Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Vaccine Candidates
Abstract
1. Introduction
2. Materials and Methods
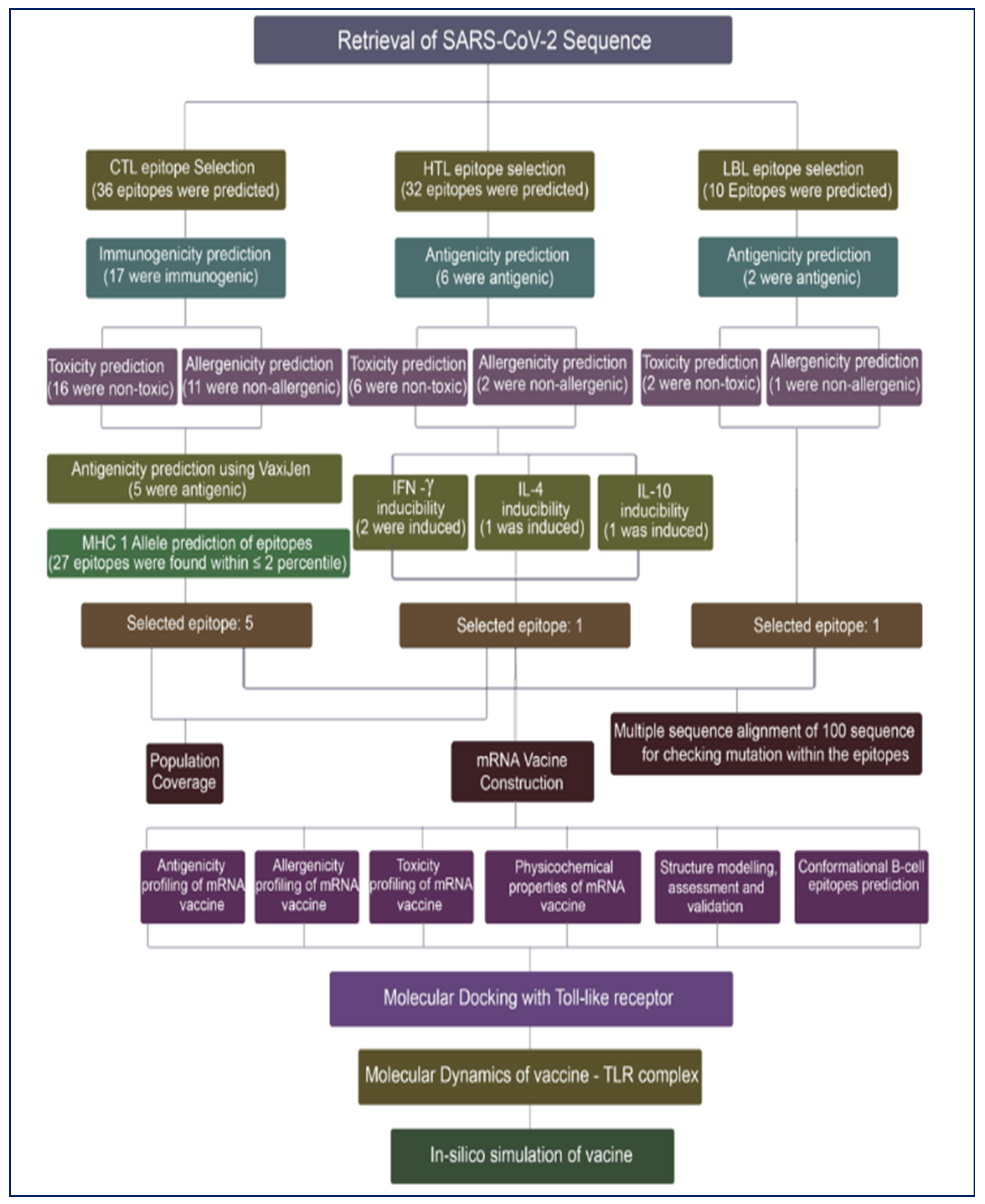
2.1. Study Design
2.2. Retrieval of SARS-CoV-2 Nucleotide Sequence
2.3. Prediction and Evaluation of Cytotoxic T-Lymphocytes (CTL) Epitopes
2.4. Prediction and Evaluation of Helper T-Lymphocytes (HTL) Epitopes
2.5. Projection and Evaluation of Linear B-Cells Lymphocytes (LBL) Epitopes
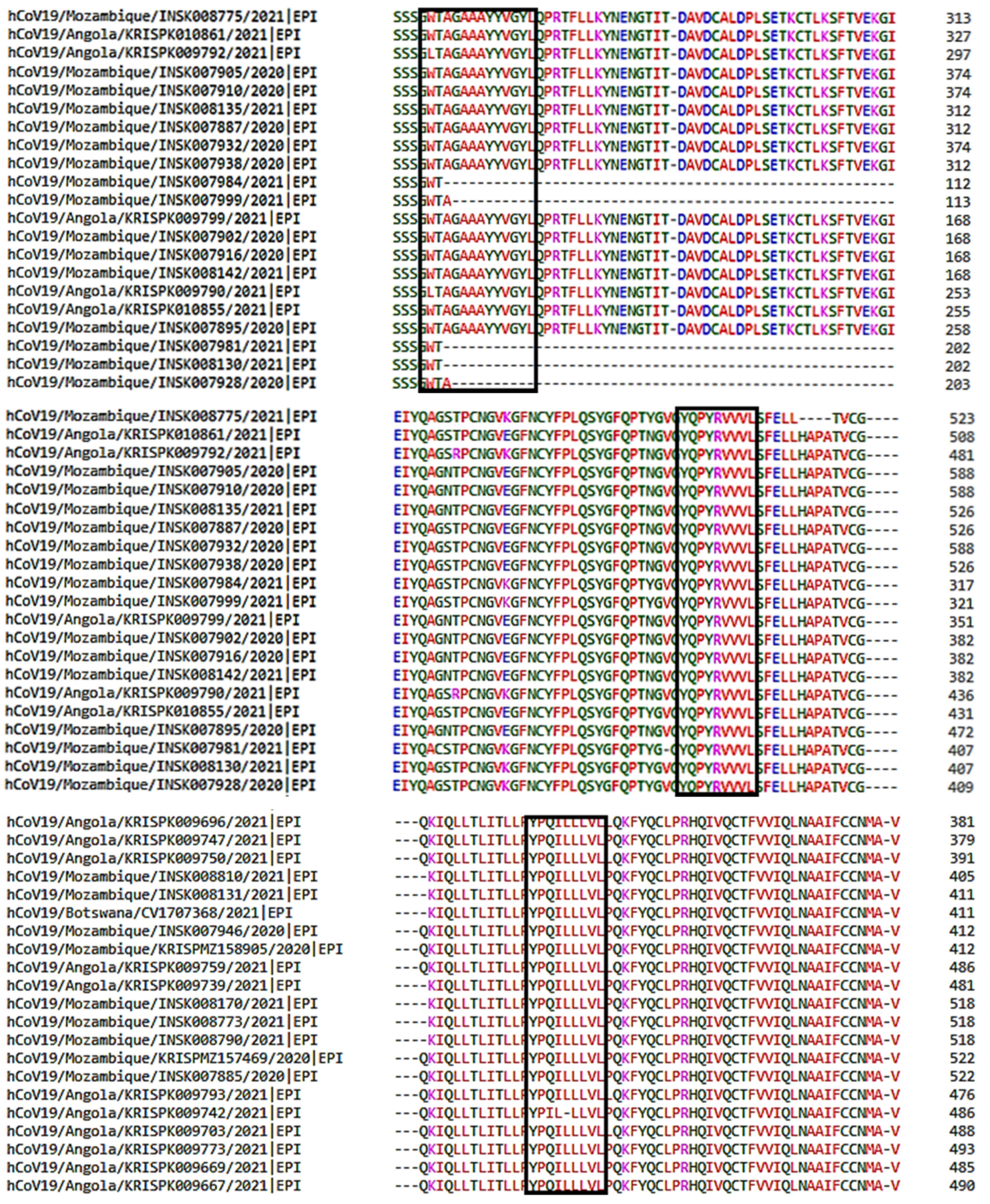
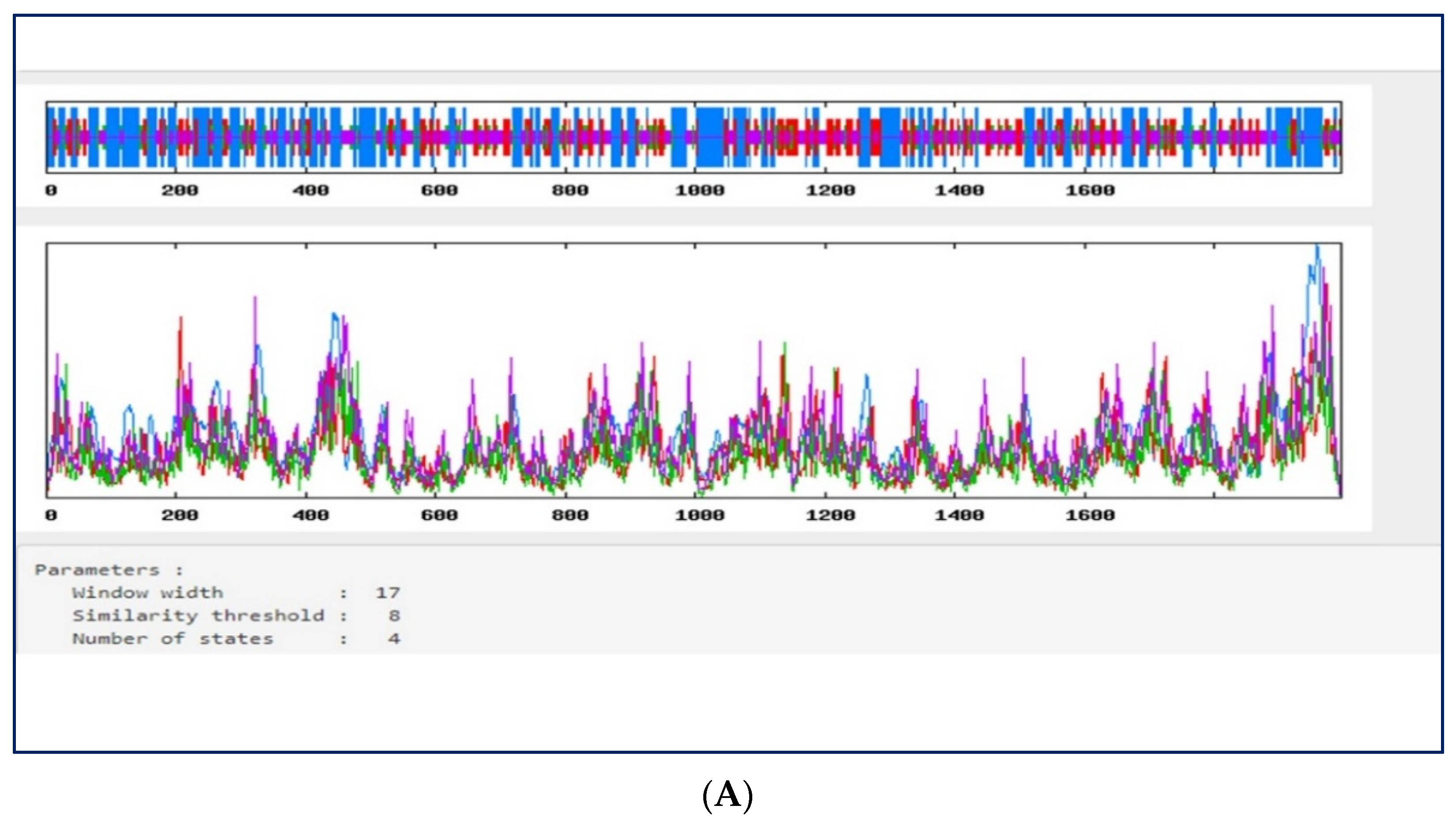
2.6. Multiple Sequence Alignment of SARS-CoV-2 Nucleotide Sequence
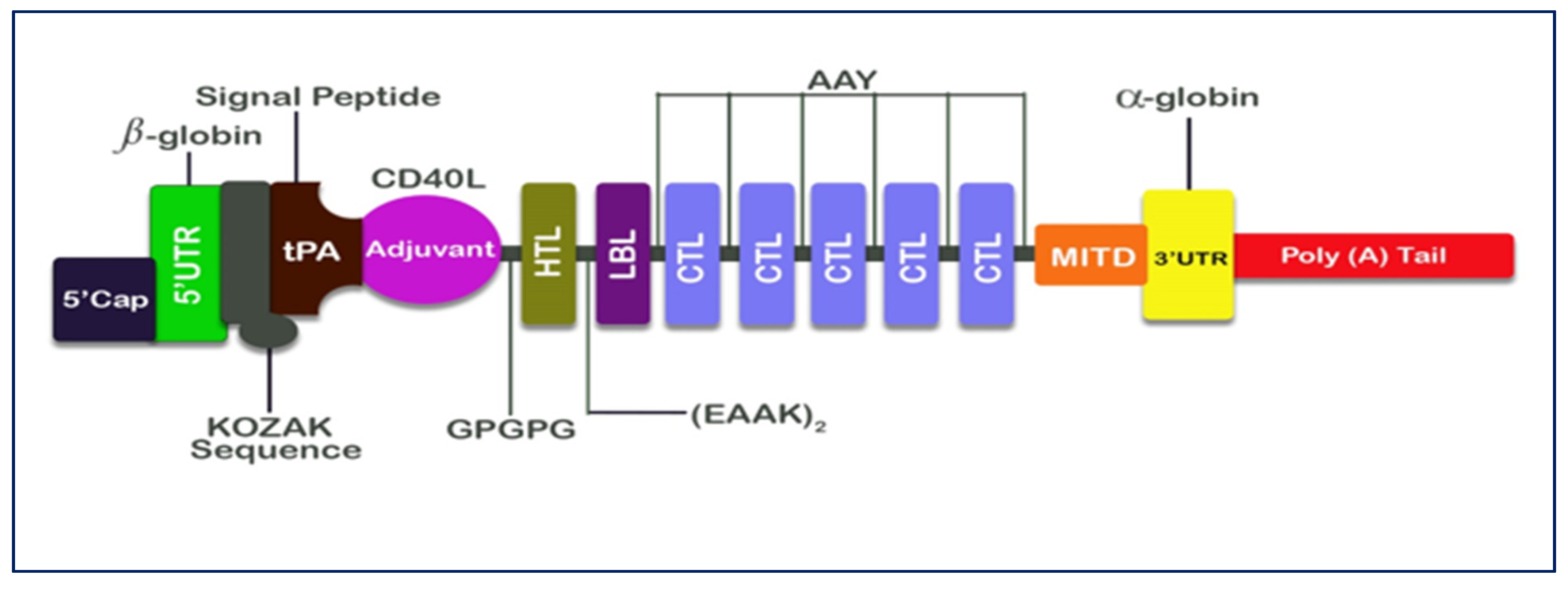
2.7. Designing of mRNA Vaccine
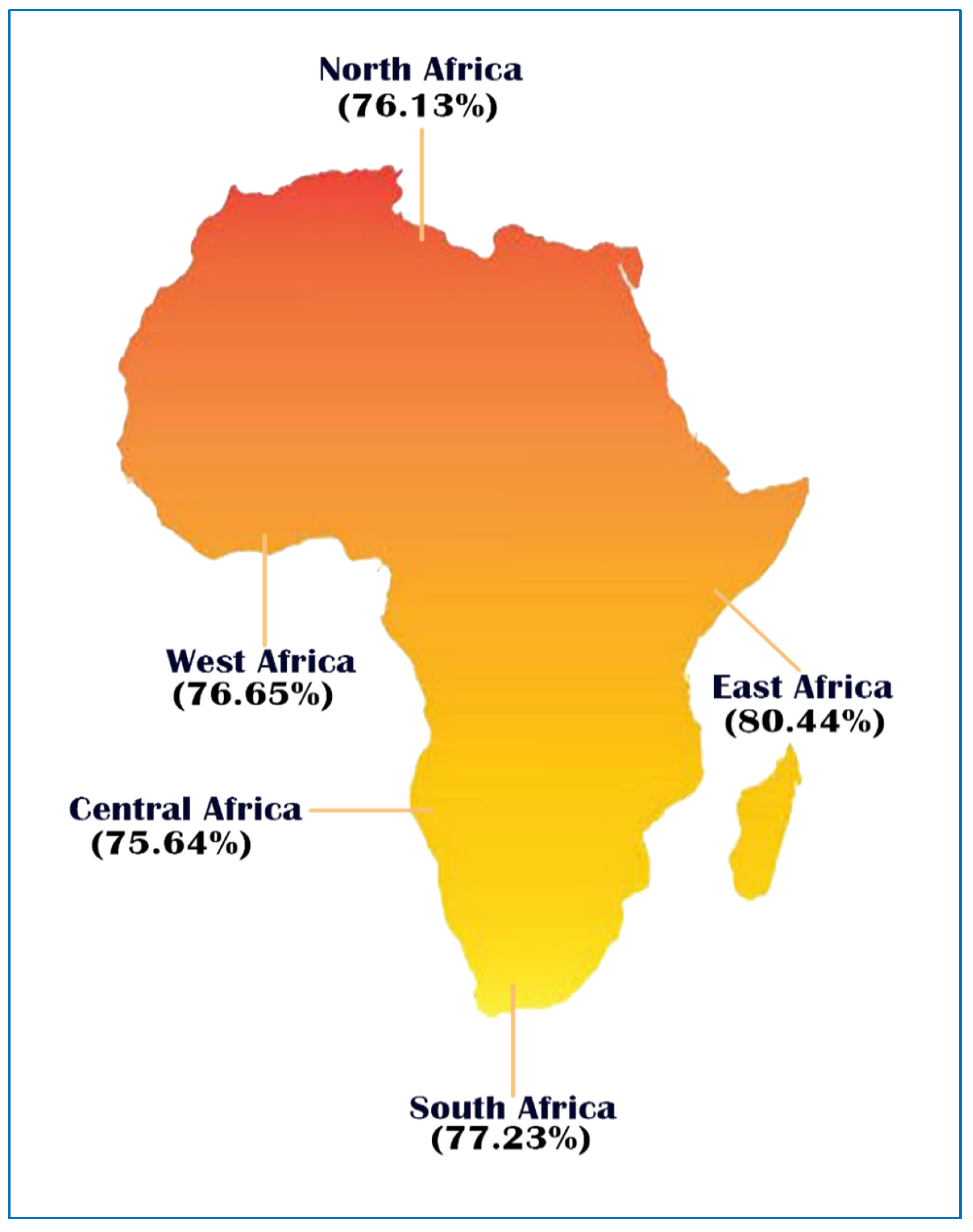
2.8. Prediction of Class I and Class II Epitopes’ Population Coverage
2.9. Vaccine Construct’s Predicted Antigenicity, Toxicity, Physicochemical Properties, and Allergenicity
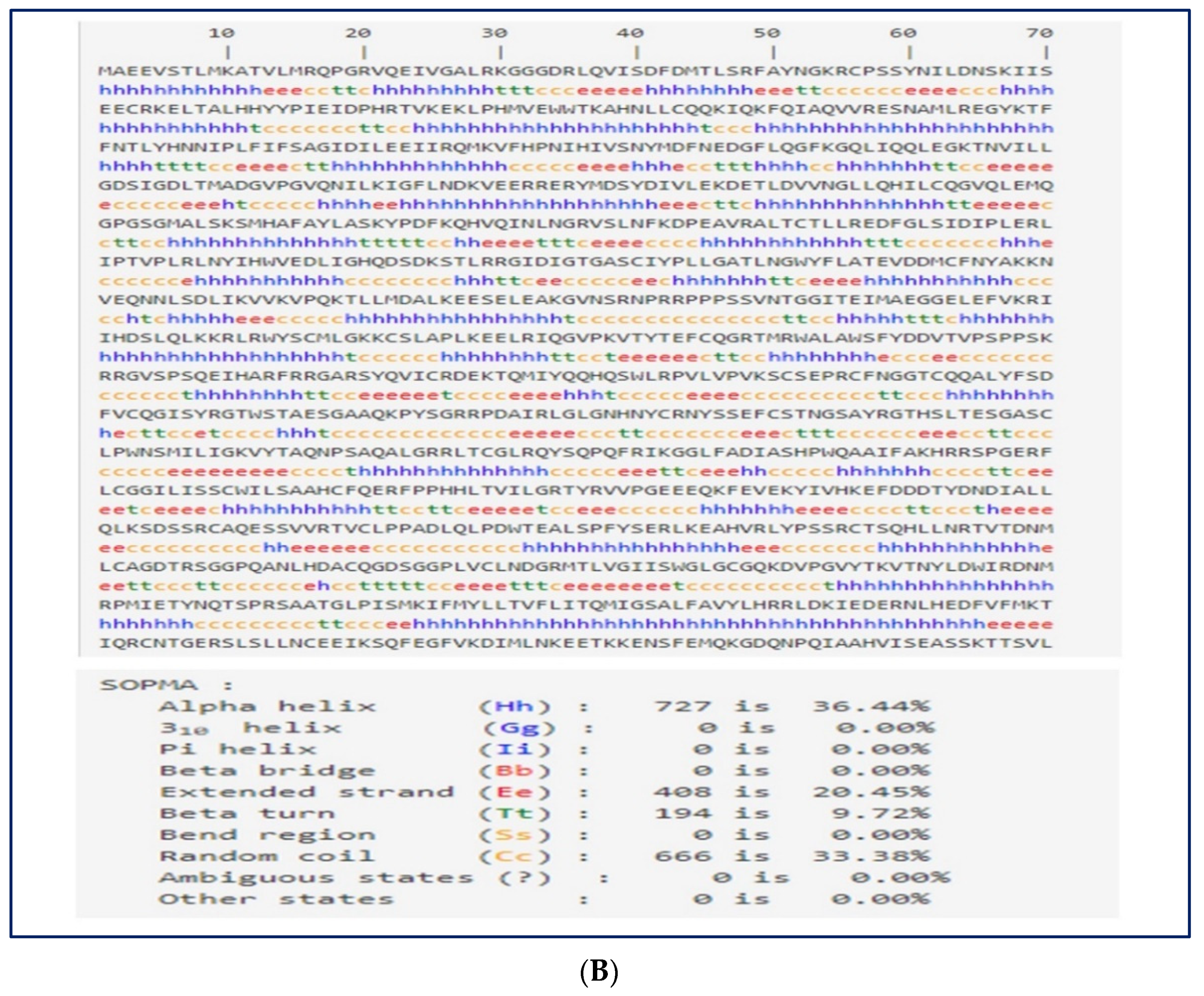
2.10. Prediction of the Secondary Structure of the Vaccine Construct
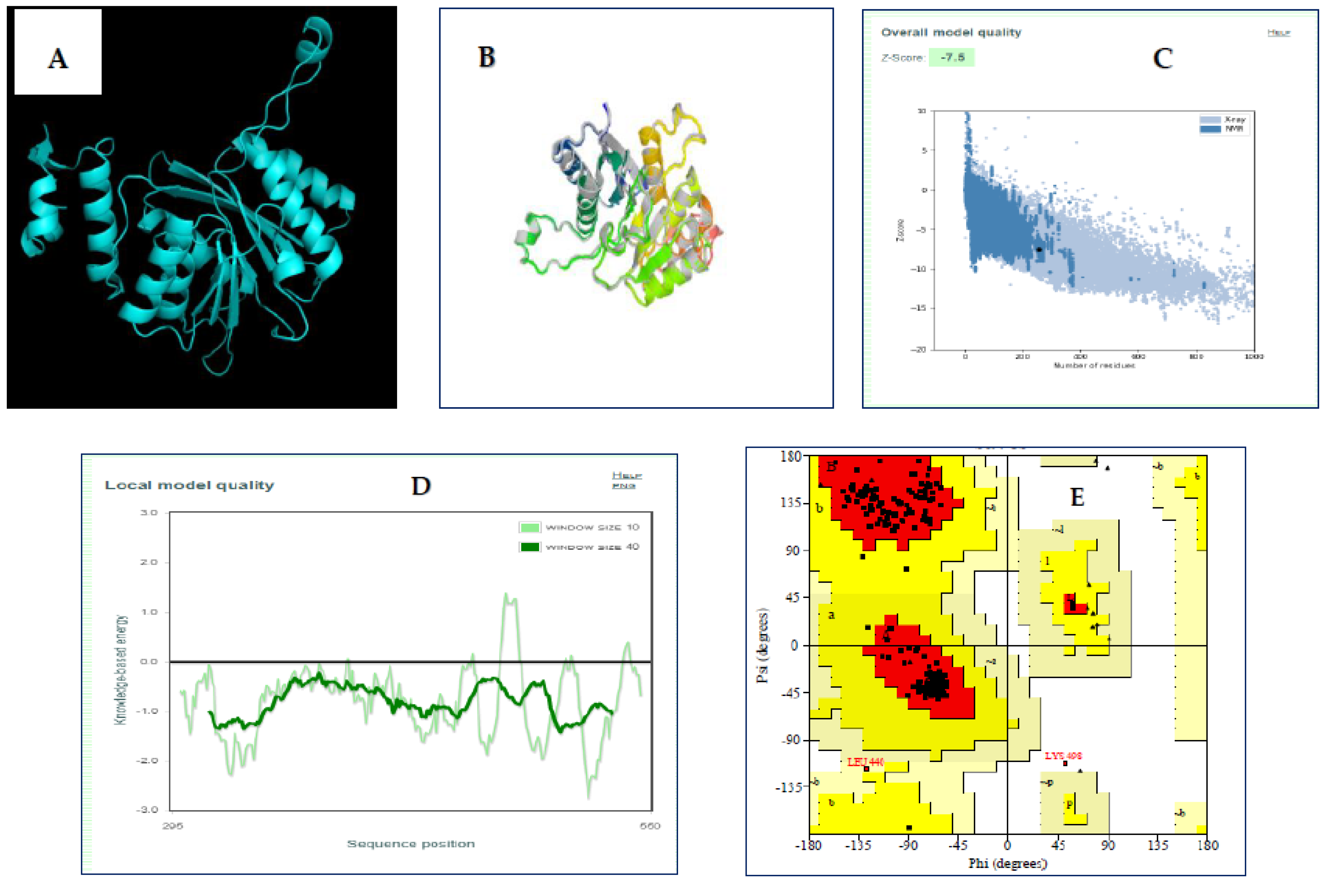
2.11. 3D Structural Modeling, Assessment, and Validation
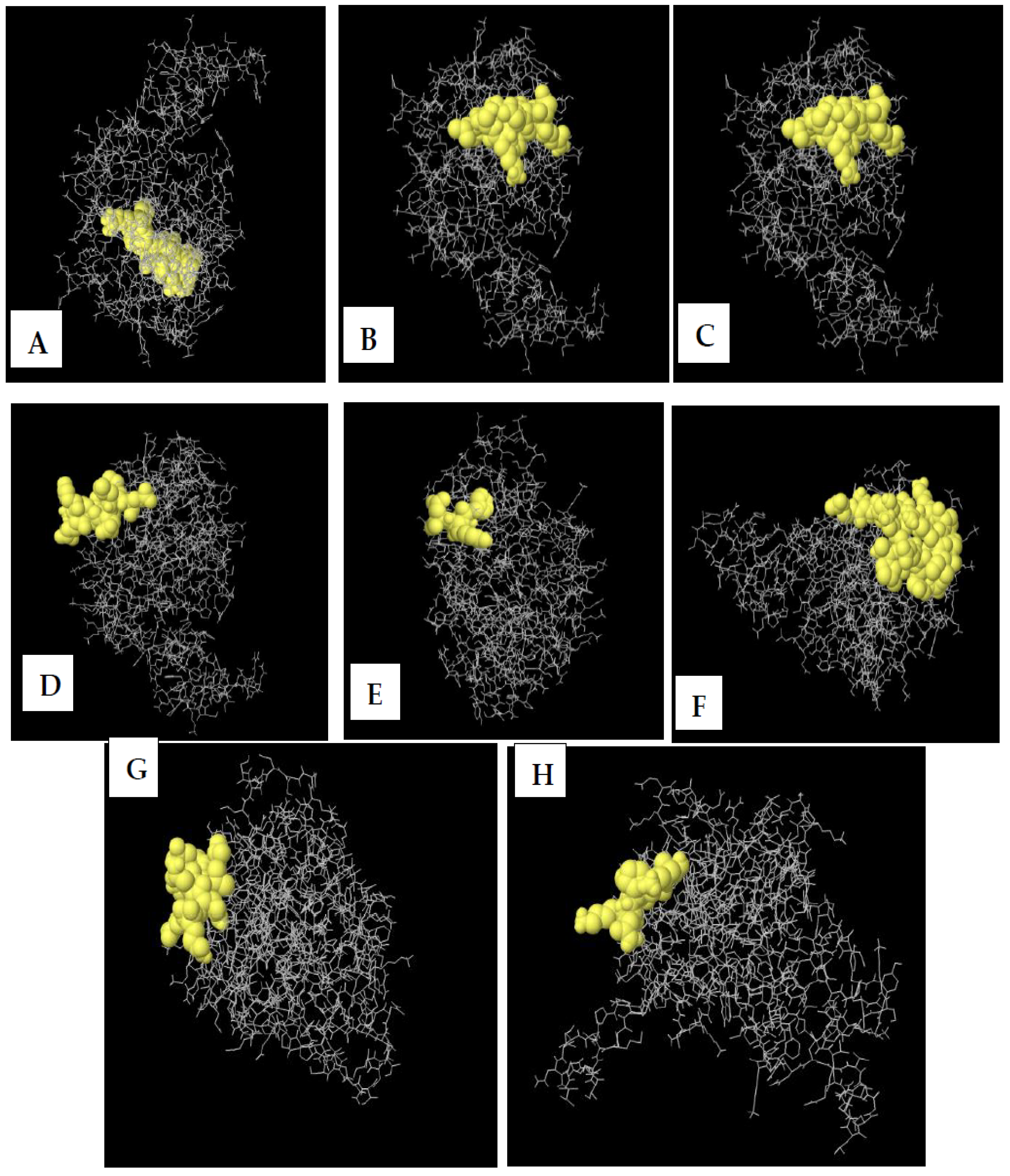
2.12. Prediction of Conformational B-Cell Epitope
2.13. Molecular Docking of Vaccine with TLR Receptor
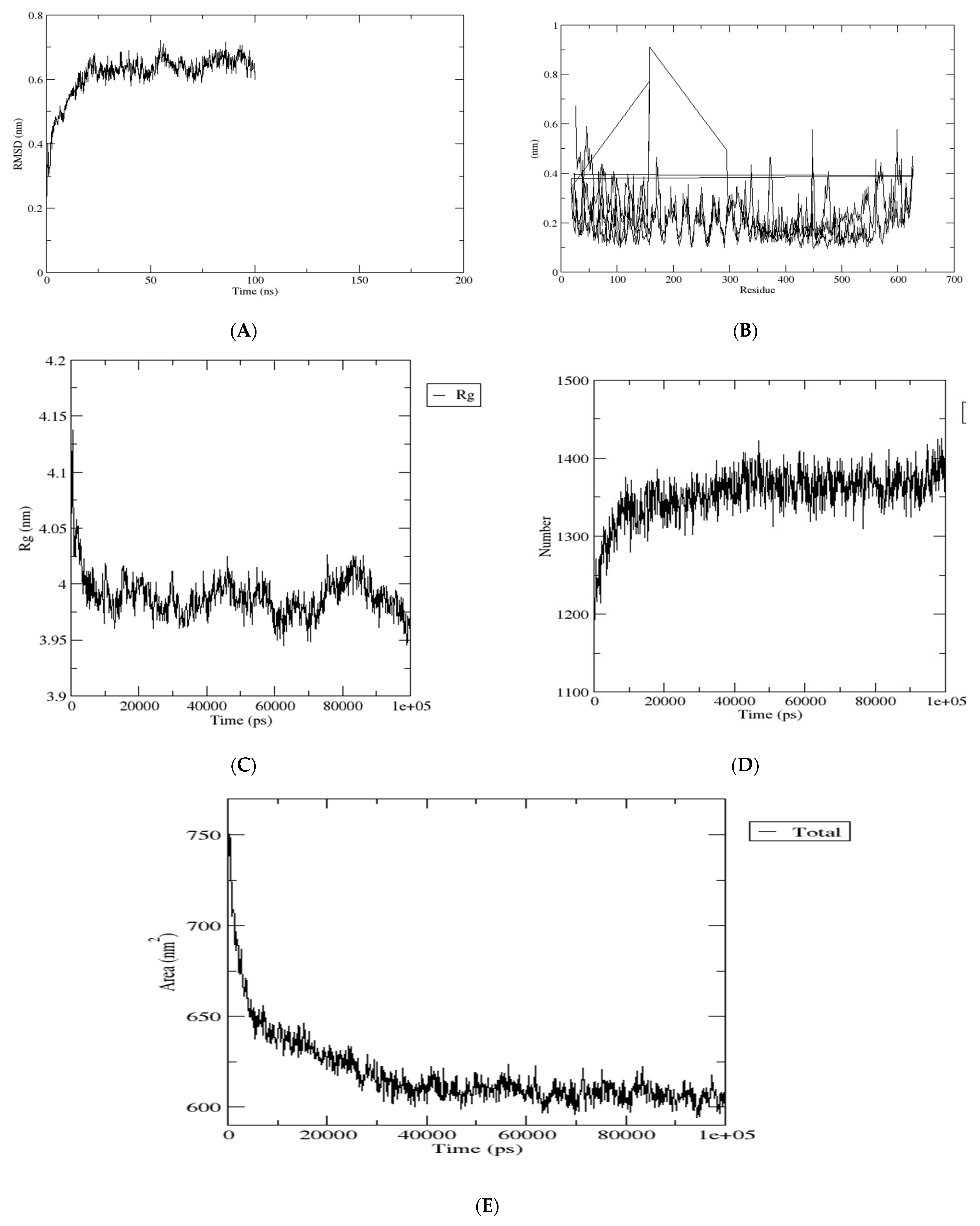
2.14. Molecular Dynamics Simulations
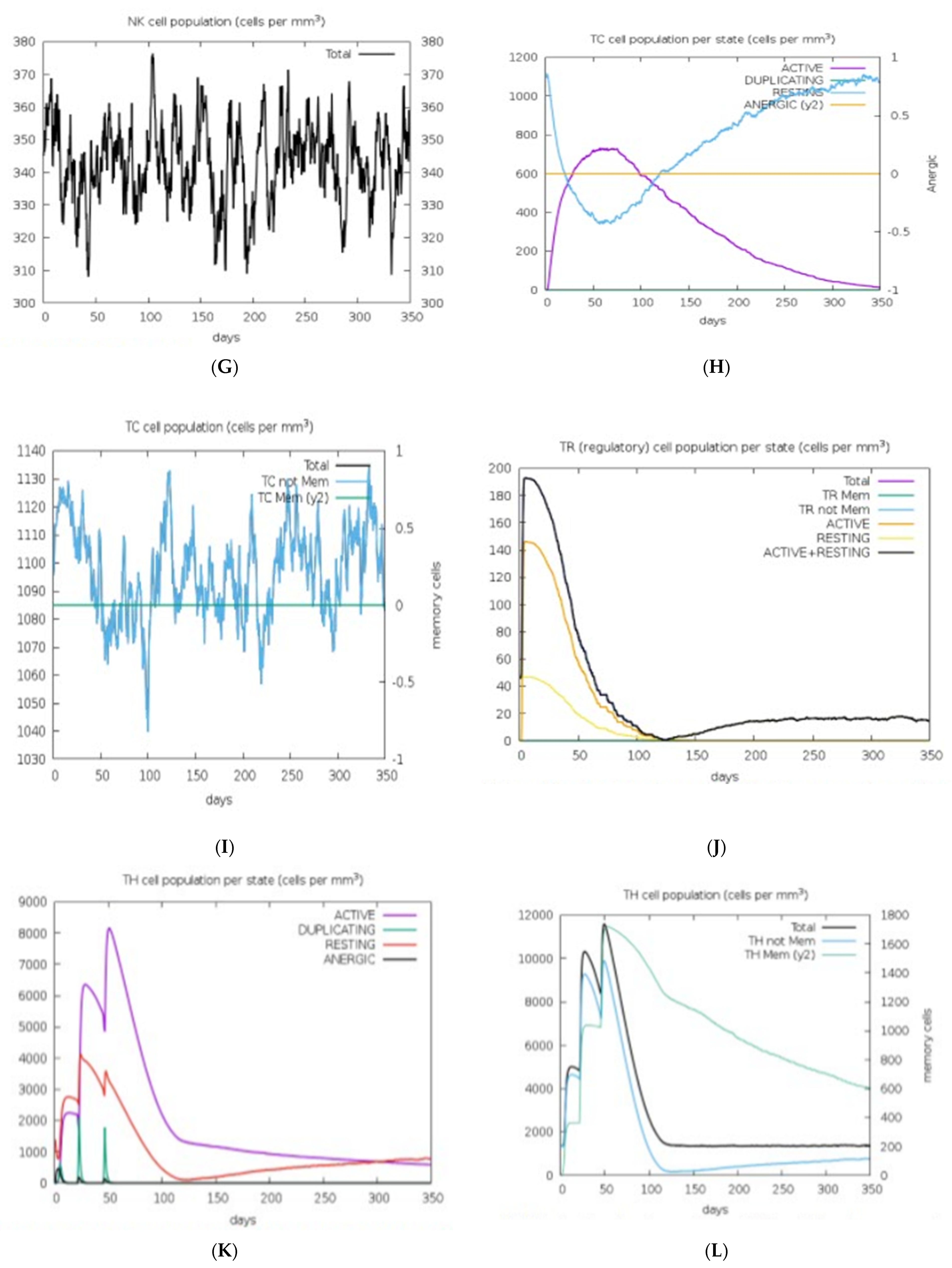
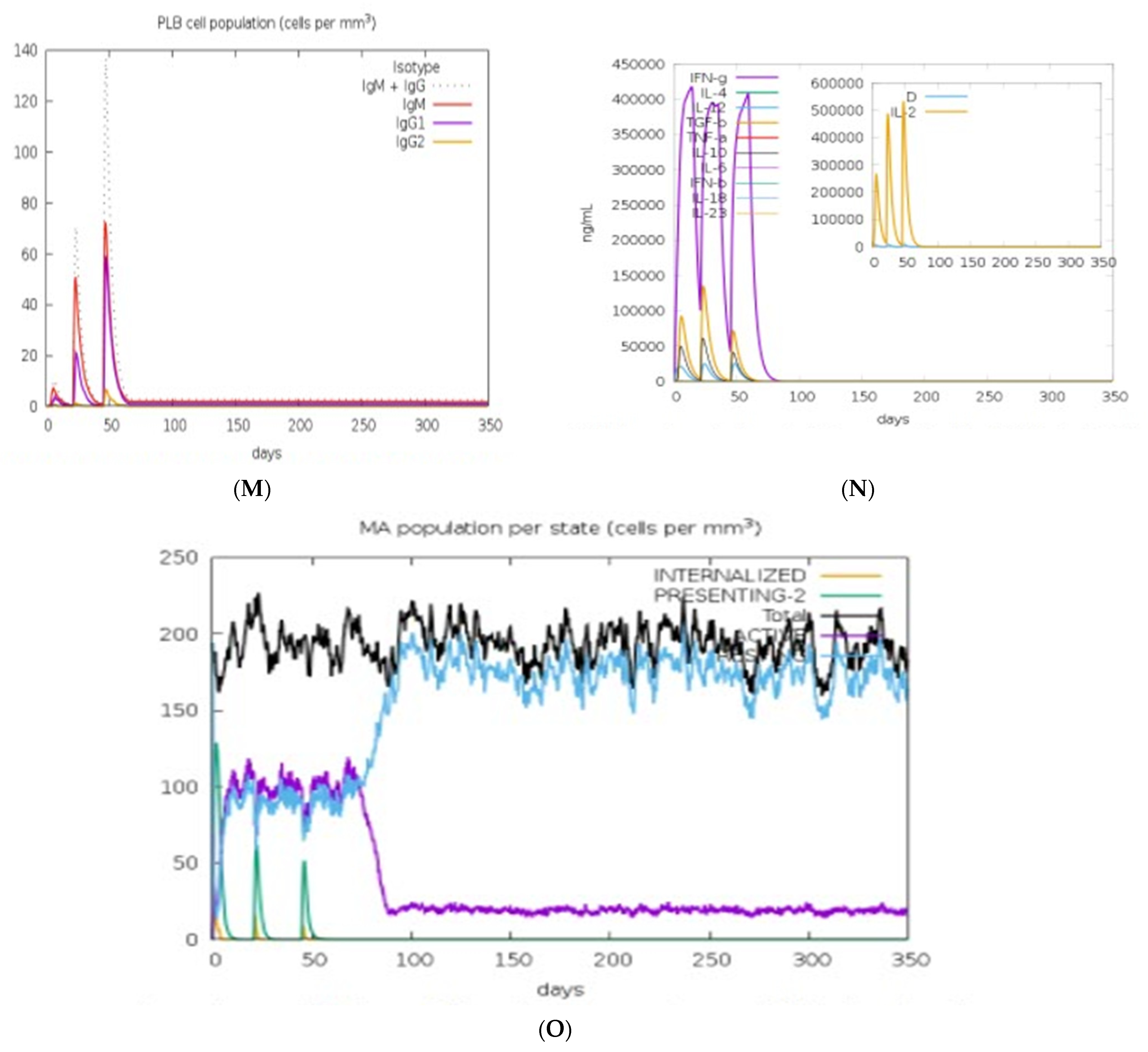
2.15. Immune Response Simulation
3. Results
3.1. Prediction and Evaluation of CTL Epitopes
3.2. Prediction and Evaluation of HTL Epitopes
3.3. Assessment and Prediction of LBL Epitopes
3.4. Multiple Sequence Alignment of SARS-CoV-2 Sequences
3.5. Designing of mRNA Vaccine
3.6. Population Coverage
3.7. Prediction of Allergenicity, Antigenicity, Physicochemical Properties, and Toxicity of the Vaccine Construct
3.8. Secondary Structure Prediction
3.9. Three-Dimensional Structural Modeling, Refining, and Validation
3.10. Conformational B-Cell Epitopes Prediction
3.11. Molecular Docking of Vaccine with TLR Receptor
3.12. Molecular Dynamics Simulations
3.13. Immune Response Simulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safavi, A.; Kefayat, A.; Mahdevar, E.; Abiri, A.; Ghahremani, F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine 2020, 38, 7612–7628. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int (accessed on 20 January 2022).
- Oladipo, E.K.; Ajayi, A.F.; Ariyo, O.E.; Onile, S.O.; Jimah, E.M.; Ezediuno, L.O.; Adebayo, O.I.; Adebayo, E.T.; Odeyemi, A.N.; Oyeleke, M.O.; et al. Exploration of surface glycoprotein to design multi-epitope vaccine for the prevention of COVID-19. Inform. Med. Unlocked 2020, 21, 100438. [Google Scholar] [CrossRef] [PubMed]
- Oluwagbemi, O.O.; Oladipo, E.K.; Dairo, E.O.; Ayeni, A.E.; Irewolede, B.A.; Jimah, E.M.; Oyewole, M.P.; Olawale, B.M.; Adegoke, H.M.; Ogunleye, A.J. Computational construction of a glycoprotein multi-epitope subunit vaccine candidate for old and new South-African SARS-CoV-2 virus strains. Inform. Med. Unlocked 2022, 28, 100845. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, B.P. In silico study to predict and characterize of SARS CoV 2 Surface glycoprotein. Vaccine Res. 2020, 7, 10–16. [Google Scholar] [CrossRef]
- WHO Africa. Eight in 10 African Countries to Miss Crucial COVID-19 Vaccination Goal. World Health Organization. 2021. Available online: https://www.afro.who.int/news/eight-10-african-countries-miss-crucial-covid-19-vaccination-goal (accessed on 3 September 2021).
- United Nations. Record Weekly COVID-19 Deaths in Africa. African Renewal (2021). Available online: https://www.un.org/africarenewal/news/record-weekly-covid-19-deaths-Africa (accessed on 6 August 2021).
- Faria, J. Number of COVID-19 Delta Variant Cases in Africa 2021, by Country. Statista. Available online: https://www.statista.com/statistics/1249798/number-of-sars-cov-2-delta-variant-cases-in-africa-by-country/ (accessed on 28 March 2021).
- Park, J.W.; Lagniton, P.N.; Liu, Y.; Xu, R.-H. mRNA vaccines for COVID-19: What, why and how. Int. J. Biol. Sci. 2021, 17, 1446–1460. [Google Scholar] [CrossRef]
- Schmidt, S.T.; Foged, C.; Korsholm, K.S.; Rades, T.; Christensen, D. Liposome-Based Adjuvants for Subunit Vaccines: Formulation Strategies for Subunit Antigens and Immunostimulators. Pharmaceutics 2016, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Blakney, A.; Ip, S.; Geall, A. An Update on Self-Amplifying mRNA Vaccine Development. Vaccines 2021, 9, 97. [Google Scholar] [CrossRef]
- Schlake, T.; Thess, A.; Fotin-Mleczek, M.; Kallen, K. Developing mRNA-vaccine technologies. RNA Biol. 2012, 9, 1319–1330. [Google Scholar] [CrossRef]
- Ho, W.; Gao, M.; Li, F.; Li, Z.; Zhang, X.Q.; Xu, X. Next-generation vaccines nanoparticle-mediated DNA and mRNA delivery. Adv. Healthc. Mater. 2021, 10, e2001812. [Google Scholar] [CrossRef]
- Xu, S.; Yang, K.; Li, R.; Zhang, L. mRNA vaccine era-mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Stahel, V.P. The safety of Covid-19 mRNA vaccines: A review. Patient Saf. Surg. 2021, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Zhang, C.; Walker, P.G.; Dong, Y. Formulation and Delivery Technologies for mRNA Vaccines. Curr. Top. Microbiol. Immunol. 2020; Epub ahead of print. [Google Scholar] [CrossRef]
- Kowalzik, F.; Schreiner, D.; Jensen, C.; Teschner, D.; Gehring, S.; Zepp, F. mRNA-Based Vaccines. Vaccines 2021, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- The Global Initiative for Sharing All Influenza Data (GISAID) Database. Available online: https://www.gisaid.org/ (accessed on 30 March 2021).
- Maarouf, M.; Rai, K.R.; Goraya, M.U.; Chen, J.-L. Immune Ecosystem of Virus-Infected Host Tissues. Int. J. Mol. Sci. 2018, 19, 1379. [Google Scholar] [CrossRef]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v.2—A server for in silico prediction of allergens. J. Mol. Model. 2014, 20, 2278. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef]
- Nagpal, G.; Usmani, S.S.; Dhanda, S.; Kaur, H.; Singh, S.; Sharma, M.; Raghava, G.P.S. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci. Rep. 2017, 7, srep42851. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Govindaraj, R.G.; Shin, T.H.; Kim, M.O.; Lee, G. iBCE-EL: A New Ensemble Learning Framework for Improved Linear B-Cell Epitope Prediction. Front. Immunol. 2018, 9, 1695. [Google Scholar] [CrossRef] [PubMed]
- Abdelmageed, M.I.; Abdelmoneim, A.H.; Mustafa, M.I.; Elfadol, N.M.; Murshed, N.S.; Shantier, S.W.; Makhawi, A.M. Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: An Immunoinformatics Approach. BioMed Res. Int. 2020, 2020, 2683286. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 1–16. [Google Scholar] [CrossRef]
- Ahammad, I.; Lira, S.S. Designing a novel mRNA vaccine against SARS-CoV-2: An immunoinformatics approach. Int. J. Biol. Macromol. 2020, 162, 820–837. [Google Scholar] [CrossRef]
- Bibi, S.; Ullah, I.; Zhu, B.; Adnan, M.; Liaqat, R.; Kong, W.-B.; Niu, S. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci. Rep. 2021, 11, 1249. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef]
- Zahroh, H.; Ma’Rup, A.; Tambunan, U.S.F.; Parikesit, A.A. Immunoinformatics Approach in Designing Epitope-based Vaccine against Meningitis-inducing Bacteria (Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae Type b). Drug Target Insights 2016, 10, DTI-S38458. [Google Scholar] [CrossRef]
- Kedzierska, K.; Thomas, P.G. Count on us: T cells in SARS-CoV-2 infection and vaccination. Cell Rep. Med. 2022, 3, 100562. [Google Scholar] [CrossRef]
- Bui, H.-H.; Sidney, J.; Dinh, K.; Southwood, S.; Newman, M.J.; Sette, A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinform. 2006, 7, 153. [Google Scholar] [CrossRef]
- Magnan, C.N.; Zeller, M.; Kayala, M.A.; Vigil, A.; Randall, A.; Felgner, P.L.; Baldi, P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 2010, 26, 2936–2943. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, I.; Naneva, L.; Doytchinova, I.; Bangov, I. AllergenFP: Allergenicity prediction by descriptor fingerprints. Bioinformatics 2013, 30, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; John, W.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. Available online: http://www.expasy.org/tools/protparam.html; (accessed on 2 July 2021).
- Rehman, A.; Ahmad, S.; Shahid, F.; Albutti, A.; Alwashmi, A.; Aljasir, M.; Alhumeed, N.; Qasim, M.; Ashfaq, U.; Qamar, M.T.U. Integrated Core Proteomics, Subtractive Proteomics, and Immunoinformatics Investigation to Unveil a Potential Multi-Epitope Vaccine against Schistosomiasis. Vaccines 2021, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvement in protein secondary structure prediction by consensus prediction from multiple alignments. C2abios 1995, 11, 681–684. Available online: https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html (accessed on 5 August 2021). [CrossRef] [PubMed]
- Deléage, G. ALIGNSEC: Viewing protein secondary structure predictions within large multiple sequence alignments. Bioinformatics 2017, 33, 3991–3992. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Lee, G.R.; Heo, L.; Seok, C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins Struct. Funct. Bioinform. 2015, 84, 293–301. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Ponomarenko, J.V.; Bui, H.-H.; Li, W.; Fusseder, N.; Bourne, P.E.; Sette, A.; Peters, B. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2020, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLoS ONE 2010, 5, e9862. [Google Scholar] [CrossRef] [PubMed]
- Chukwudozie, O.S.; Gray, C.M.; Fagbayi, T.A.; Chukwuanukwu, R.C.; Oyebanji, V.O.; Bankole, T.T.; Adewole, R.A.; Daniel, E.M. Immuno-informatics design of a multimeric epitope peptide-based vaccine targeting SARS-CoV-2 spike glycoprotein. PLoS ONE 2021, 16, e0248061. [Google Scholar] [CrossRef]
- Chen, J.; Liu, H.; Yang, J.; Chou, K. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef]
- Roush, S.W.; Murphy, T.V.; The Vaccine-Preventable Disease Table Working Group. Historical Comparisons of Morbidity and Mortality for Vaccine-Preventable Diseases in the United States. J. Am. Med Assoc. 2007, 298, 2155–2163. [Google Scholar] [CrossRef]
- Yang, Z.; Bogdan, P.; Nazarian, S. An in silico deep learning approach to multi-epitope vaccine design: A SARS-CoV-2 case study. Sci. Rep. 2021, 11, 3238. [Google Scholar] [CrossRef]
- Kalita, P.; Padhi, A.K.; Zhang, K.Y.; Tripathi, T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020, 145, 104236. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Alameh, M.-G.; Weissman, D.; Pardi, N. Messenger RNA-Based Vaccines Against Infectious Diseases; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–35. [Google Scholar] [CrossRef]
- Kozlova, E.E.G.; Cerf, L.; Schneider, F.S.; Viart, B.T.; Nguyen, C.; Steiner, B.T.; Lima, S.D.A.; Molina, F.; Guerra-Duarte, C.; Felicori, L.; et al. Computational B-cell epitope identification and production of neutralizing murine antibodies against Atroxlysin-I. Sci. Rep. 2018, 8, 14904. [Google Scholar] [CrossRef] [PubMed]
- Bettini, E.; Locci, M. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism and Beyond. Vaccines 2021, 9, 147. [Google Scholar] [CrossRef]
- Pellegrino, P.; Clementi, E.; Radice, S. On vaccine’s adjuvants and autoimmunity: Current evidence and future perspectives. Autoimmun. Rev. 2015, 14, 880–888. [Google Scholar] [CrossRef]
- Bastola, R.; Noh, G.; Keum, T.; Bashyal, S.; Seo, J.-E.; Choi, J.; Oh, Y.; Cho, Y.; Lee, S. Vaccine adjuvants: Smart components to boost the immune system. Arch. Pharmacal Res. 2017, 40, 1238–1248. [Google Scholar] [CrossRef]
- Daoussis, D.; Andonopoulos, A.P.; Liossis, S.-N.C. Targeting CD40L: A Promising Therapeutic Approach. Clin. Vaccine Immunol. 2004, 11, 635–641. [Google Scholar] [CrossRef]
- Tsui, N.B.; Ng, E.K.; Lo, Y.D. Stability of Endogenous and Added RNA in Blood Specimens, Serum, and Plasma. Clin. Chem. 2002, 48, 1647–1653. [Google Scholar] [CrossRef]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef]
- Adibzadeh, S.; Fardaei, M.; Takhshid, M.A.; Miri, M.R.; Dehbidi, G.R.; Farhadi, A.; Ranjbaran, R.; Alavi, P.; Nikouyan, N.; Seyyedi, N.; et al. Enhancing Stability of Destabilized Green Fluorescent Protein Using Chimeric mRNA Containing Human Beta-Globin 5′ and 3′ Untranslated Regions. Avicenna J. Med Biotechnol. 2019, 11, 112–117. [Google Scholar]
- Oyarzun, P.; Kobe, B. Computer-aided design of T-cell epitope-based vaccines: Addressing population coverage. Int. J. Immunogenet. 2015, 42, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef] [PubMed]
- Ferrel, M.N.; Ryan, J.J. The Impact of COVID-19 on Medical Education. Cureus 2020, 12, e7492. [Google Scholar] [CrossRef]
- Liu, H.; Manzoor, A.; Wang, C.; Zhang, L.; Manzoor, Z. The COVID-19 Outbreak and Affected Countries Stock Markets Response. Int. J. Environ. Res. Public Health 2020, 17, 2800. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.; Zhao, H.; Zhang, H.; Yang, H.; Shah, M.H.; Jahanger, A. The Impact of COVID-19 Pandemic on Stock Markets: An Empirical Analysis of World Major Stock Indices. J. Asian Financ. Econ. Bus. 2020, 7, 463–474. [Google Scholar] [CrossRef]
- Caparros-Gonzalez, R.A.; Ganho-Ávila, A.; De La Torre-Luque, A. The COVID-19 Pandemic Can Impact Perinatal Mental Health and the Health of the Offspring. Behav. Sci. 2020, 10, 162. [Google Scholar] [CrossRef]
- Bell, D.; Hansen, K.S.; Kiragga, A.N.; Kambugu, A.; Kissa, J.; Mbonye, A.K. Predicting the Impact of COVID-19 and the Potential Impact of the Public Health Response on Disease Burden in Uganda. Am. J. Trop. Med. Hyg. 2020, 103, 1191–1197. [Google Scholar] [CrossRef]
- Coccia, M. The impact of first and second wave of the COVID-19 pandemic in society: Comparative analysis to support control measures to cope with negative effects of future infectious diseases. Environ. Res. 2021, 197, 111099. [Google Scholar] [CrossRef]
- Malesza, M.; Kaczmarek, M.C. Predictors of anxiety during the COVID-19 pandemic in Poland. Pers. Individ. Differ. 2020, 170, 110419. [Google Scholar] [CrossRef]
- Vasudevan, M.; Mehrolia, S.; Alagarsamy, S. Battle fatigue of Covid 19 warriors—Heal the healers. J. Affect. Disord. 2021, 294, 477–478. [Google Scholar] [CrossRef]
- Koçak, O.; Koçak, Ö.; Younis, M. The Psychological Consequences of COVID-19 Fear and the Moderator Effects of Individuals’ Underlying Illness and Witnessing Infected Friends and Family. Int. J. Environ. Res. Public Health 2021, 18, 1836. [Google Scholar] [CrossRef] [PubMed]
- Usher, K.; Durkin, J.; Bhullar, N. The COVID-19 pandemic and mental health impacts. Int. J. Ment. Health Nurs. 2020, 29, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Jungmann, S.M.; Witthöft, M. Health anxiety, cyberchondria, and coping in the current COVID-19 pandemic: Which factors are related to coronavirus anxiety? J. Anxiety Disord. 2020, 73, 102239. [Google Scholar] [CrossRef]
- Oluwagbemi, O. Development of a prototype hybrid-grid-based computing framework for accessing bioinformatics databases and resources. Sci. Res. Essays 2012, 7, 730–739. [Google Scholar] [CrossRef]
- Oluwagbemi, O.; Adeoye, E.T.; Fatumo, S. Building a Computer-Based Expert System for Malaria Environmental Diagnosis: An Alternative Malaria Control Strategy. Egypt. Comput. Sci. J. 2009, 33, 55–69. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Oluwagbemi, O.; Oluwagbemi, F.; Abimbola, O. Ebinformatics: Ebola fuzzy informatics systems on the diagnosis, prediction and recommendation of appropriate treatments for Ebola virus disease (EVD). Inform. Med. Unlocked 2016, 2, 12–37. [Google Scholar] [CrossRef]
- Oluwagbemi, O.O.; Fornadel, C.M.; Adebiyi, E.F.; Norris, D.E.; Rasgon, J. AnoSpEx: A Stochastic, Spatially -Explicit Computational Model for Studying Anopheles Metapopulation Dynamics. PLoS ONE 2013, 8, e68040. [Google Scholar] [CrossRef]
- Oluwagbemi, O.; Oluwagbemi, F.; Ughamadu, C. Android Mobile Informatics Application for some Hereditary Diseases and Disorders (AMAHD): A complementary framework for medical practitioners and patients. Inform. Med. Unlocked 2016, 2, 38–69. [Google Scholar] [CrossRef]
- Oluwagbemi, O.; Oluwagbemi, F.; Fagbore, O. Malavefes: A computational fuzzy voice-enabled anti-malarial drug informatics software for correct dosage prescription of anti-malaria drugs. J. King Saud Univ.—Comput. Inf. Sci. 2017, 30, 185–197. [Google Scholar] [CrossRef]
- Oluwagbemi, O.; Awe, O. A comparative computational genomics of Ebola Virus Disease strains: In-silico Insight for Ebola control. Inform. Med. Unlocked 2018, 12, 106–119. [Google Scholar] [CrossRef]
- Oluwagbemi, O.; Jatto, A. Implementation of a TCM-based computational health informatics diagnostic tool for Sub-Saharan African students. Inform. Med. Unlocked 2019, 14, 43–58. [Google Scholar] [CrossRef]
- Oluwagbemi, O.O.; Oluwagbemi, F.E.; Jatto, A.; Hui, C. MAVSCOT: A fuzzy logic-based HIV diagnostic system with indigenous multi-lingual interfaces for rural Africa. PLoS ONE 2020, 15, e0241864. [Google Scholar] [CrossRef]
- Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available online: https://coronavirus.jhu.edu/map.html (accessed on 15 May 2022).
- Japan COVID-19 Coronavirus Tracker. Available online: https://covid19japan.com/ (accessed on 3 April 2022).
- China COVID-19 Dashboard. Available online: https://www.zoho.com/covid/china/ (accessed on 3 April 2022).
- European COVID-19 Data Portal. Available online: https://www.covid19dataportal.org/ (accessed on 15 May 2022).
- Cushnan, D.; Bennett, O.; Berka, R.; Bertolli, O.; Chopra, A.; Dorgham, S.; Favaro, A.; Ganepola, T.; Halling-Brown, M.; Imreh, G.; et al. An overview of the National COVID-19 Chest Imaging Database: Data quality and cohort analysis. GigaScience 2021, 10, giab076. [Google Scholar] [CrossRef] [PubMed]
- Noordzij, M.; Duivenvoorden, R.; Pena, M.; De Vries, H.; Kieneker, L.M.; Franssen, C.F.M.; Hemmelder, M.H.; Hilbrands, L.B.; Jager, K.J.; Gansevoort, R.T.; et al. ERACODA: The European database collecting clinical information of patients on kidney replacement therapy with COVID-19. Nephrol. Dial. Transplant. 2020, 35, 2023–2025. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Venugopal, V.K.; Agarwal, V.K.; Malhotra, M.; Chatha, J.S.; Kapur, S.; Gupta, A.; Batra, V.; Majumdar, P.; Malhotra, A.; et al. A Novel Abnormality Annotation Database for COVID-19 Affected Frontal Lung X-rays. MedRxiv 2021. [Google Scholar] [CrossRef]
- Latz, C.A.; DeCarlo, C.; Boitano, L.; Png, C.Y.M.; Patell, R.; Conrad, M.F.; Eagleton, M.; Dua, A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020, 99, 2113–2118. [Google Scholar] [CrossRef]
- Wu, Y.; Feng, Z.; Li, P.; Yu, Q. Relationship between ABO blood group distribution and clinical characteristics in patients with COVID-19. Clin. Chim. Acta 2020, 509, 220–223. [Google Scholar] [CrossRef]
- Xie, G.; Ding, F.; Han, L.; Yin, D.; Lu, H.; Zhan, M. The role of peripheral blood eosinophil counts in COVID-19 patients. Allergy 2021, 76, 471–482. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, K.; Zuo, P.; Liu, Y.; Zhang, M.; Xie, S.; Zhang, H.; Chen, X.; Liu, C. Early decrease in blood platelet count is associated with poor prognosis in COVID-19 patients—indications for predictive, preventive, and personalized medical approach. EPMA J. 2020, 11, 139–145. [Google Scholar] [CrossRef]
- Brinati, D.; Campagner, A.; Ferrari, D.; Locatelli, M.; Banfi, G.; Cabitza, F. Detection of COVID-19 Infection from Routine Blood Exams with Machine Learning: A Feasibility Study. J. Med. Syst. 2020, 44, 135. [Google Scholar] [CrossRef] [PubMed]
- Joob, B.; Wiwanitkit, V. Blood viscosity of COVID-19 patient: A preliminary report. Am. J. Blood Res. 2021, 11, 93–95. [Google Scholar] [PubMed]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR Test Results in Patients Recovered From COVID-19. J. Am. Med Assoc. 2020, 323, 1502. [Google Scholar] [CrossRef]
- Elezkurtaj, S.; Greuel, S.; Ihlow, J.; Michaelis, E.G.; Bischoff, P.; Kunze, C.A.; Sinn, B.V.; Gerhold, M.; Hauptmann, K.; Ingold-Heppner, B.; et al. Causes of death and comorbidities in hospitalized patients with COVID-19. Sci. Rep. 2021, 11, 4263. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 21 May 2022).
- EMBL’s European Bioinformatics Institute (EMBL-EBI). Available online: https://www.ebi.ac.uk/ (accessed on 21 May 2022).
- WebGRO. Available online: https://simlab.uams.edu/index.php. (accessed on 21 May 2022).
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Kalimuthu, A.K.; Panneerselvam, T.; Pavadai, P.; Pandian, S.R.K.; Sundar, K.; Murugesan, S.; Ammunje, D.N.; Kumar, S.; Arunachalam, S.; Kunjiappan, S. Pharmacoinformatics-based investigation of bioactive compounds of Rasam (South Indian recipe) against human cancer. Sci. Rep. 2021, 11, 21488. [Google Scholar] [CrossRef]
- Vishvakarma, V.K.; Singh, M.B.; Jain, P.; Kumari, K.; Singh, P. Hunting the main protease of SARS-CoV-2 by plitidepsin: Molecular docking and temperature-dependent molecular dynamics simulations. Amino Acids 2021, 54, 205–213. [Google Scholar] [CrossRef]
- Gorai, S.; Junghare, V.; Kundu, K.; Gharui, S.; Kumar, M.; Patro, B.S.; Nayak, S.K.; Hazra, S.; Mula, S. Synthesis of Dihydrobenzofuro [3, 2-b] chromenes as Potential 3CLpro Inhibitors of SARS-CoV-2: A Molecular Docking and Molecular Dynamics Study. ChemMedChem 2022, 17, e202100782. [Google Scholar] [CrossRef]
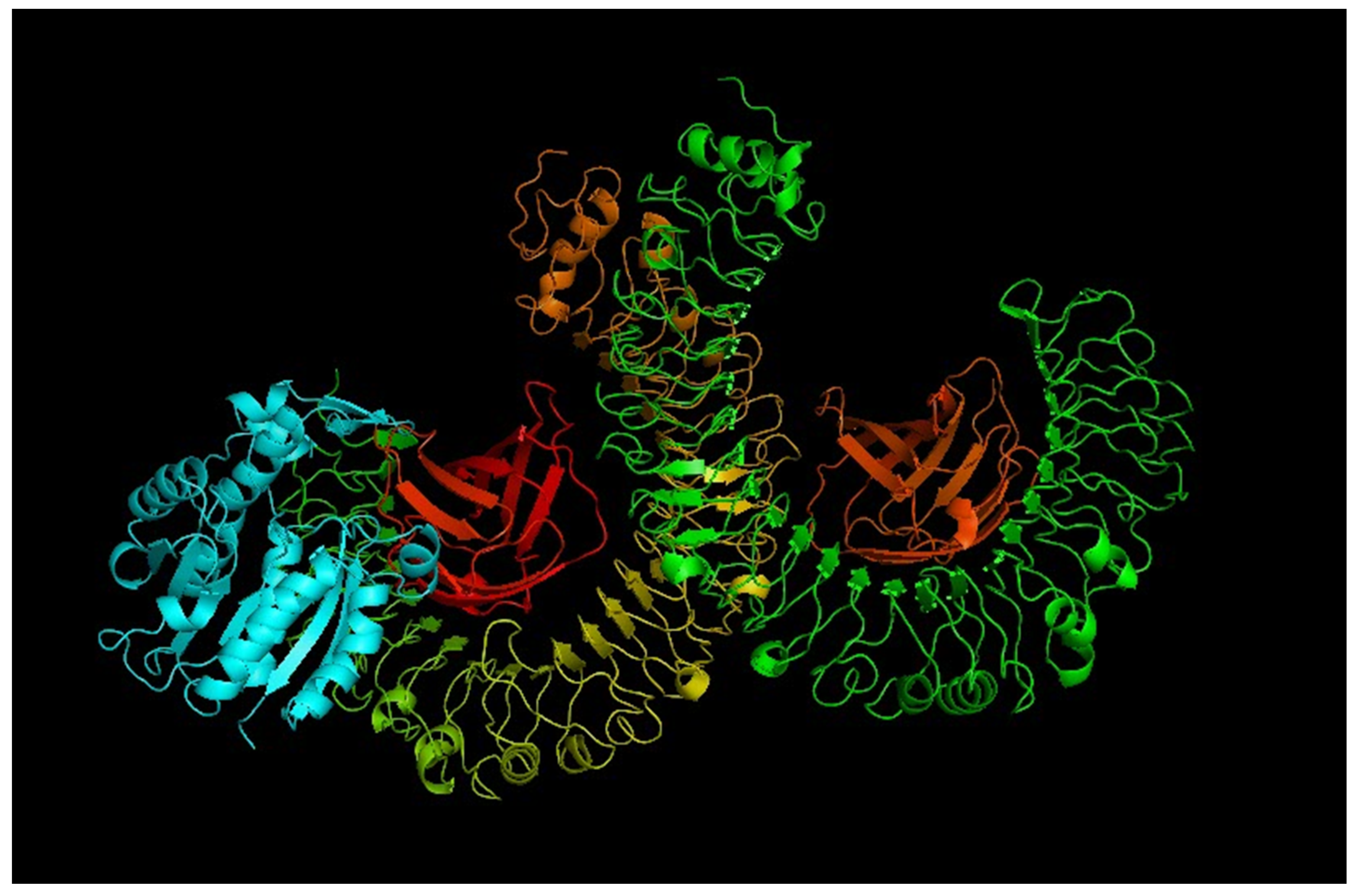
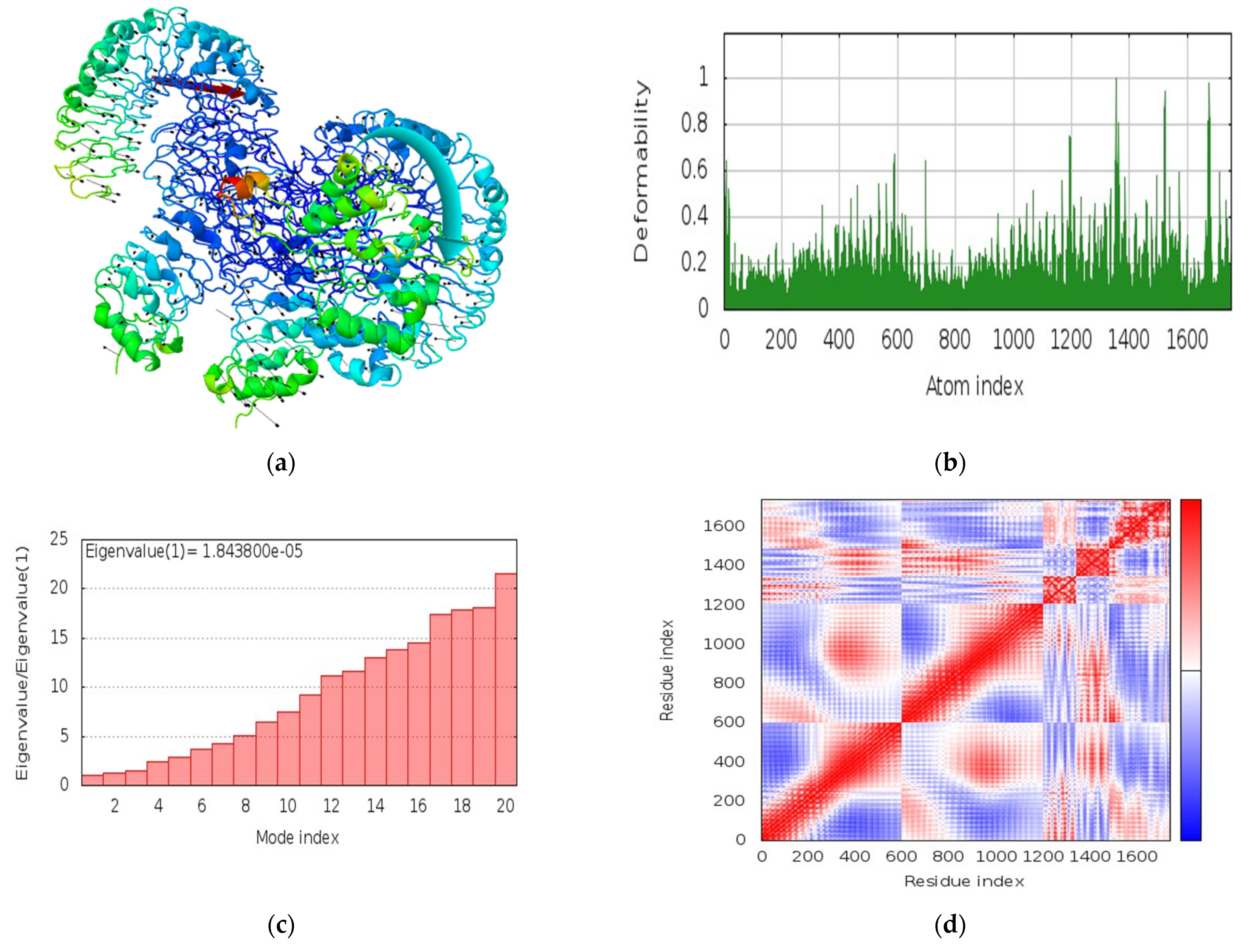
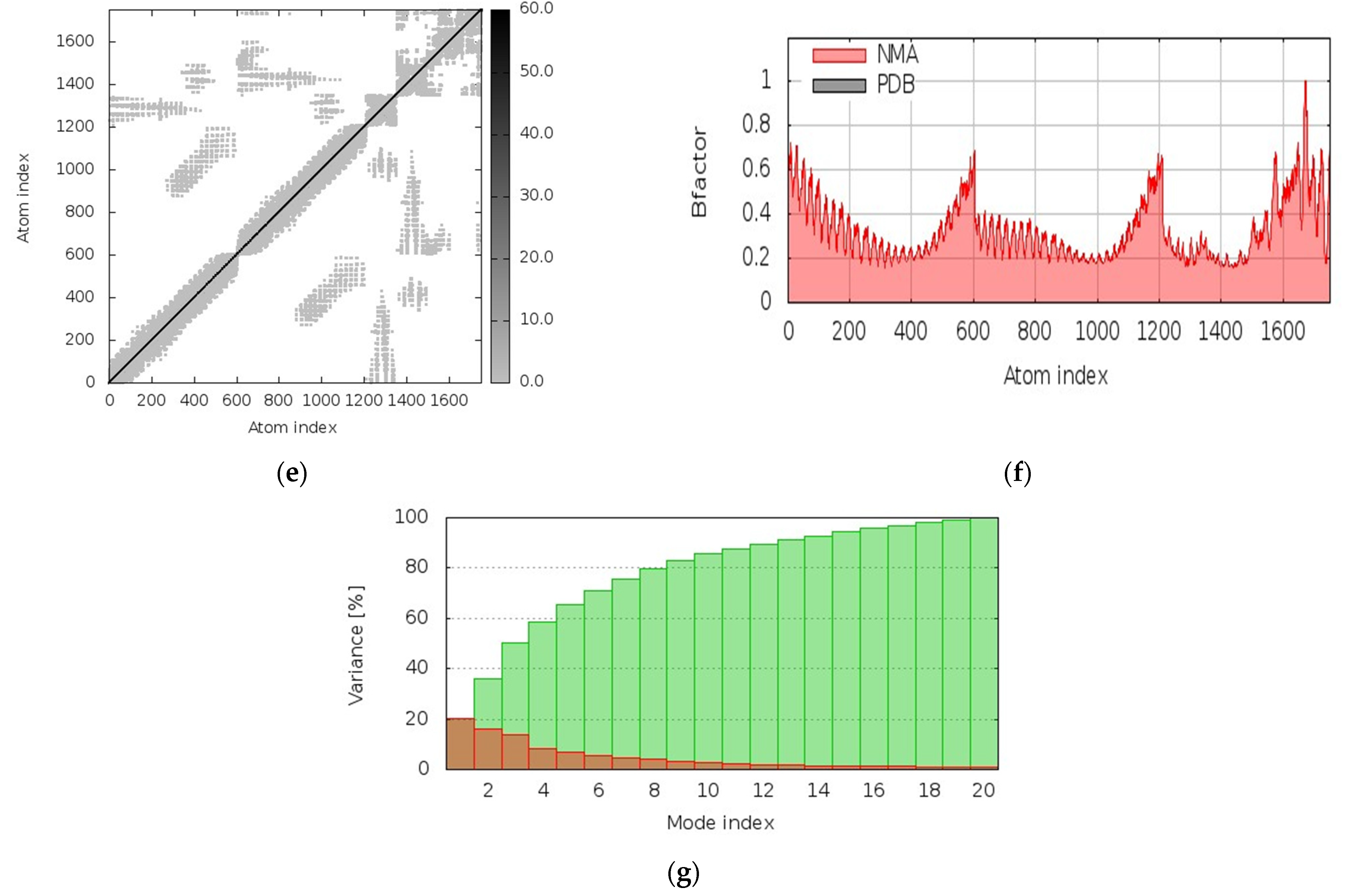
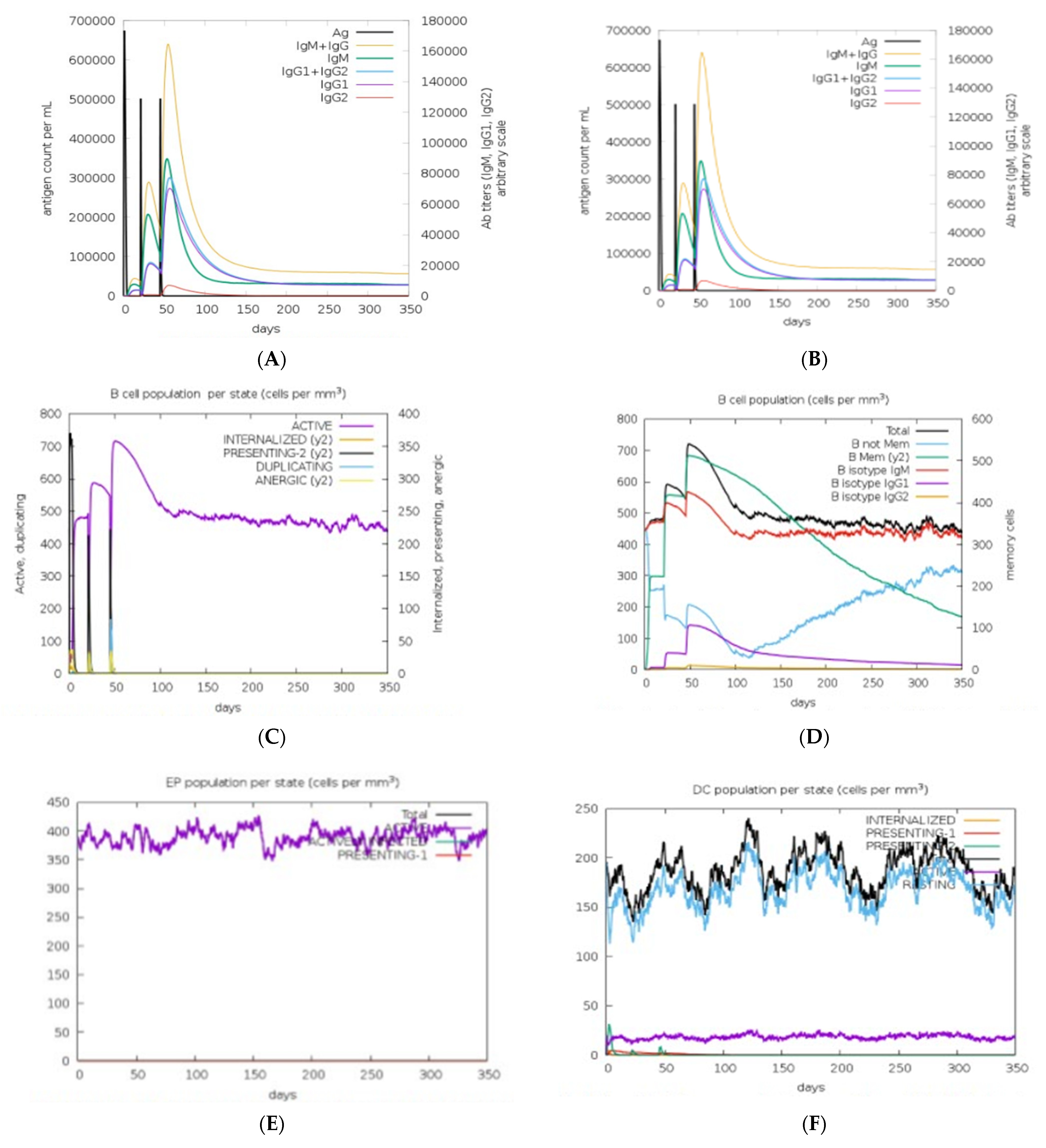
| Recognizing Cell | Epitope Sequence |
|---|---|
| Cytotoxic T lymphocyte | WTAGAAAYY HRHLRFLTL YQPYRVVVL YPQILLLVL SPRRARSVA |
| Helper T lymphocyte | ISFHVLTKLRLKCKL |
| B lymphocyte | WVFITTKTTKVGWKVSSEF |
| T-Lymphocyte Type | CTL Epitopes | MHC Binding ALLELES |
|---|---|---|
| CTL | WTAGAAAYY | HLA-A*29:02, HLA-A*30:02, HLA-B*15:01, HLA-B*46:01, HLA-B*58:01, HLA-B*53:01, HLA-B*35:01, HLA-C*07:01, HLA-C*03:03 |
| HRHLRFLTL | HLA-B*48:01, HLA-C*06:02, HLA-C*07:01 | |
| YQPYRVVVL | HLA-A*02:06, HLA-A*32:01, HLA-B*48:01, HLA-B*46:01, HLA-C*06:02, HLA-C*07:01, HLA-C*03:03 | |
| YPQILLLVL | HLA-B*51:01, HLA-B*53:01, HLA-B*35:01 | |
| SPRRARSVA | HLA-B*51:01 | |
| HTL | ISFHVLTKLRLKCKL | HLA-DRB1*11:01 |
| Population/Region | MHC Class Combined | ||
|---|---|---|---|
| Coverage Area | Average Hit | PC90 | |
| Central Africa | 75.64% | 2.21 | 0.41 |
| East Africa | 80.44% | 2.33 | 0.51 |
| North Africa | 76.13% | 2.29 | 0.42 |
| South Africa | 77.23% | 2.23 | 0.44 |
| West Africa | 76.65% | 2.22 | 0.43 |
| Average | 77.22 | 2.26 | 0.44 |
| Standard deviation | 1.7 | 0.05 | 0.04 |
| Features | Result | Assessment |
|---|---|---|
| Number of amino acids | 1995 | Suitable |
| Molecular weight | 223.1 kDa | Average |
| Theoretical pI | 8.69 | Slightly basic |
| Total number of negatively charged residues (Asp + Glu) | 196 | - |
| Total number of positively charged residues (Arg + Lys) | 223 | - |
| Total number of atoms | 312178 | - |
| Chemical formula | C9908H15532N2774O2906S97 | - |
| Instability index (II) | 48.78 | Unstable |
| Aliphatic index | 82.13 | Thermostable |
| Grand average of hydropathicity (GRAVY) | −0.296 | Hydrophilic |
| Antigenicity | 0.5059 (VaxiJen) 0.7334 (ANTIGENPro) | Antigenic Antigenic |
| Allergenicity | Probable non-allergen (AllerTOP 2.0 and AllergenFP) | Non-allergen |
| Toxicity | Non-toxin (ToxinPred) | Non-toxic |
| Model | GDT-HA | RMSD | MolProbity | Clash Score | Poor Rotamers | Rama Favored |
|---|---|---|---|---|---|---|
| Initial | 1.0000 | 0.000 | 2.457 | 48.0 | 0.0 | 95.7 |
| Model 1 | 0.9707 | 0.353 | 1.527 | 10.2 | 0.4 | 98.4 |
| Model 2 | 0.9717 | 0.364 | 1.733 | 10.6 | 1.8 | 98.8 |
| Model 3 | 0.9658 | 0.371 | 1.570 | 11.3 | 0.4 | 98.8 |
| Model 4 | 0.9707 | 0.365 | 1.594 | 12.1 | 0.0 | 98.4 |
| Model 5 | 0.9697 | 0.359 | 1.545 | 10.6 | 0.4 | 98.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oluwagbemi, O.O.; Oladipo, E.K.; Kolawole, O.M.; Oloke, J.K.; Adelusi, T.I.; Irewolede, B.A.; Dairo, E.O.; Ayeni, A.E.; Kolapo, K.T.; Akindiya, O.E.; et al. Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Vaccine Candidates. Computation 2022, 10, 117. https://doi.org/10.3390/computation10070117
Oluwagbemi OO, Oladipo EK, Kolawole OM, Oloke JK, Adelusi TI, Irewolede BA, Dairo EO, Ayeni AE, Kolapo KT, Akindiya OE, et al. Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Vaccine Candidates. Computation. 2022; 10(7):117. https://doi.org/10.3390/computation10070117
Chicago/Turabian StyleOluwagbemi, Olugbenga Oluseun, Elijah K. Oladipo, Olatunji M. Kolawole, Julius K. Oloke, Temitope I. Adelusi, Boluwatife A. Irewolede, Emmanuel O. Dairo, Ayodele E. Ayeni, Kehinde T. Kolapo, Olawumi E. Akindiya, and et al. 2022. "Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Vaccine Candidates" Computation 10, no. 7: 117. https://doi.org/10.3390/computation10070117
APA StyleOluwagbemi, O. O., Oladipo, E. K., Kolawole, O. M., Oloke, J. K., Adelusi, T. I., Irewolede, B. A., Dairo, E. O., Ayeni, A. E., Kolapo, K. T., Akindiya, O. E., Oluwasegun, J. A., Oluwadara, B. F., & Fatumo, S. (2022). Bioinformatics, Computational Informatics, and Modeling Approaches to the Design of mRNA COVID-19 Vaccine Candidates. Computation, 10(7), 117. https://doi.org/10.3390/computation10070117








