Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques
Abstract
:1. Introduction
2. Related Work
- The proposed system in this work dealt with two different types of databases for evaluating two different diseases and determining the performance of DL and ML methods, while all previous studies dealt with one database for diagnosing one type of disease for evaluating the performance of DL and ML methods. A continuous comparison between the application of the methods on both datasets has been made, emphasizing the fact that the type of evaluated disease matters.
- In comparison with [11], we note that the authors used a dull-razor tool to remove hair from skin images, while we suggested in our work an accurate algorithm to remove hair from skin images while preserving the shape of the lesion and the quality of the image.
- We noticed that most of the previous studies [10,11,12] used the object-oriented image method in the segmentation stage to extract the ROI from the images. In our study, we proposed methods for extracting the ROI (lung and skin lesions) that depend on the threshold techniques, binarization, negation, and morphological operations to segment the colored and gray level images. In addition to this, ref. [14] does not mention any image-segmentation method.
- Compared with previous studies, our study focused on extracting hybrid features from images, which included most types of features (texture, color, shape, geometry, and intensity), and different methods for extracting features were addressed.
- In the classification stage, we noticed that our study dealt with most of the methods of machine learning (nine methods) for an advanced comparison, while the rest of the studies dealt with a limited number of machine-learning methods, and may be limited to one [15], two [10,11], or three methods [12].
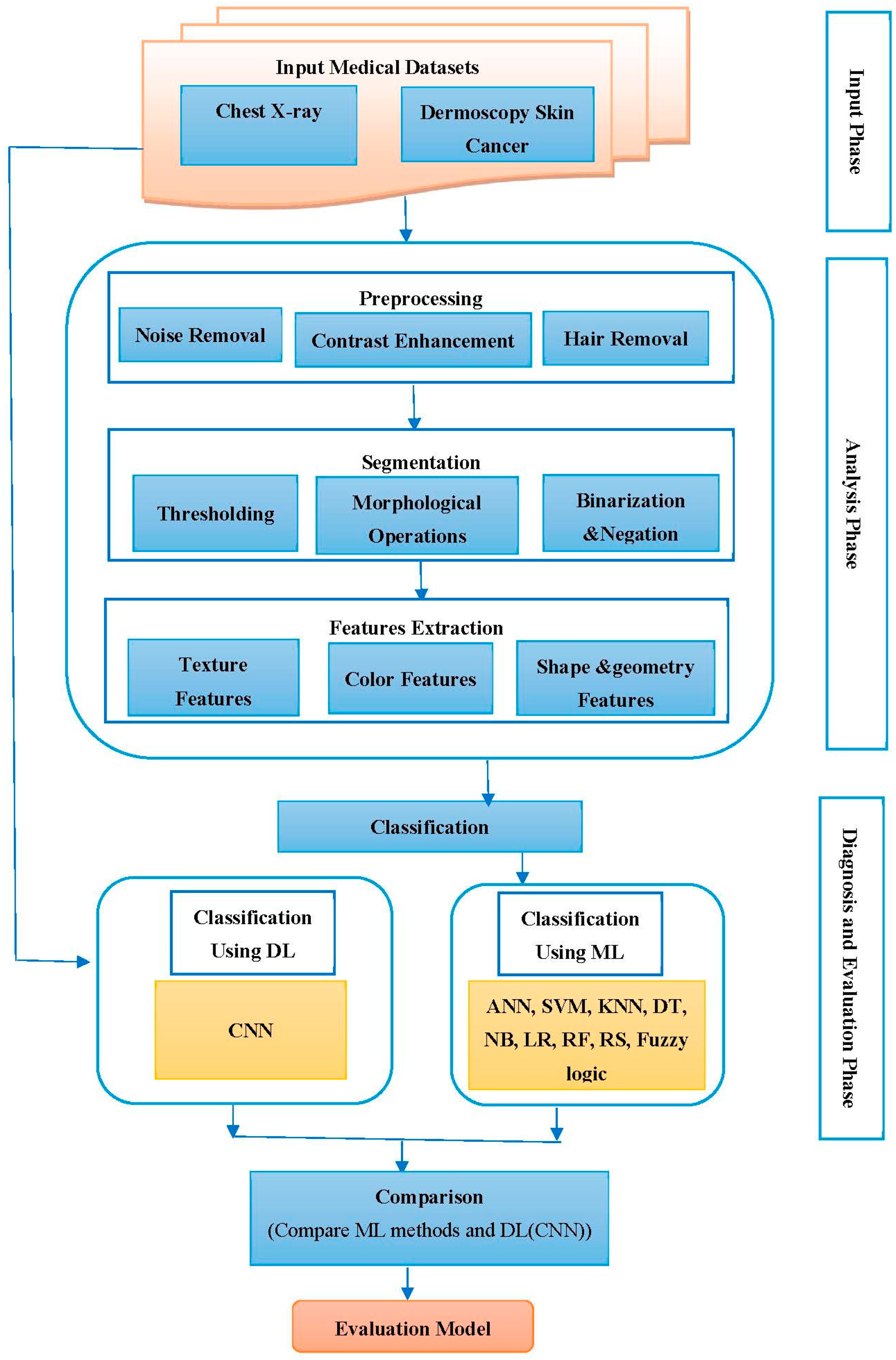
3. Workflow Design
4. Methods
4.1. Medical Datasets
4.1.1. The Chest X-ray Dataset
4.1.2. The Dermoscopy Melanoma Skin Cancer Dataset
4.2. Datasets Analysis
4.2.1. Image Preprocessing
4.2.1.1. Image Cropping
4.2.1.2. Noise Removal
4.2.1.3. Contrast Enhancement
- The color image is converted into a grayscale image;
- Black top-hat transformation is utilized for the detection of dark and thick hairs and is represented as the following equation:where ● denotes the closing operation, is the local contrasted image, and b is a grayscale structuring element.
- By filling the regions in the image that the mask specifies, we can use region fill to remove items from the image or to replace invalid pixel values with their neighbors. The mask’s nonzero pixels specify the image pixels to be filled.
- The result is a fully preprocessed image maintained throughout the subsequent phases.
4.2.2. Image Segmentation
4.2.3. Feature Extraction
- A.
- Texture features set
- 1.
- Gray-Level Co-occurrence Matrix (GLCM)
- 2.
- Gray-Level Run-Length Matrix (GLRLM)
- B.
- Shape features set
- 1.
- Color features set
- 2.
- Texture features set
- 3.
- Geometry features set
4.3. Diagnosis and Evaluation
4.3.1. Classification
- 1.
- Artificial Neural Network (ANN) Classifier
- 2.
- K Nearest Neighbor (K-NN) Classifier
- 3.
- Support Vector Machine (SVM) Classifier
- 4.
- Naïve Bayes (NB) Classifier
- 5.
- Decision Tree (DT) Classifier
- 6.
- Random Forest (RF) Classifier
- 7.
- Random Subspace (RS) Classifier
- 8.
- Logistic Regression (LR) Classifier
- 9.
- Fuzzy logic Classifier
- 10.
- CNN of DL Classifier
- Learning rate (LR): The network’s learning rate is inversely proportional to convergence speed. We experimented with a large spectrum of values; however, their effect on overall performance was insignificant and the model did not exhibit pathological behavior. The training of the proposed CNN network was realized with a learning rate of 1 × 10−2.
- Epochs: We selected 25 epochs for training the network because training over many epochs is common in applications and often results in greater potential for overfitting, 25 epochs were enough to train the datasets and obtain good accuracy.
- Activation function: We conducted several experiments on choosing the activation functions and changing the function in each experiment (we used several activation functions commonly used like the sigmoid function, and the hyperbolic tangent tanh(x)), but we did not obtain satisfactory results for the proposed network except in the case of activation functions for the ReLU and softmax, because it is considered the most effective activation function. In comparison to sigmoid and tanh, ReLU is more trustworthy and speeds up convergence by six times.
- Test different topologies: Some advanced CNNs have more complicated topologies and network architecture for different tasks, for example, GoogLeNet, ResNet, AlexNet, VGGNet, and inception modules. In this work, we tested the ResNet18 model to compare the result with the proposed CNN structure result. The ResNet model’s architecture is shown in Figure 16.

4.3.2. Model Evaluation and Validation
5. Results and Comparison
6. Contributions
- We exploited ML and DL to find the most precise techniques for diagnosis to provide directions for future research.
- We analyzed more than one medical image database to evaluate more than one disease using the proposed system.
- By improving the raw images, finding the ROI (lung and lesion), extracting ROI-specific features, and applying ML and DL algorithms for automatic classification, we present an integrated framework for identifying lung disease utilizing chest X-ray scans and melanoma skin cancer using skin dermoscopy.
- We suggest an algorithm for image preprocessing, where the raw X-ray images were processed and their quality was improved. Additionally, an algorithm was proposed to remove hair from dermoscopy skin images to enhance them and obtain a precise diagnosis. The proposed preprocessing algorithms provided good results in the work.
- We suggest an algorithm for image segmentation to separate the ROI from the image to extract only lung regions from chest X-ray images and lesion regions from dermoscopy skin images. The proposed segmentation algorithms achieved good results in the work.
- We extracted a robust collection of features from ROI (lung and skin lesion) images, including color, texture, shape, and geometry features to help us achieve satisfactory results in the classification.
- Good results were obtained for the proposed system, utilizing two scalable datasets and an appropriate training-to-testing ratio of 70% to 30%. The CNN model and machine-learning techniques such as SVM, KNN, ANN, NB, LR, RF, RS, and fuzzy logic were trained for assessment. In the end, the results of the suggested model methods were compared.
7. Concluded Discussion and Future Directions
7.1. Discussion
7.2. Future Directions
- Increase the number of diseases that are diagnosed and employ other classifiers.
- We also plan to work with more sophisticated medical image data.
- Employ new sets of features for more medical images, to improve performance.
- Although good findings were produced in this work, more research should be conducted by merging the algorithms employed in classification or by adding optimization tools.
- There is a need to develop or create a new classification system for the diagnosis of diseases based on medical image databases.
- Further extensive studies or experiments with vast datasets and hybrid or optimized classification approaches are necessary.
8. Conclusions
- Most of the classification algorithms based on machine learning that were applied to the two selected databases provided good results in terms of various classification performance metrics such as accuracy, sensitivity, specificity, precision, recall, F-measure, and AUC.
- The deep-learning-based convolutional neural network algorithm outperformed in others when applied to the two selected medical databases, as it provided high classification accuracy, reaching 95% in classifying the lung dataset into normal and abnormal, and 93% in classifying the melanoma skin cancer dataset into benign and malignant.
- Additionally, the outcomes varied from one dataset to another, according to the type of medical dataset, the type of medical imaging, and the efficiency of the methods applied in the preprocessing, segmentation, and feature extraction to classify the medical dataset; whenever the methods that were applied to a dataset to train the model were accurate and worked well, the performance of the classification model was better.
- The work provides some crucial insights into modern ML/DL methodologies in the medical field that are applied in disease research nowadays.
- Better outcomes are anticipated with the usage of hybrid algorithms and combined ML and DL techniques. Even minor adjustments can sometimes yield good results. We found that training data quality is an important consideration when creating ML- and DL-based systems.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
Appendix A
| Samples | GLCM Features | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contrast | Correlation | Energy | Homogeneity | |||||||||||||
| 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | |
| Image 1 | 0.23 | 0.31 | 0.15 | 0.28 | 0.931 | 0.933 | 0.96 | 0.951 | 0.294 | 0.241 | 0.25 | 0.23 | 0.68 | 0.65 | 0.69 | 0.61 |
| Image 22 | 0.18 | 0.34 | 0.23 | 0.18 | 0.933 | 0.924 | 0.94 | 0.942 | 0.259 | 0.243 | 0.24 | 0.24 | 0.69 | 0.66 | 0.67 | 0.62 |
| Image 69 | 0.25 | 0.23 | 0.17 | 0.27 | 0.94 | 0.931 | 0.95 | 0.953 | 0.281 | 0.252 | 0.23 | 0.22 | 0.66 | 0.64 | 0.63 | 0.65 |
| Image 80 | 0.26 | 0.33 | 0.21 | 0.19 | 0.921 | 0.913 | 0.961 | 0.939 | 0.278 | 0.251 | 0.25 | 0.24 | 0.62 | 0.63 | 0.68 | 0.64 |
| Image 187 | 0.14 | 0.208 | 0.15 | 0.24 | 0.924 | 0.915 | 0.952 | 0.941 | 0.284 | 0.247 | 0.24 | 0.23 | 0.67 | 0.62 | 0.69 | 0.66 |
| Samples | GLCM Features | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Contrast | Correlation | Energy | Homogeneity | |||||||||||||
| 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | |
| Image 1 | 0.504 | 0.61 | 0.37 | 0.54 | 0.809 | 0.87 | 0.82 | 0.86 | 0.196 | 0.168 | 0.15 | 0.14 | 0.77 | 0.701 | 0.75 | 0.701 |
| Image 22 | 0.47 | 0.53 | 0.35 | 0.48 | 0.806 | 0.89 | 0.83 | 0.85 | 0.201 | 0.147 | 0.14 | 0.16 | 0.76 | 0.703 | 0.76 | 0.702 |
| Image 69 | 0.44 | 0.49 | 0.38 | 0.55 | 0.807 | 0.84 | 0.86 | 0.88 | 0.188 | 0.156 | 0.12 | 0.15 | 0.74 | 0.702 | 0.73 | 0.703 |
| Image 80 | 0.43 | 0.63 | 0.31 | 0.49 | 0.804 | 0.86 | 0.84 | 0.87 | 0.202 | 0.138 | 0.13 | 0.13 | 0.72 | 0.711 | 0.72 | 0.711 |
| Image 187 | 0.502 | 0.54 | 0.43 | 0.51 | 0.803 | 0.88 | 0.81 | 0.89 | 0.191 | 0.127 | 0.16 | 0.14 | 0.71 | 0.712 | 0.76 | 0.721 |
| Samples | GLRLM Features | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRE | LRE | RP | LGRE | |||||||||||||
| 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | |
| Image 1 | 0.17 | 0.33 | 0.31 | 0.32 | 221.9 | 191.2 | 313.2 | 173.2 | 0.15 | 0.105 | 0.12 | 0.104 | 91.4 | 86.9 | 85.2 | 88.1 |
| Image 22 | 0.23 | 0.28 | 0.28 | 0.36 | 189.2 | 188.6 | 298.3 | 177.3 | 0.13 | 0.104 | 0.14 | 0.112 | 87.6 | 86.5 | 85.6 | 77.8 |
| Image 69 | 0.28 | 0.31 | 0.26 | 0.29 | 196.9 | 196.5 | 322.5 | 191.5 | 0.17 | 0.106 | 0.16 | 0.115 | 79.1 | 88.5 | 77.8 | 76.9 |
| Image 80 | 0.15 | 0.27 | 0.33 | 0.38 | 203.7 | 189.6 | 389.7 | 188.4 | 0.14 | 0.103 | 0.13 | 0.108 | 94.1 | 79.8 | 79.3 | 85.8 |
| Image 187 | 0.29 | 0.25 | 0.32 | 0.28 | 206.9 | 186.9 | 299.5 | 169.9 | 0.11 | 0.102 | 0.11 | 0.103 | 89.5 | 83.8 | 83.9 | 79.2 |
| Samples | GLRLM Features | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRE | LRE | RP | LGRE | |||||||||||||
| 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | 0° | 45° | 90° | 135° | |
| Image 1 | 0.48 | 0.51 | 0.501 | 0.51 | 396.3 | 288.5 | 675.7 | 357.1 | 0.25 | 0.216 | 0.21 | 0.214 | 60.8 | 65.8 | 53.4 | 59.1 |
| Image 22 | 0.52 | 0.48 | 0.46 | 0.46 | 323.8 | 292.5 | 586.8 | 287.5 | 0.23 | 0.218 | 0.19 | 0.215 | 59.8 | 58.7 | 58.8 | 57.2 |
| Image 69 | 0.46 | 0.46 | 0.45 | 0.54 | 391.5 | 253.7 | 564.9 | 267.6 | 0.24 | 0.215 | 0.23 | 0.206 | 61.8 | 44.8 | 61.1 | 61.2 |
| Image 80 | 0.39 | 0.45 | 0.44 | 0.48 | 378.6 | 304.5 | 621.3 | 311.5 | 0.27 | 0.211 | 0.201 | 0.209 | 58.8 | 49.7 | 59.4 | 49.8 |
| Image 187 | 0.45 | 0.52 | 0.503 | 0.52 | 369.4 | 312.2 | 584.7 | 312.4 | 0.21 | 0.214 | 0.212 | 0.211 | 60.8 | 52.1 | 66.3 | 55.8 |
| Samples | MI Features | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal | Abnormal | |||||||||||||
| I1 | I2 | I3 | I4 | I5 | I6 | I7 | I1 | I2 | I3 | I4 | I5 | I6 | I7 | |
| Image 1 | 2.08 | 7.02 | 9.08 | 8.56 | −15.78 | −13.65 | 18.11 | 2.74 | 4.5 | 7.6 | 9.9 | −18.5 | −9.9 | 15.4 |
| Image 22 | 2.04 | 7.01 | 8.99 | 8.44 | −16.65 | −12.88 | 18.15 | 2.66 | 4.9 | 6.9 | 10.1 | −18.6 | −10.9 | 14.8 |
| Image 69 | 1.81 | 6.98 | 9.06 | 7.64 | −14.12 | −13.81 | 17.8 | 2.82 | 5.9 | 7.4 | 9.8 | −19.1 | −10.2 | 14.9 |
| Image 80 | 1.99 | 6.44 | 8.87 | 8.88 | −16.76 | −12.76 | 17.5 | 2.65 | 4.8 | 6.8 | 10.07 | −18.7 | −9.7 | 15.2 |
| Image 187 | 1.92 | 7.04 | 9.11 | 7.89 | −16.82 | −13.32 | 18.2 | 2.52 | 5.8 | 7.1 | 9.08 | −19.3 | −11.1 | 13.9 |
| Samples | CM Features | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Benign | Malignant | |||||||||||||||||
| Mean (H) | Mean (S) | Mean (V) | STD (H) | STD (S) | STD (V) | Skewness (H) | Skewness (S) | Skewness (V) | Mean (H) | Mean (S) | Mean (V) | STD (H) | STD (S) | STD (V) | Skewness (H) | Skewness (S) | Skewness (V) | |
| Image 1 | 0.11 | 0.16 | 0.62 | 0.02 | 0.03 | 0.04 | 3.09 | 1.26 | 1.28 | 0.36 | 0.38 | 0.78 | 0.16 | 0.107 | 0.102 | 1.52 | 0.49 | 0.88 |
| Image 33 | 0.13 | 0.12 | 0.59 | 0.01 | 0.02 | 0.03 | 2.08 | 1.37 | 1.25 | 0.49 | 0.4 | 0.76 | 0.13 | 0.104 | 0.101 | 1.44 | 0.39 | 0.87 |
| Image 88 | 0.12 | 0.15 | 0.55 | 0.01 | 0.02 | 0.05 | 3.07 | 1.4 | 1.32 | 0.32 | 0.28 | 0.8 | 0.14 | 0.103 | 0.101 | 1.48 | 0.45 | 0.78 |
| Image 101 | 0.13 | 0.14 | 0.66 | 0.02 | 0.03 | 0.03 | 3.08 | 1.32 | 1.31 | 0.45 | 0.35 | 0.69 | 0.17 | 0.108 | 0.103 | 1.58 | 0.51 | 0.83 |
| Image 203 | 0.12 | 0.13 | 0.57 | 0.01 | 0.02 | 0.06 | 3.06 | 1.41 | 1.35 | 0.5 | 0.27 | 0.79 | 0.15 | 0.104 | 0.101 | 1.44 | 0.42 | 0.79 |
| Samples | Tamura Features | |||||
|---|---|---|---|---|---|---|
| Benign | Malignant | |||||
| Coarseness | Contrast | Directionality | Coarseness | Contrast | Directionality | |
| Image 1 | 14.1 | 14.2 | 0.06 | 23.2 | 31.8 | 0.02 |
| Image 33 | 12.2 | 10.9 | 0.05 | 22.6 | 25.1 | 0.01 |
| Image 88 | 12.8 | 16.8 | 0.05 | 21.8 | 32.8 | 0.03 |
| Image 101 | 13.5 | 11.5 | 0.04 | 24.4 | 27.3 | 0.02 |
| Image 203 | 11.9 | 12.1 | 0.06 | 20.5 | 30.9 | 0.03 |
| Sample | Geometry Features | |||||||
|---|---|---|---|---|---|---|---|---|
| Benign | Malignant | |||||||
| Area | Perimeter | Eccentricity | Diameter | Area | Perimeter | Eccentricity | Diameter | |
| Image 1 | 421 | 166.09 | 0.51 | 28.8 | 842 | 289.2 | 0.82 | 49.8 |
| Image 33 | 511 | 107.7 | 0.46 | 33.6 | 721 | 301.1 | 0.71 | 44.9 |
| Image 88 | 399 | 133.09 | 0.43 | 22.9 | 711 | 302.5 | 0.85 | 48.6 |
| Image 101 | 451 | 196.02 | 0.54 | 30.5 | 802 | 299.4 | 0.79 | 56.1 |
| Image 203 | 411 | 145.02 | 0.49 | 34.1 | 741 | 284.2 | 0.74 | 53.08 |
References
- Tripathi, S.; Shetty, S.; Jain, S.; Sharma, V. Lung disease detection using deep learning. Int. J. Innov. Technol. Explor. Eng. 2021, 10, 154–159. [Google Scholar]
- Saba, T.; Javed, R.; Rahim, M.; Rehman, A.; Bahaj, S. IoMT Enabled Melanoma Detection Using Improved Region Growing Lesion Boundary Extraction. Comput. Mater. Contin. 2022, 71, 6219–6237. [Google Scholar] [CrossRef]
- Usama, M.; Naeem, M.A.; Mirza, F. Multi-Class Skin Lesions Classification Using Deep Features. Sensors 2022, 22, 8311. [Google Scholar] [CrossRef] [PubMed]
- Chola, C.; Mallikarjuna, P.; Muaad, A.Y.; Bibal Benifa, J.; Hanumanthappa, J.; Al-antari, M.A. A hybrid deep learning approach for COVID-19 diagnosis via CT and X-ray medical images. Comput. Sci. Math. Forum 2022, 2, 13. [Google Scholar]
- Canayaz, M.; Şehribanoğlu, S.; Özdağ, R.; Demir, M. COVID-19 diagnosis on CT images with Bayes optimization-based deep neural networks and machine learning algorithms. Neural Comput. Appl. 2022, 34, 5349–5365. [Google Scholar] [CrossRef] [PubMed]
- Clement, J.C.; Ponnusamy, V.; Sriharipriya, K.; Nandakumar, R. A survey on mathematical, machine learning and deep learning models for COVID-19 transmission and diagnosis. IEEE Rev. Biomed. Eng. 2021, 15, 325–340. [Google Scholar]
- Varoquaux, G.; Cheplygina, V. Machine learning for medical imaging: Methodological failures and recommendations for the future. Npj Digit. Med. 2022, 5, 48. [Google Scholar] [CrossRef]
- Sujatha, R.; Chatterjee, J.M.; Jhanjhi, N.Z.; Brohi, S.N. Performance of deep learning vs machine learning in plant leaf disease detection. Microprocess. Microsyst. 2021, 80, 103615. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- Allugunti, V.R. Breast cancer detection based on thermographic images using machine learning and deep learning algorithms. Int. J. Eng. Comput. Sci. 2022, 4, 49–56. [Google Scholar]
- Abunadi, I.; Senan, E.M. Deep learning and machine learning techniques of diagnosis dermoscopy images for early detection of skin diseases. Electronics 2021, 10, 3158. [Google Scholar] [CrossRef]
- Goyal, S.; Singh, R. Detection and classification of lung diseases for pneumonia and COVID-19 using machine and deep learning techniques. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 1–21. [Google Scholar] [CrossRef]
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of heart disease using a combination of machine learning and deep learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef]
- Mamlook, R.E.A.; Chen, S.; Bzizi, H.F. Investigation of the performance of Machine Learning Classifiers for Pneumonia Detection in Chest X-ray Images. In Proceedings of the 2020 IEEE International Conference on Electro Information Technology (EIT), Chicago, IL, USA, 31 July–1 August 2020; pp. 98–104. [Google Scholar]
- Narayanan, B.N.; Ali, R.; Hardie, R.C. Performance analysis of machine learning and deep learning architectures for malaria detection on cell images. Appl. Mach. Learn. 2019, 11139, 240–247. [Google Scholar]
- Data Availability: Data Available for Free at the Kaggle Repository. Available online: www.kaggle.com/amanullahasraf/covid19-pneumonia-normal-chest-xray-pa-dataset; https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia (accessed on 10 August 2022).
- The Lloyd Dermatology and Laser Center. Available online: https://lloyd-derm.com/searchresults.php?search=images&sort=score (accessed on 9 August 2022).
- Dermatology Online Atlas. Available online: http://homepages.inf.ed.ac.uk/rbf/DERMOFIT/ (accessed on 9 August 2022).
- Roy, A.; Maity, P. A Comparative Analysis of Various Filters to Denoise Medical X-ray Images. In Proceedings of the 2020 4th International Conference on Electronics, Materials Engineering & Nano-Technology (IEMENTech), Kolkata, India, 2–4 October 2020; pp. 1–5. [Google Scholar]
- Pitoya, P.A.; Suputraa, I.P.G.H. Dermoscopy image segmentation in melanoma skin cancer using Otsu thresholding method. J. Elektron. Ilmu Komput. Udayana 2021, 2301, 5373. [Google Scholar] [CrossRef]
- Ashraf, H.; Waris, A.; Ghafoor, M.F.; Gilani, S.O.; Niazi, I.K. Melanoma segmentation using deep learning with test-time augmentations and conditional random fields. Sci. Rep. 2022, 12, 3948. [Google Scholar] [CrossRef] [PubMed]
- Sivaraj, S.; Malmathanraj, R. Detection and Classification of Skin Lesions using Probability Map based Region Growing with BA-KNN Classifier. JMIR Pubilications 2021. [Google Scholar] [CrossRef]
- Zafar, K.; Gilani, S.O.; Waris, A.; Ahmed, A.; Jamil, M.; Khan, M.N.; Sohail Kashif, A. Skin lesion segmentation from dermoscopic images using convolutional neural network. Sensors 2020, 20, 1601. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, S.; Tauqeer, A.; Bhatti, R.; Ali, S.B. Improved lung segmentation based on U-Net architecture and morphological operations. arXiv 2022, arXiv:2210.10545. [Google Scholar]
- Khairnar, S.; Thepade, S.D.; Gite, S. Effect of image binarization thresholds on breast cancer identification in mammography images using OTSU, Niblack, Burnsen, Thepade’s SBTC. Intell. Syst. Appl. 2021, 10–11, 200046. [Google Scholar] [CrossRef]
- Park, Y.; Guldmann, J.-M. Measuring continuous landscape patterns with Gray-Level Co-Occurrence Matrix (GLCM) indices: An alternative to patch metrics? Ecol. Indic. 2020, 109, 105802. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, M. Bone region segmentation in medical images based on improved watershed algorithm. Comput. Intell. Neurosci. 2022, 2022, 3975853. [Google Scholar] [CrossRef]
- Venkatesh, U.; Balachander, B. Analysis of Textural Variations in Cerebellum in Brain to Identify Alzheimers by using Haralicks in Comparison with Gray Level Co-occurrence Matrix (GLRLM). In Proceedings of the 2022 2nd International Conference on Innovative Practices in Technology and Management (ICIPTM), Gautam Buddha Nagar, India, 23–25 February 2022; pp. 549–556. [Google Scholar]
- Chandraprabha, K.; Akila, S. Texture Feature Extraction for Batik Images Using GLCM and GLRLM with Neural Network Classification. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2019, 5, 6–15. [Google Scholar] [CrossRef]
- Khan, S.; Kaklis, P.; Serani, A.; Diez, M.; Kostas, K. Shape-supervised Dimension Reduction: Extracting Geometry and Physics Associated Features with Geometric Moments. Comput. Aided Des. 2022, 150, 103327. [Google Scholar] [CrossRef]
- Zhang, H.; Hung, C.-L.; Min, G.; Guo, J.-P.; Liu, M.; Hu, X. GPU-accelerated GLRLM algorithm for feature extraction of MRI. Sci. Rep. 2019, 9, 1–13. [Google Scholar]
- Vishnoi, V.K.; Kumar, K.; Kumar, B. A comprehensive study of feature extraction techniques for plant leaf disease detection. Multimed. Tools Appl. 2022, 81, 367–419. [Google Scholar] [CrossRef]
- Hammad, B.T.; Jamil, N.; Ahmed, I.T.; Zain, Z.M.; Basheer, S. Robust Malware Family Classification Using Effective Features and Classifiers. Appl. Sci. 2022, 12, 7877. [Google Scholar] [CrossRef]
- Khan, P.; Kader, M.F.; Islam, S.R.; Rahman, A.B.; Kamal, M.S.; Toha, M.U.; Kwak, K.-S. Machine learning and deep learning approaches for brain disease diagnosis: Principles and recent advances. IEEE Access 2021, 9, 37622–37655. [Google Scholar] [CrossRef]
- Hariraj, V.; Khairunizam, W.; Vikneswaran, V.; Ibrahim, Z.; Shahriman, A.; Zuradzman, M.; Rajendran, T.; Sathiyasheelan, R. Fuzzy multi-layer SVM classification of breast cancer mammogram images. Int. J. Mech. Eng. Tech. 2018, 9, 1281–1299. [Google Scholar]
- Tripathi, M. Analysis of Convolutional Neural Network based Image Classification Techniques. J. Innov. Image Process. 2021, 3, 100–117. [Google Scholar] [CrossRef]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar]
- Sarwar, A.; Ali, M.; Manhas, J.; Sharma, V. Diagnosis of diabetes type-II using hybrid machine learning based ensemble model. Int. J. Inf. Technol. 2020, 12, 419–428. [Google Scholar]
- Suri, J.S.; Puvvula, A.; Biswas, M.; Majhail, M.; Saba, L.; Faa, G.; Singh, I.M.; Oberleitner, R.; Turk, M.; Chadha, P.S.; et al. COVID-19 pathways for brain and heart injury in comorbidity patients: A role of medical imaging and artificial intelligence-based COVID severity classification: A review. Comput. Biol. Med. 2020, 124, 103960. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Sharma, V.; Singh, D. Comparative analysis of proficiencies of various textures and geometric features in breast mass classification using k-nearest neighbor. Vis. Comput. Ind. Biomed. Art 2022, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Houssein, E.H.; Emam, M.M.; Ali, A.A.; Suganthan, P.N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2021, 167, 114161. [Google Scholar]
- Rezaei, K.; Agahi, H.; Wyld, D.C. Segmentation and Classification of Brain Tumor CT Images Using SVM with Weighted Kernel Width. Comput. Sci. Inf. Technol. 2017, 7, 39–50. [Google Scholar]
- Ahsan, M.M.; Luna, S.A.; Siddique, Z. Machine-Learning-Based Disease Diagnosis: A Comprehensive Review. Healthcare 2022, 10, 541. [Google Scholar]
- Arumugam, K.; Naved, M.; Shinde, P.P.; Leiva-Chauca, O.; Huaman-Osorio, A.; Gonzales-Yanac, T. Multiple disease prediction using Machine learning algorithms. Mater. Today Proc. 2021, in press. [Google Scholar] [CrossRef]
- Balaji, V.R.; Suganthi, S.T.; Rajadevi, R.; Krishna Kumar, V.; Saravana Balaji, B.; Pandiyan, S. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 2020, 163, 107922. [Google Scholar] [CrossRef]
- Hazra, R.; Banerjee, M.; Badia, L. Machine Learning for Breast Cancer Classification with ANN and Decision Tree. In Proceedings of the 2020 11th IEEE Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON), Vancouver, BC, Canada, 4–7 November 2020; pp. 0522–0527. [Google Scholar]
- Subudhi, A.; Dash, M.; Sabut, S. Automated segmentation and classification of brain stroke using expectation-maximization and random forest classifier. Biocybern. Biomed. Eng. 2020, 40, 277–289. [Google Scholar] [CrossRef]
- Amini, N.; Shalbaf, A. Automatic classification of severity of COVID-19 patients using texture feature and random forest based on computed tomography images. Int. J. Imaging Syst. Technol. 2022, 32, 102–110. [Google Scholar] [PubMed]
- Assam, M.; Kanwal, H.; Farooq, U.; Shah, S.K.; Mehmood, A.; Choi, G.S. An efficient classification of MRI brain images. IEEE Access 2021, 9, 33313–33322. [Google Scholar] [CrossRef]
- Deegalla, S.; Walgama, K.; Papapetrou, P.; Boström, H. Random subspace and random projection nearest neighbor ensembles for high dimensional data. Expert Syst. Appl. 2022, 191, 116078. [Google Scholar] [CrossRef]
- Almeida, M.A.; Santos, I.A. Classification models for skin tumor detection using texture analysis in medical images. J. Imaging 2020, 6, 51. [Google Scholar] [CrossRef]
- Ali, N.M.; Aziz, N.; Besar, R. Comparison of microarray breast cancer classification using support vector machine and logistic regression with LASSO and boruta feature selection. Indones. J. Electr. Eng. Comput. Sci. 2020, 20, 712–719. [Google Scholar]
- Roy, S.; Chandra, A. On the detection of Alzheimer’s disease using fuzzy logic based majority voter classifier. Multimed. Tools Appl. 2022, 81, 43145–43161. [Google Scholar] [CrossRef]
- Maqsood, S.; Damasevicius, R.; Shah, F.M. An efficient approach for the detection of brain tumor using fuzzy logic and U-NET CNN classification. In Proceedings of the Computational Science and Its Applications–ICCSA 2021: 21st International Conference, Cagliari, Italy, 13–16 September 2021; Part V 21. pp. 105–118. [Google Scholar]
- Sarvamangala, D.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar]
- Mijwil, M.M.; Al-Zubaidi, E.A. Medical image classification for coronavirus disease (COVID-19) using convolutional neural networks. Iraqi J. Sci. 2021, 62, 2740–2747. [Google Scholar]
- Ashraf, R.; Habib, M.A.; Akram, M.; Latif, M.A.; Malik, M.S.A.; Awais, M.; Dar, S.H.; Mahmood, T.; Yasir, M.; Abbas, Z. Deep convolution neural network for big data medical image classification. IEEE Access 2020, 8, 105659–105670. [Google Scholar]
- Ramzan, F.; Khan, M.U.G.; Rehmat, A.; Iqbal, S.; Saba, T.; Rehman, A.; Mehmood, Z. A deep learning approach for automated diagnosis and multi-class classification of Alzheimer’s disease stages using resting-state fMRI and residual neural networks. J. Med. Syst. 2020, 44, 1–16. [Google Scholar]
- Sai Abhishek, A.V. Resnet18 Model with Sequential Layer for Computing Accuracy on Image Classification Dataset. Int. J. Creat. Res. Thoughts 2022, 10, 2320–2882. [Google Scholar]
- Sarwinda, D.; Paradisa, R.H.; Bustamam, A.; Anggia, P. Deep learning in image classification using residual network (ResNet) variants for detection of colorectal cancer. Procedia Comput. Sci. 2021, 179, 423–431. [Google Scholar]


























| GLCM Feature | Description | Equation |
|---|---|---|
| Contrast | It measures the extreme difference in grayscale between adjacent pixels. | |
| Correlation | It examines the linear dependency between the gray levels of adjacent pixels. | |
| Energy | It measures texture uniformity or pixel-pair repetitions. | |
| Homogeneity | It measures the homogeneity of the image and the degree of local uniformity that is present in the image. |
| GLRLM Feature | Description | Equation |
|---|---|---|
| SRE | It measures the distribution of small run lengths, with a higher value indicating shorter run lengths and finer textures. | |
| LRE | It measures the distribution of lengthy run lengths, with higher values indicating longer run lengths and coarser structural textures. | |
| RP | It measures the coarseness of the texture by comparing the number of runs to the number of voxels in the ROI. | |
| LGRE | It measures the distribution of low grayscale values in an image, with a larger value denoting a higher concentration of low grayscale values. |
| MI Feature | Equation |
|---|---|
| I1 | |
| I2 | |
| I3 | |
| I4 | |
| I5 | |
| I6 | |
| I7 |
| CM Feature | Equation |
|---|---|
| Mean | |
| STD | |
| Skewness |
| Tamura Features | Description | Equation |
|---|---|---|
| Coarseness | It represents the size and number of textures primitives. It seeks to find the maximum size at which a texture exists. | |
| Contrast | It indicates the difference in intensity between adjacent pixels. | |
| Directionality | It is used to calculate directionality. The frequency distribution of oriented local edges against their directional angles is used to calculate an image’s directionality. |
| Geometry Feature | Description | Equation |
|---|---|---|
| Area (A) | It is the real number of pixels in the region which is returned as a scalar. The lesion area can be represented by the region of the lesion containing the total number of pixels. | |
| Perimeter (P) | It is a distance around the boundary of a region which is returned as a scalar by computing the distance between every contiguous pair of pixels around the border of the region. | |
| Eccentricity (Ecc) | It is the ratio of the length of the short (minor) axis to the length of an object’s long (major) axis; it is defined as the proportion of eigenvalues of the covariance matrix that matches a binary image of the shape. | |
| Diameter (D) | The diameter is identified by calculating the distance between every pair of points in a binary image and taking the maximum of these distances. |
| Test No. | Number of Layers | Result | |
|---|---|---|---|
| Lung Dataset | Skin Cancer Dataset | ||
| 1 | Two convolution layers, two max-pooling layers, two batch-normalization layers | 81.5% | 80.6% |
| 2 | One convolution layer, one max-pooling layer, one batch-normalization layer | 73.5% | 71.5% |
| The Type of Architecture | Accuracy | |
|---|---|---|
| Lung Dataset | Skin Cancer Dataset | |
| The proposed CNN | 95.1% | 93.3% |
| ResNet18 | 94% | 91% |
| Method | Advantage | Disadvantage |
|---|---|---|
| 1. ANN | Advanced predictive ability Parallel processing ability | Computationally costly Long time to process massive amounts of data |
| 2. SVM | The ability to handle structured and semistructured data Appropriate for nonlinear problems and those with little samples and high dimensions | Decreased performance with large amounts of data Imperfect work with noisy data |
| 3. KNN | Flexibility Easy to implement | Sensitive to k-value selection Requires well-classified training data |
| 4. DT | Ease and speed in implementation The ability to generate rules easily | Difficulty controlling tree size Can suffer from overfitting |
| 5. NB | Speed in predicting the dataset category Simplicity in implementation | Accuracy decreases with a small amount of data Necessitates a vast number of records |
| 6. LR | Speed in training Ease in implementation and application | Not suited for predicting the value of a binary variable, only accepts Boolean values Unable to solve nonlinear problems |
| 7. RF | Flexibility There is no need to normalize data because it employs a rule-based approach | Takes a long time to train Takes a lot of resources and computational effort to build multiple trees and integrate their outputs |
| 8. RS | Precise and reliable predictions Implements a random subset of features to a combined group of foundation classifiers | Takes a long time to train Risk of overfitting |
| 9. Fuzzy Logic | Flexibility Active system for nonlinear problems | Necessitates a large amount of data Rules need to be updated frequently |
| 10. CNN | Effective with large amounts of data Extremely good at image identification and classification | Requires sufficient data and time for training High computational cost |
| Algorithm | Acc% | Sn% | Sp% | Pr | Recall | F-Measure | AUC |
|---|---|---|---|---|---|---|---|
| ANN | 91.1 | 94.7 | 88.4 | 0.916 | 0.911 | 0.912 | 0.945 |
| SVM | 84.4 | 84.2 | 84.6 | 0.846 | 0.844 | 0.845 | 0.844 |
| KNN | 86.6 | 84.2 | 88.4 | 0.867 | 0.867 | 0.867 | 0.800 |
| DT | 74.4 | 73.6 | 75.5 | 0.747 | 0.747 | 0.747 | 0.743 |
| NB | 81.1 | 76.3 | 84.6 | 0.811 | 0.811 | 0.811 | 0.887 |
| LR | 92 | 92.3 | 91.6 | 0.920 | 0.920 | 0.920 | 0.947 |
| RF | 93.3 | 94.7 | 92.3 | 0.935 | 0.933 | 0.934 | 0.992 |
| RS | 84.4 | 92.1 | 78.8 | 0.860 | 0.844 | 0.845 | 0.948 |
| Fuzzy Logic | 81.1 | 71 | 88.4 | 0.821 | 0.811 | 0.809 | 0.798 |
| CNN | 95.1 | 94 | 96.3 | 0.969 | 0.94 | 0.954 | 0.994 |
| Algorithm | Acc% | Sn% | Sp% | Pr | Recall | F-Measure | AUC |
|---|---|---|---|---|---|---|---|
| ANN | 96.6 | 95.4 | 97.8 | 0.967 | 0.967 | 0.967 | 0.974 |
| SVM | 84.4 | 97.7 | 71.7 | 0.871 | 0.844 | 0.842 | 0.847 |
| KNN | 95.5 | 95.4 | 95.6 | 0.956 | 0.956 | 0.956 | 0.930 |
| DT | 84.4 | 100 | 69.5 | 0.882 | 0.844 | 0.841 | 0.848 |
| NB | 80 | 84 | 76 | 0.803 | 0.800 | 0.800 | 0.874 |
| LR | 87.7 | 93.1 | 82.6 | 0.883 | 0.878 | 0.878 | 0.949 |
| RF | 94.6 | 94.8 | 94.4 | 0.947 | 0.947 | 0.947 | 0.984 |
| RS | 93.3 | 94.8 | 91.6 | 0.934 | 0.933 | 0.933 | 0.986 |
| Fuzzy Logic | 90 | 100 | 80.4 | 0.917 | 0.900 | 0.899 | 0.902 |
| CNN | 93.3 | 95.1 | 91.6 | 0.906 | 0.915 | 0.928 | 0.919 |
| Algorithm | Accuracy in the First Database (Chest X-ray) | Accuracy in the Second Database (Melanoma Skin Cancer Dermoscopy) |
|---|---|---|
| ANN | 91.1% | 96.6% |
| SVM | 84.4% | 84.4% |
| KNN | 86.6% | 95.5% |
| DT | 74.4% | 84.4% |
| NB | 81.1% | 80% |
| LR | 92% | 87.7% |
| RF | 93.3% | 94.6% |
| RS | 84.4% | 93.3% |
| Fuzzy Logic | 81.1% | 90% |
| CNN | 95.1% | 93.3% |
| Algorithm | Accuracy in the First Database (Chest X-ray) | Accuracy in the Second Database (Melanoma Skin Cancer Dermoscopy) |
|---|---|---|
| ANN | 92.% | 95.2% |
| SVM | 88.8% | 84.5% |
| KNN | 86.2% | 95.8% |
| DT | 75% | 83.8% |
| NB | 80.9% | 80.4% |
| LR | 92.8% | 88.3% |
| RF | 92.9% | 93.7% |
| RS | 85.7% | 94.8% |
| Fuzzy Logic | 80.9% | 90.6% |
| CNN | 95.08% | 92.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashed, B.M.; Popescu, N. Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques. Computation 2023, 11, 63. https://doi.org/10.3390/computation11030063
Rashed BM, Popescu N. Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques. Computation. 2023; 11(3):63. https://doi.org/10.3390/computation11030063
Chicago/Turabian StyleRashed, Baidaa Mutasher, and Nirvana Popescu. 2023. "Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques" Computation 11, no. 3: 63. https://doi.org/10.3390/computation11030063
APA StyleRashed, B. M., & Popescu, N. (2023). Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques. Computation, 11(3), 63. https://doi.org/10.3390/computation11030063





