.; Ge, W. Genome editing in fishes and their applications. Gen. Comp. Endocrinol. 2018, 257, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Jeffery, W.R.; Essner, J.J.; Kowalko, J.E. Genome Editing Using TALENs in Blind Mexican Cavefish, Astyanax mexicanus. PLoS ONE 2015, 10, e0119370. [Google Scholar] [CrossRef]
- Chakrapani, V.; Patra, S.K.; Panda, R.P.; Rasal, K.D.; Jayasankar, P.; Barman, H.K. Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. Dev. Comp. Immunol. 2016, 61, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.; Zhang, X.; Ren, J.; Dong, X.; Zhu, Z.; Jia, L.; Zhang, Q.; Li, W. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system. Sci. Rep. 2016, 6, 23496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y.; et al. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.C.; Kinoshita, M.; Ng, T.H.; Chang, Y.H.; Maekawa, S.; Chiang, Y.A.; Aoki, T.; Wang, H.C. Using CRISPR/Cas9-mediated gene editing to further explore growth and trade-off effects in myostatin mutated F4 medaka (Oryzias latipes). Sci. Rep. 2017, 7, 11435. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, K.; Washio, Y.; Yoshiura, Y.; Toyoda, A.; Ueno, T.; Fukuyama, H.; Kato, K.; Kinoshita, M. Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture 2018, 495, 415–427. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.; Du, J.; Si, Z.; Yang, H.; Xu, X.; Wang, C. ASIP disruption via CRISPR/Cas9 system induces black patches dispersion in Oujiang color common carp. Aquaculture 2019, 498, 230–235. [Google Scholar] [CrossRef]
- Sakaguchi, K.; Yoneda, M.; Sakai, N.; Nakashima, K.; Kitano, H.; Matsuyama, M. Comprehensive experimental system for a promising model organism candidate for marine teleosts. Sci. Rep. 2019, 9, 4948. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.A.; Li, T.; Yang, B.; Spalding, M.H. TALEN-mediated genome editing: Prospects and perspectives. Biochem. J. 2014, 462, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Yamamoto, T. CRISPR/Cas9: The leading edge of genome editing technology. In Targeted Genome Editing Using Site-Specific Nucleases; Yamamoto, T., Ed.; Springer: Tokyo, Japan, 2015; pp. 25–41. [Google Scholar] [CrossRef]
- Sakuma, T.; Yamamoto, T. Acceleration of cancer science with genome editing and related technologies. Cancer Sci. 2018, 109, 3679–3685. [Google Scholar] [CrossRef] [PubMed]
- Bogdanove, A.J.; Voytas, D.F. TAL Effectors: Customizable Proteins for DNA Targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef]
- Bedell, V.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.; Ii, R.G.K.; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.H.; et al. In vivo genome editing using a high-efficiency TALEN system. Nat. Cell Biol. 2012, 491, 114–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Y.; Guo, X.; Liu, Y.; Cao, Y.; Deng, Y.; Chen, X.; Cheng, C.H.K.; Dawid, I.B.; Chen, Y.; Zhao, H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. USA 2012, 109, 17484–17489. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef]
- Liu, J.; Li, C.; Yu, Z.; Huang, P.; Wu, H.; Wei, C.; Zhu, N.; Shen, Y.; Chen, Y.; Zhang, B.; et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genom. 2012, 39, 209–215. [Google Scholar] [CrossRef]
- Li, M.H.; Yang, H.H.; Li, M.R.; Sun, Y.L.; Jiang, X.L.; Xie, Q.P.; Wang, T.R.; Shi, H.J.; Sun, L.N.; Zhou, L.Y.; et al. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 2013, 154, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Ansai, S.; Sakuma, T.; Yamamoto, T.; Ariga, H.; Uemura, N.; Takahashi, R.; Kinoshita, M. Efficient targeted mutagenesis in medaka using custom-designed transcription activator-like effector nucleases. Genetics 2013, 193, 739–749. [Google Scholar] [CrossRef] [Green Version]
- Ansai, S.; Inohaya, K.; Yoshiura, Y.; Schartl, M.; Uemura, N.; Takahashi, R.; Kinoshita, M. Design, evaluation, and screening methods for efficient targeted mutagenesis with transcription activator-like effector nucleases in medaka. Dev. Growth Differ. 2014, 56, 98–107. [Google Scholar] [CrossRef] [Green Version]
- Bannister, S.; Antonova, O.; Polo, A.; Lohs, C.; Hallay, N.; Valinciute, A.; Raible, F.; Tessmar-Raible, K. TALENs mediate efficient and heritable mutation of endogenous genes in the marine annelid Platynereis dumerilii. Genetics 2014, 197, 77–89. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Ge, J.; Xu, Z.; Dong, X.; Cao, S.; Pan, J.; Zhao, Q. Generation of myostatin B knockout yellow catfish (Tachysurus fulvidraco) using transcription activator-like effector nucleases. Zebrafish 2014, 11, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barman, H.K.; Rasal, K.D.; Chakrapani, V.; Ninawe, A.S.; Vengayil, D.T.; Asrafuzzaman, S.; Sundaray, J.K.; Jayasankar, P. Gene editing tools: State-of-the-art and the road ahead for the model and non-model fishes. Transgenic Res. 2017, 26, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Irion, U.; Krauss, J.; Nüsslein-Volhard, C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development 2014, 141, 4827–4830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edvardsen, R.B.; Leininger, S.; Kleppe, L.; Skaftnesmo, K.O.; Wargelius, A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS ONE 2014, 9, e108622. [Google Scholar] [CrossRef]
- Williams, R.M.; Winkfein, R.J.; Ginger, R.S.; Green, M.R.; Schnetkamp, P.P.; Wheeler, G.N. A functional approach to understanding the role of NCKX5 in Xenopus pigmentation. PLoS ONE 2017, 12, e0180465. [Google Scholar] [CrossRef] [Green Version]
- Parichy, D.M. Pigment patterns: Fish in stripes and spots. Curr. Biol. 2003, 13, 947–950. [Google Scholar] [CrossRef]
- Horstadius, S. The Neural Crest: Its properties and Derivatives in the Light of Experimental Research; Oxford University Press: Oxford, UK, 1950. [Google Scholar]
- Weston, J.A. A radio autographic analysis of the migration and localization of trunk neural crest cells in the chick. Dev. Biol. 1963, 6, 279–310. [Google Scholar] [CrossRef]
- Bronner-Fraser, M.; Fraser, S. Developmental potential of avian trunk neural crest cells in situ. Neuron 1989, 3, 755–766. [Google Scholar] [CrossRef]
- Braasch, I.; Schartl, M.; Volff, J.N. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 2007, 7, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamason, R.L.; Mohideen, M.P.K.; Mest, J.R.; Wong, A.C.; Norton, H.L.; Aros, M.C.; Jurynec, M.J.; Mao, X.; Humphreville, V.R.; Humbert, J.E.; et al. Slc24a5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005, 310, 1782–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streisinger, G.; Walker, C.; Dower, N.; Knauber, D.; Singer, F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 1981, 291, 293–296. [Google Scholar] [CrossRef]
- Nicolaus, R.A. Melanins; Lederer, E., Ed.; Chemistry of Natural Products Series; Hermann: Paris, France, 1968. [Google Scholar]
- Prota, G. Melanins and Melanogenesis; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- D’Ischia, M.; Wakamatsu, K.; Napolitano, A.; Briganti, S.; Borron, J.C.G.; Kovacs, D.; Meredith, P.; Pezzella, A.; Picardo, M.; Sarna, T.; et al. Melanins and melanogenesis: Methods, standards, protocols. Pigment. Cell Melanoma Res. 2013, 26, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Solano, F. Melanins: Skin pigments and much more-types, structural models, biological functions, and formation routes. New J. Sci. 2014, 28, 498276. [Google Scholar] [CrossRef] [Green Version]
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspengren, S.; Hedberg, D.; Sköld, H.N.; Wallin, M. New insights into melanosome transport in vertebrate pigment cells. Int. Rev. Cell Mol. Biol. 2009, 272, 245–302. [Google Scholar] [CrossRef]
- Hinaux, H.; Bachem, K.; Battistara, M.; Rossi, M.; Xin, Y.; Jaenichen, R.; Yann Le Poul, Y.L.; Arnoult, L.; Kobler, J.M.; Kadow, I.C.G.; et al. Revisiting the developmental and cellular role of the pigmentation gene yellow in Drosophila using a tagged allele. Dev. Biol. 2018, 438, 111–123. [Google Scholar] [CrossRef]
- Sakuma, T.; Ochiai, H.; Kaneko, T.; Mashimo, T.; Tokumasu, D.; Sakane, Y.; Suzuki, K.I.; Miyamoto, T.; Sakamoto, N.; Matsuura, S.; et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci. Rep. 2013, 3, 3379. [Google Scholar] [CrossRef] [Green Version]
- Havelka, M.; Sawayama, E.; Saito, T.; Yoshitake, K.; Saka, D.; Ineno, T.; Asakawa, S.; Takagi, M.; Goto, R.; Matsubara, T. Chromosome-Scale Genome Assembly and Transcriptome Assembly of Kawakawa Euthynnus affinis: A Tuna-Like Species. Front. Genet. 2021, 12, 739781. [Google Scholar] [CrossRef]
- Doyle, E.L.; Booher, N.J.; Standage, D.S.; Voytas, D.S.; Brendel, V.P.; VanDyk, J.K.; Bagdanove, A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0, Tools for TAL effector design and target prediction. Nucleic Acids Res. 2012, 40, W117–W122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, T.; Hosoi, S.; Woltjen, K.; Suzuki, K.; Kashiwagi, K.; Wada, H.; Ochiai, H.; Miyamoto, T.; Kawai, N.; Sasakura, Y.; et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 2013, 18, 315–326. [Google Scholar] [CrossRef]
- Goto, R.; Saito, T.; Matsubara, T.; Yamaha, E. Microinjection of marine fish eggs. In Microinjection. Methods in Molecular Biology; Liu, C., Du, Y., Eds.; Humana Press: New York, NY, USA, 2019; Volume 1874. [Google Scholar] [CrossRef]
- Sander, J.D.; Cade, L.; Khayter, C.; Reyon, D.; Peterson, R.T.; Joung, J.K.; Yeh, J.R. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011, 29, 697–698. [Google Scholar] [CrossRef]
- Ota, S.; Hisano, Y.; Muraki, M.; Hoshijima, K.; Dahlem, T.J.; Grunwald, D.J.; Okada, Y.; Kawahara, A. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 2013, 1818, 450–458. [Google Scholar] [CrossRef]
- Darias, M.J.; Andree, K.B.; Boglino, A.; Fernandez, I.; Estevez, A.; Gisbert, E. Coordinated regulation of chromatophore differentiation and melanogenesis during the ontogeny of skin pigmentation of solea senegalensis (Kaup, 1858). PLoS ONE 2013, 8, e63005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. J. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef] [Green Version]
- Dahlem, T.J.; Hoshijima, K.; Jurynec, M.J.; Gunther, D.; Starker, C.G.; Locke, A.S.; Weis, A.M.; Voytas, D.F.; Grunwald, D.J. Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome. PLoS Genet. 2012, 8, e1002861. [Google Scholar] [CrossRef] [Green Version]
- Juan, L.; Wei, Z.; Bing, Y.; Xiao-Xiang, H.; Ning, L. cDNA cloning, expression analysis of slc24a5 and its relationship with melanin deposition in chicken. Prog. Biochem. Biophys. 2008, 35, 69–76. [Google Scholar]
- Liu, Y.; Luo, D.; Lei, Y.; Hu, W.; Zhao, H.; Cheng, C.H. A highly effective TALEN-mediated approach for targeted gene disruption in Xenopus tropicalis and zebrafish. Methods 2014, 69, 58–66. [Google Scholar] [CrossRef]
- Higuchi, K.; Kazeto, Y.; Ozaki, Y.; Yamaguchi, T.; Shimada, Y.; Ina, Y.; Soma, S.; Sakakura, Y.; Goto, R.; Matsubara, T.; et al. Targeted mutagenesis of the ryanodine receptor by platinum TALENs causes slow swimming behavior in Pacific bluefin tuna (Thunnus orientalis). Sci. Rep. 2019, 9, 13871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | Direction | Nucleotide Sequence | TM | Use and Its Target |
|---|---|---|---|---|
| t_F1 | Sense | 5′-GAGTTTCCTGACGGCTTCTTC-3′ | 58 | bluefin tuna specific for slc24a5 |
| t_F2 | Sense | 5′-CAGGAGCAACATTTATGGCAG-3′ | 56 | bluefin tuna specific for slc24a5 |
| t_R1 | Anti-sense | 5′-CACCGACAACAGTGACCATC-3′ | 57 | bluefin tuna specific for slc24a5 |
| t_R2 | Anti-sense | 5′-GCAGGCTGAGTGAGAGATGAG-3′ | 60 | bluefin tuna specific for slc24a5 |
| 5′RACE_R1 | Anti-sense | 5′-AGTACCCCGAGTCGTCTTGGAAGATC-3′ | 63 | 5′-Race for slc24a5 cDNA |
| 5′RACE_Rn | Anti-sense | 5′-CTGGCAGCTTCATACCAATACACCTTG-3′ | 62 | 5′-Race for slc24a5 cDNA |
| 3′RACE_F1 | Sense | 5′-GCTTATTTTCTACATGCTTTTGGCTGTCG-3′ | 63 | 3′-Race for slc24a5 cDNA |
| 3′RACE_Fn | Sense | 5′-GTGGGGTCAGCAGTCTACAACCTCTTAG-3′ | 64 | 3′-Race for slc24a5 cDNA |
| ORF_F | Sense | 5′-GCATGCTTGCCTATGATTCC-3′ | 57 | ORF of slc24a5 cDNA |
| ORF_R | Anti-sense | 5′-GTCCTGGGAAGGCAAGTTTC-3′ | 56 | ORF of slc24a5 cDNA |
| TD_F | Sense | 5′-GAGTTTCCTGACGGCTTCTTC-3′ | 59 | RT-PCR of slc24a5 |
| TD_R | Anti-sense | 5′-GCGAAGGTCAAAGCACAGAAC-3′ | 60 | RT-PCR of slc24a5 |
| genome_F | Sense | 5′-CACACAGCTTCAAGCCACAT-3′ | 55 | exon1 of slc24a5 |
| genome_R | Anti-sense | 5′-AGAAGCCGTCAGGAAACTCA-3′ | 55 | exon1 of slc24a5 |
| HMA_F | Sense | 5′-CTTTGCATGCTTGCCTATGA-3′ | 58 | RT-PCR, HMA assay |
| HMA_R | Anti-sense | 5′-TGACAGAGTGGGTTTCCTGA-3′ | 58 | RT-PCR, HMA assay |
| qPCR_F | Sense | 5′-GCTGGGAGTTCTGCACCTGA-3′ | 60 | qPCR of slc24a5 |
| qPCR_R | Anti-sense | 5′-GCCATAGAGGCCAAGAGTCC-3′ | 60 | qPCR of slc24a5 |
| β-act_F1 | Sense | 5′-CAGGCATCAGGGAGTGATGG-3′ | 57 | primer for internal control |
| β-act_R1 | Anti-sense | 5′-GCTCGATGGGGTACTTCAGG-3′ | 58 | primer for internal control |
| Number of Embryo | |||||
|---|---|---|---|---|---|
| Experimental Group | Total | Survived (%) | Normal Development (%) | Abnormal Development (%) ‘Type IV’ | |
| slc24a5 47A | Control | 270 | 242 (89.6) | 242 (100.0) | 0 (00.0) |
| Injected | 78 | 68 (87.2) | 68 (100.0) | 0 (00.0) | |
| slc24a5 63A | Control | 149 | 142 (95.3) | 142 (100.0) | 0 (00.0) |
| Injected | 140 | 130 (92.9) | 110 (084.6) | 20 (15.4) | |
| slc24a5 47B | Control | 113 | 108 (95.6) | 113 (100.0) | 0 (00.0) |
| Injected | 67 | 61 (91.0) | 60 (098.4) | 1 (01.6) | |
| slc24a5 63B | Control | 304 | 285 (93.8) | 285 (100.0) | 0 (00.0) |
| Injected | 122 | 87 (71.3) | 59 (067.8) | 28 (32.2) | |
| Target Group | Sequences (5′-3′) | Genotype (%) | |
|---|---|---|---|
| Group A | Wild | tctgcagaaaaagaaaagaAAAGATTTTATTCCTtacttcctgggatttgta | (10/10) |
| slc24a5, 47A | Type II | TCTGCAGAAAAAGAAAAGAAAAGAWTTTATTCCTTACTTCCTGGGATTTGTA | 4/15 (27%) |
| slc24a5, 63A | Type II | TCTGCAGAAAAAGAAAAGAWAARWTTTTATTCCTTACTTCCTGGGATTTGTA | 7/15 (47%) |
| TCTGCAGAAAAAGAAAAGAAAAR----------TTACTTCCTGGGATTTGTA | |||
| Group B | Wild | tcctgggatttgtactattTTTATATTGCACRGYCcatcttgtatcattcaca | (10/10) |
| slc24a5, 47B | Type I | TCCTGGGATTTGTACTATTTTTWccaTATTGCAYRGYCCATCTTGTATCATTCACA | 7/10 (70%) |
| Type I | TCCTGGGATTTGTACTATTTT-------CACG---CATCTTGTATCATTCACA | ||
| Type II | TCCTGGGATTTGTACTATTTTTWTATWGCtatttACGGTCCATCTTGTATCATTCACA | 9/10 (90%) | |
| Type II | TCCTGGGATTTGTACTATT----------------CATCTTGTATCATTCACA | ||
| Type III | TCCTGGGATTTGTACTATTTTTATActatttTTGCACRGYCCATCTTGTATCATTCACA | 10/10 (100%) | |
| Type III | TCCTGGGATTTGTACTATT------------------TCTTGTATCATTCACA | ||
| slc24a5, 63B | Type I | TCCTGGGATTTGTACTATTTTTWTATTGCACRGYCCATCTTGTATCATTCACA | 6/10 (60%) |
| Type I | TCCTGGGATTTGTACTATTTT-------------CCATCTTGTATCATTCACA | ||
| Type II | TCCTGGGATTTGTACTATTTTTAYDWWGCACSGTCCATCTTGTATCATTCACA | 7/10 (70%) | |
| Type II | TCCTGGGATTTGTACTATT----------------CATCTTGTATCATTCACA | ||
| Type III | TCCTGGGATTTGTACTATT----------------CATCTTGTATCATTCACA | 7/10 (70%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, D.; Matsubara, T.; Saito, T.; Kazeto, Y.; Gen, K.; Sakuma, T.; Yamamoto, T.; Mekuchi, M.; Goto, R. TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis. J. Mar. Sci. Eng. 2021, 9, 1378. https://doi.org/10.3390/jmse9121378
Pandey D, Matsubara T, Saito T, Kazeto Y, Gen K, Sakuma T, Yamamoto T, Mekuchi M, Goto R. TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis. Journal of Marine Science and Engineering. 2021; 9(12):1378. https://doi.org/10.3390/jmse9121378
Chicago/Turabian StylePandey, Dipak, Takahiro Matsubara, Taiju Saito, Yukinori Kazeto, Koichiro Gen, Tetsushi Sakuma, Takashi Yamamoto, Miyuki Mekuchi, and Rie Goto. 2021. "TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis" Journal of Marine Science and Engineering 9, no. 12: 1378. https://doi.org/10.3390/jmse9121378
APA StylePandey, D., Matsubara, T., Saito, T., Kazeto, Y., Gen, K., Sakuma, T., Yamamoto, T., Mekuchi, M., & Goto, R. (2021). TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa, Euthynnus affinis. Journal of Marine Science and Engineering, 9(12), 1378. https://doi.org/10.3390/jmse9121378








