From 2D to 3D Cancer Cell Models—The Enigmas of Drug Delivery Research
Abstract
:1. Introduction
2. From 2D to 3D Cancer Models
2.1. Scaffold-Free 3D Model
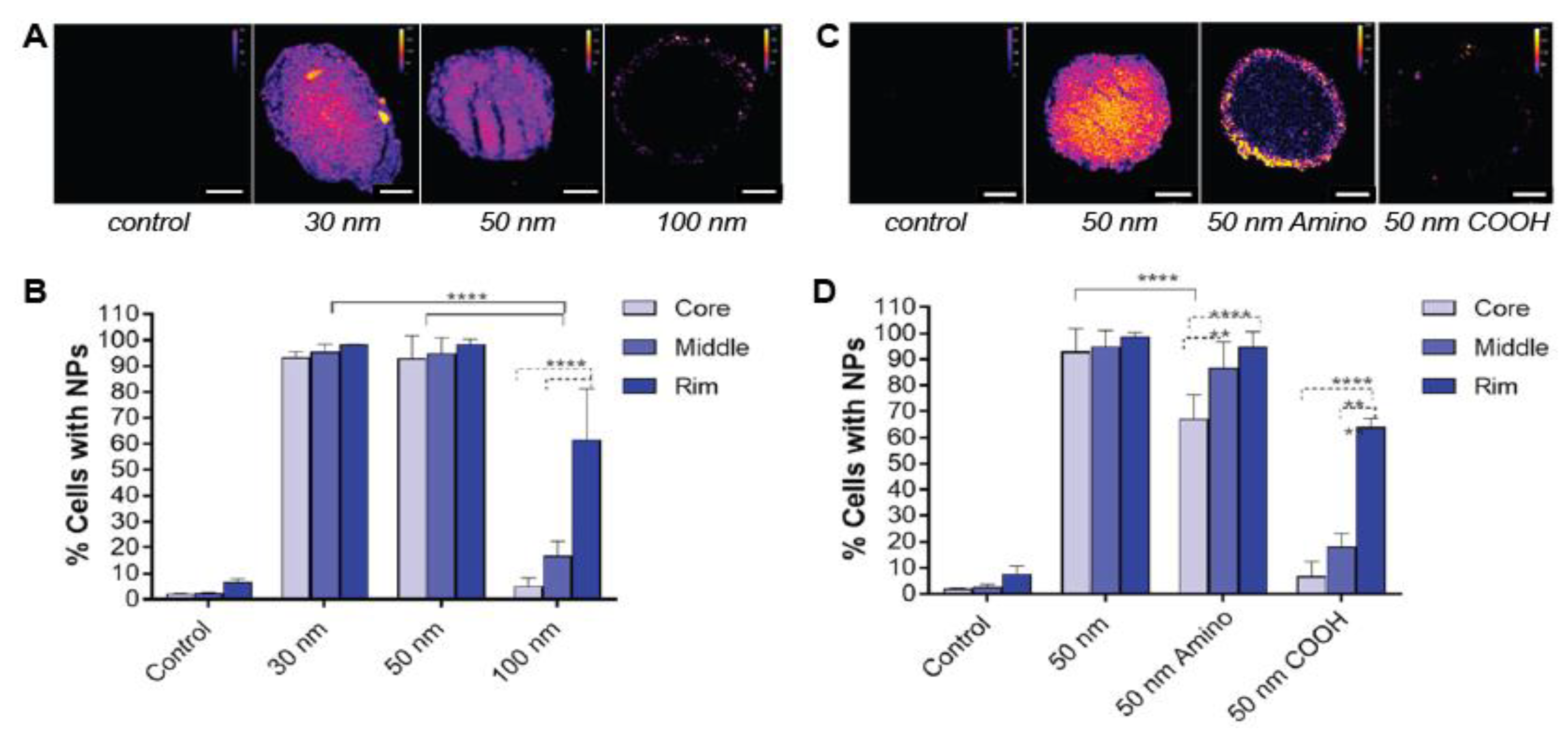
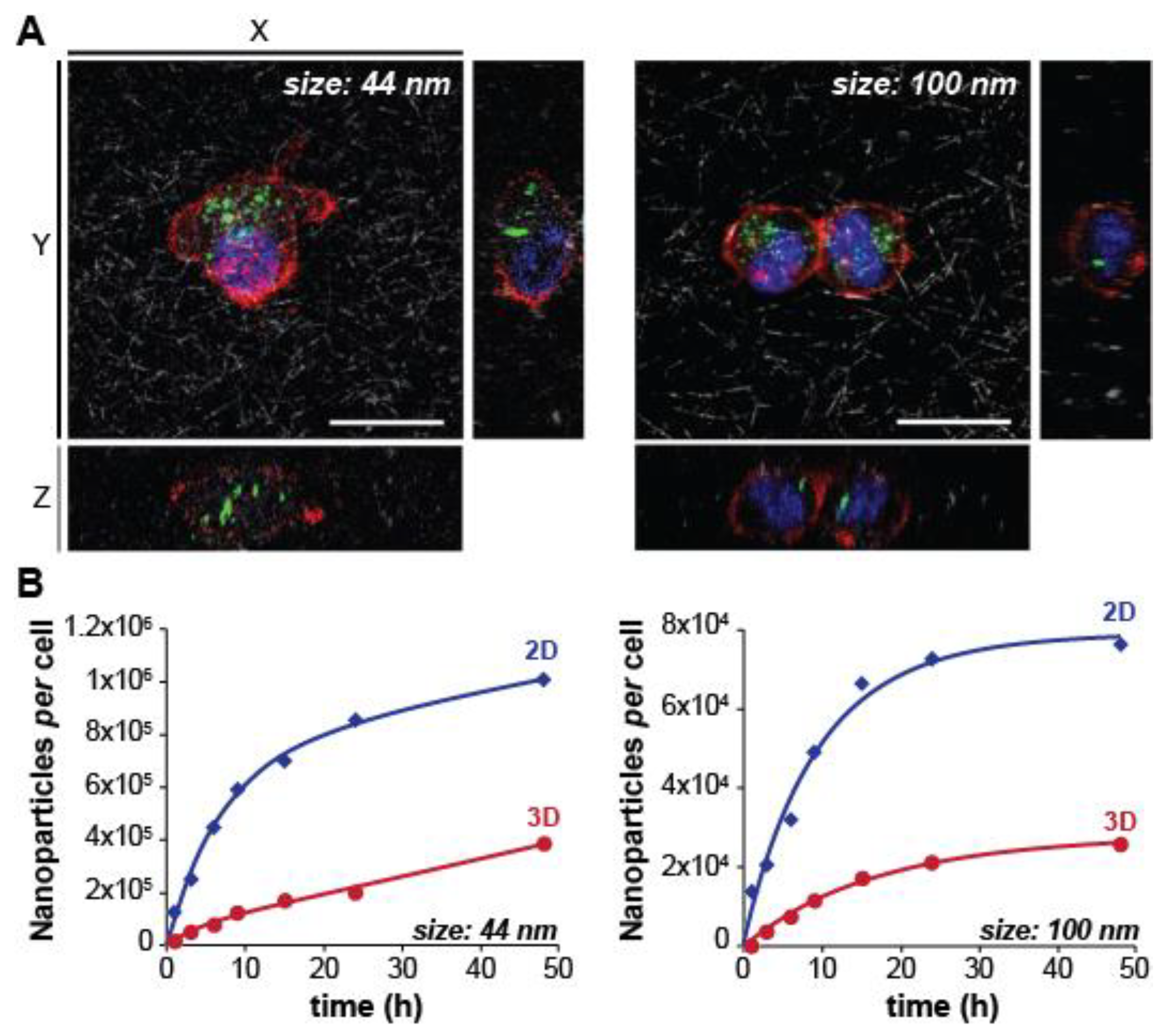
2.1.1. Nanoparticle Size
2.1.2. Surface Charge
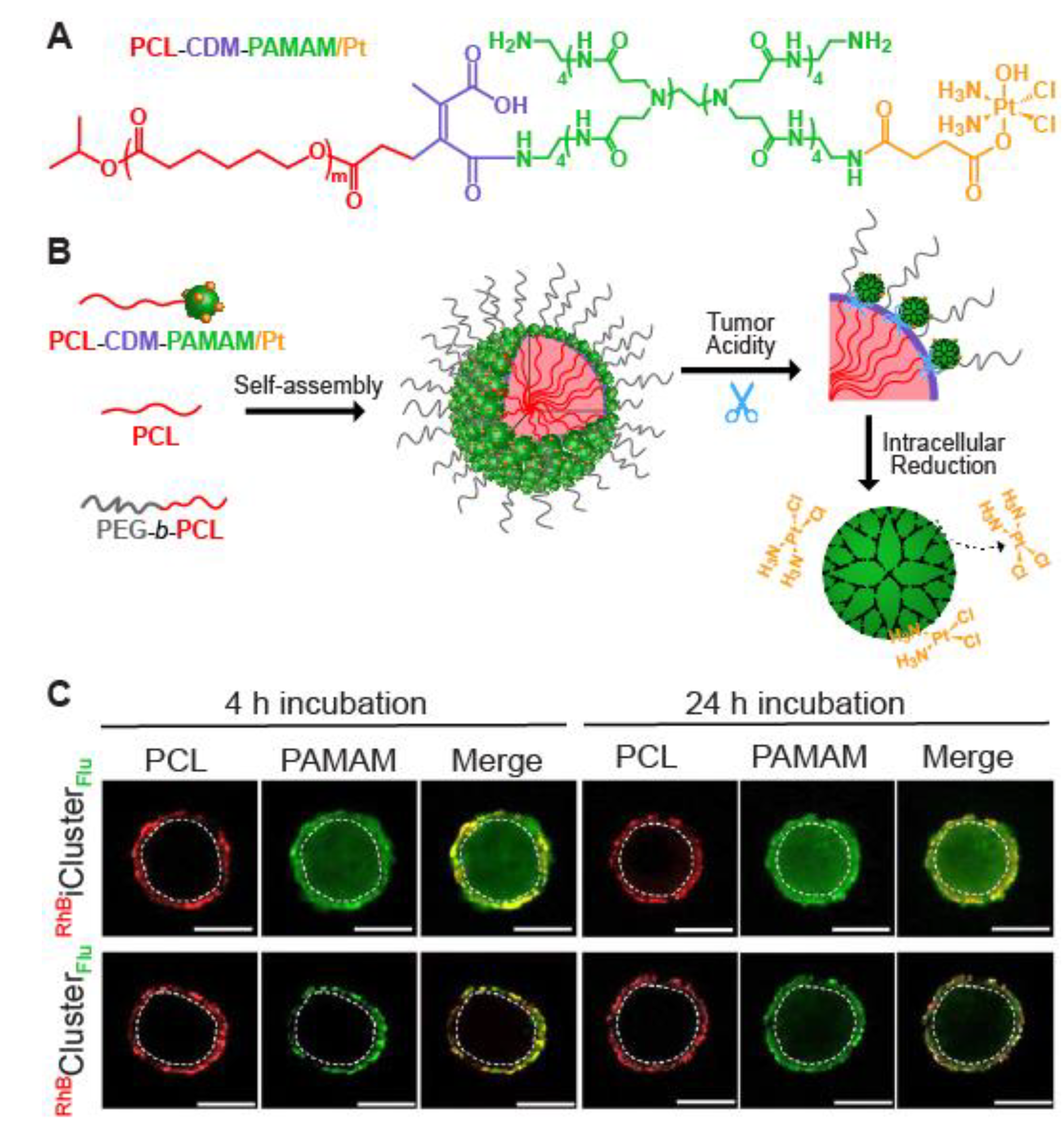
2.1.3. Surface Functionalization
2.2. Scaffold-Embedded 3D Models
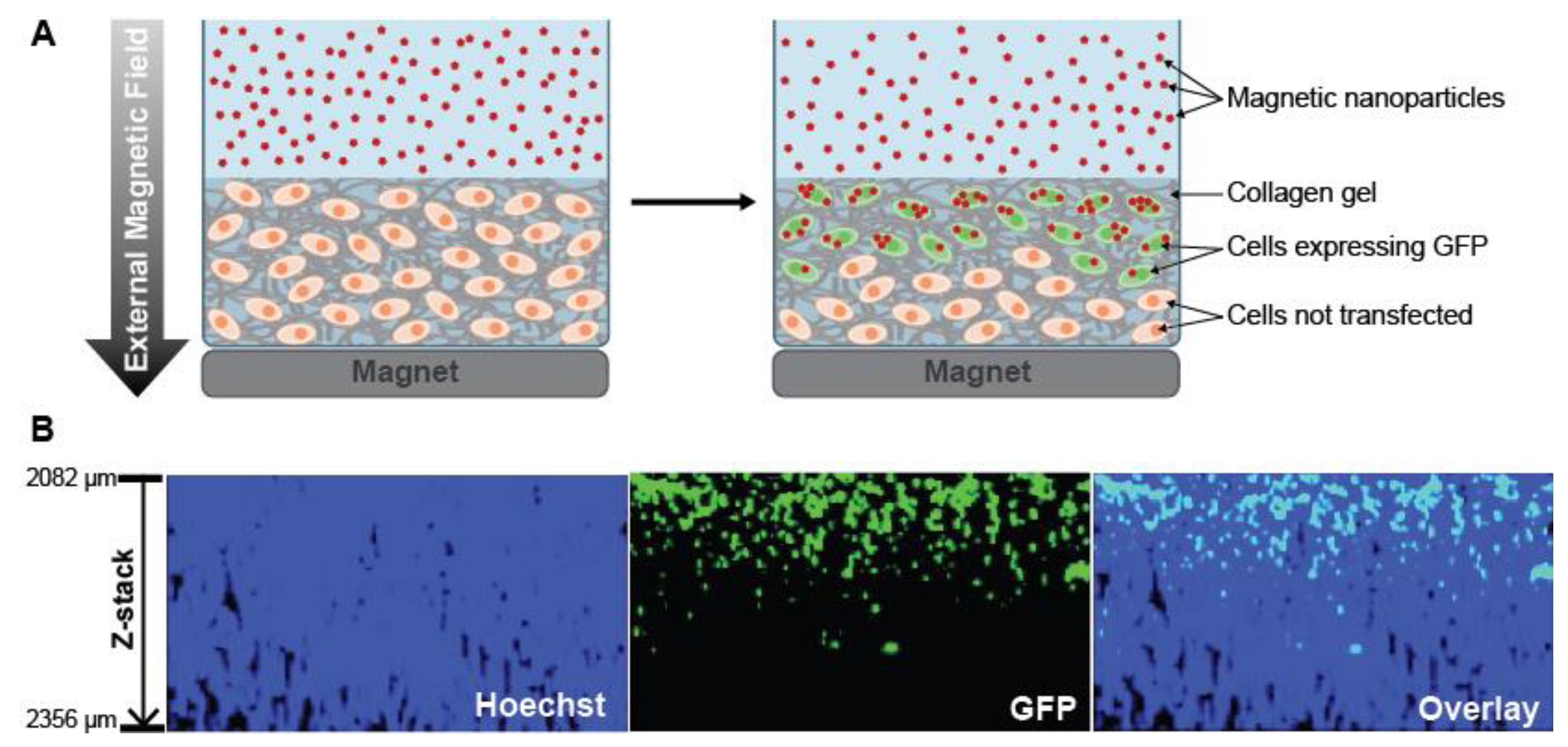
2.2.1. Scaffold-Embedded Cells
2.2.2. Human-Derived Cancer Organoids
2.3. Microfluidic Platforms
3. Characterization and Biological Assays for 3D Models
4. Summary and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO | Publications on Cancer. Available online: http://www.who.int/cancer/publications/en/ (accessed on 17 October 2020).
- Sriraman, S.K.; Aryasomayajula, B.; Torchilin, V.P. Barriers to drug delivery in solid tumors. Tissue Barriers 2014, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Li, J.; Wang, Y.; Koenig, L.; Gjyrezi, A.; Giannakakou, P.; Shin, E.H.; Tighiouart, M.; Chen, Z.; Nie, S.; et al. A folate receptor-targeting nanoparticle minimizes drug resistance in a human cancer model. ACS Nano 2011, 5, 6184–6194. [Google Scholar] [CrossRef] [PubMed]
- Zwicke, G.L.; Mansoori, G.A.; Jeffery, C.J. Utilizing the folate receptor for active targeting of cancer nanotherapeutics. Nano Rev. 2012, 3, 18496. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, B.; Hojjat-Farsangi, M.; Mohammadi, H.; Anvari, E.; Ghalamfarsa, G.; Yousefi, M.; Jadidi-Niaragh, F. Nanoparticles and targeted drug delivery in cancer therapy. Immunol. Lett. 2017, 190, 64–83. [Google Scholar] [CrossRef]
- Dadwal, A.; Baldi, A.; Narang, R.K. Nanoparticles as carriers for drug delivery in cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y. (Chezy) Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Miele, E.; Spinelli, G.P.; Miele, E.; Tomao, F.; Tomao, S. Albumin-bound formulation of paclitaxel (Abraxane® ABI-007) in the treatment of breast cancer. Int. J. Nanomed. 2009, 4, 99–105. [Google Scholar]
- Goodman, T.T.; Ng, C.P.; Pun, S.H. 3-D Tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjugate Chem. 2008, 19, 1951–1959. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.K.; Stylianopoulos, T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010, 7, 653–664. [Google Scholar] [CrossRef] [Green Version]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Foglietta, F.; Canaparo, R.; Muccioli, G.; Terreno, E.; Serpe, L. Methodological aspects and pharmacological applications of three-dimensional cancer cell cultures and organoids. Life Sci. 2020, 254, 117784. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzari, G.; Couvreur, P.; Mura, S. Multicellular tumor spheroids: A relevant 3D model for the in vitro preclinical investigation of polymer nanomedicines. Polym. Chem. 2017. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Stenzel, M.H. Multicellular Tumor Spheroids (MCTS) as a 3D in vitro evaluation tool of nanoparticles. Small 2018, 14, e1702858. [Google Scholar] [CrossRef]
- Mapanao, A.K.; Voliani, V. Three-dimensional tumor models: Promoting breakthroughs in nanotheranostics translational research. Appl. Mater. Today 2020, 19, 100552. [Google Scholar] [CrossRef]
- Santini, M.T.; Rainaldi, G.; Indovina, P.L. Apoptosis, cell adhesion and the extracellular matrix in the three-dimensional growth of multicellular tumor spheroids. Crit. Rev. Oncol. Hematol. 2000, 36, 75–87. [Google Scholar] [CrossRef]
- Djordjevic, B.; Lange, C.S. Cell-cell interactions in spheroids maintained in suspension. Acta Oncol. 2006, 45, 412–420. [Google Scholar] [CrossRef]
- Oktem, G.; Bilir, A.; Ayla, S.; Yavasoglu, A.; Goksel, G.; Saydam, G.; Uysal, A. Role of intercellular communications in breast cancer multicellular tumor spheroids after chemotherapy. Oncol. Res. 2006, 16, 225–233. [Google Scholar] [CrossRef]
- Metzger, W.; Sossong, D.; Bächle, A.; Pütz, N.; Wennemuth, G.; Pohlemann, T.; Oberringer, M. The liquid overlay technique is the key to formation of co-culture spheroids consisting of primary osteoblasts, fibroblasts and endothelial cells. Cytotherapy 2011, 13, 1000–1012. [Google Scholar] [CrossRef]
- Foty, R. A Simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011. [Google Scholar] [CrossRef]
- Souza, G.R.; Molina, J.R.; Raphael, R.M.; Ozawa, M.G.; Stark, D.J.; Levin, C.S.; Bronk, L.F.; Ananta, J.S.; Mandelin, J.; Georgescu, M.-M.; et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat. Nanotechnol. 2010, 5, 291–296. [Google Scholar] [CrossRef] [Green Version]
- Velasco, V.; Shariati, S.A.; Esfandyarpour, R. Microtechnology-based methods for organoid models. Microsyst. Nanoeng. 2020, 6, 1–13. [Google Scholar] [CrossRef]
- Liao, W.; Wang, J.; Xu, J.; You, F.; Pan, M.; Xu, X.; Weng, J.; Han, X.; Li, S.; Li, Y.; et al. High-throughput three-dimensional spheroid tumor model using a novel stamp-like tool. J. Tissue Eng. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.; Carlsson, J.; Durand, R.; Yuhas, J. Spheroids in cancer research. Cancer Res 1981, 41, 2980–2984. [Google Scholar]
- Ware, M.J.; Colbert, K.; Keshishian, V.; Ho, J.; Corr, S.J.; Curley, S.A.; Godin, B. Generation of homogenous three-dimensional pancreatic cancer cell spheroids using an improved hanging drop technique. Tissue Eng. Part C Methods 2016, 22, 312–321. [Google Scholar] [CrossRef] [Green Version]
- Franko, A.J.; Freedman, H.I. Model of diffusion of oxygen to spheroids grown in stationary medium—I. Complete spherical symmetry. Bull. Math. Biol. 1984, 46, 205–217. [Google Scholar]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Pérez, J.; Ballesteros, P.; Cerdán, S. Microscopic images of intraspheroidal pH by 1H magnetic resonance chemical shift imaging of pH sensitive indicators. MAGMA 2005, 18, 293–301. [Google Scholar] [CrossRef]
- Görlach, A.; Acker, H. pO2- and pH-gradients in multicellular spheroids and their relationship to cellular metabolism and radiation sensitivity of malignant human tumor cells. BBA Mol. Basis Dis. 1994, 1227, 105–112. [Google Scholar] [CrossRef]
- Tevis, K.M.; Cecchi, R.J.; Colson, Y.L.; Grinstaff, M.W. Mimicking the tumor microenvironment to regulate macrophage phenotype and assessing chemotherapeutic efficacy in embedded cancer cell/macrophage spheroid models. Acta Biomater. 2017, 50, 271–279. [Google Scholar] [CrossRef] [Green Version]
- Rebelo, S.P.; Pinto, C.; Martins, T.R.; Harrer, N.; Estrada, M.F.; Loza-Alvarez, P.; Cabeçadas, J.; Alves, P.M.; Gualda, E.J.; Sommergruber, W.; et al. 3D-3-culture: A tool to unveil macrophage plasticity in the tumour microenvironment. Biomaterials 2018, 163, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Nederman, T.; Norling, B.; Glimelius, B.; Carlsson, J.; Brunk, U. Demonstration of an extracellular matrix in multicellular tumor spheroids. Cancer Res. 1984, 44, 3090–3097. [Google Scholar] [PubMed]
- Goodman, T.T.; Olive, P.L.; Pun, S.H. Increased nanoparticle penetration in collagenase-treated multicellular spheroids. Int. J. Nanomed. 2007, 2, 265–274. [Google Scholar]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Hinger, D.; Navarro, F.; Käch, A.; Thomann, J.-S.; Mittler, F.; Couffin, A.-C.; Maake, C. Photoinduced effects of m-tetrahydroxyphenylchlorin loaded lipid nanoemulsions on multicellular tumor spheroids. J. Nanobiotechnol. 2016, 14, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchoryk, A.; Taresco, V.; Argent, R.H.; Ashford, M.; Gellert, P.R.; Stolnik, S.; Grabowska, A.; Garnett, M.C. Penetration and uptake of nanoparticles in 3D tumor spheroids. Bioconjugate Chem. 2019, 30, 1371–1384. [Google Scholar] [CrossRef]
- Bugno, J.; Hsu, H.-J.; Pearson, R.M.; Noh, H.; Hong, S. Size and Surface Charge of engineered poly (amidoamine) dendrimers modulate tumor accumulation and penetration: A model study using multicellular tumor spheroids. Mol. Pharm. 2016, 13, 2155–2163. [Google Scholar] [CrossRef]
- Huo, S.; Ma, H.; Huang, K.; Liu, J.; Wei, T.; Jin, S.; Zhang, J.; He, S.; Liang, X.-J. Superior penetration and retention behavior of 50 nm gold nanoparticles in tumors. Cancer Res. 2013, 73, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Ma, H.; Liu, J.; Huo, S.; Kumar, A.; Wei, T.; Zhang, X.; Jin, S.; Gan, Y.; Wang, P.C.; et al. Size-dependent localization and penetration of ultrasmall gold nanoparticles in cancer cells, multicellular spheroids and tumors in vivo. ACS Nano 2012, 6, 4483–4493. [Google Scholar] [CrossRef] [Green Version]
- Priwitaningrum, D.L.; Blondé, J.-B.G.; Sridhar, A.; van Baarlen, J.; Hennink, W.E.; Storm, G.; Le Gac, S.; Prakash, J. Tumor stroma-containing 3D spheroid arrays: A tool to study nanoparticle penetration. J. Control Release 2016, 244, 257–268. [Google Scholar] [CrossRef]
- Jin, S.; Ma, X.; Ma, H.; Zheng, K.; Liu, J.; Hou, S.; Meng, J.; Wang, P.C.; Wu, X.; Liang, X.-J. Surface chemistry-mediated penetration and gold nanorod thermotherapy in multicellular tumor spheroids. Nanoscale 2012, 5, 143–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.-J.; Du, J.-Z.; Du, X.-J.; Xu, C.-F.; Sun, C.-Y.; Wang, H.-X.; Cao, Z.-T.; Yang, X.-Z.; Zhu, Y.-H.; Nie, S.; et al. Stimuli-responsive clustered nanoparticles for improved tumor penetration and therapeutic efficacy. Proc. Natl. Acad. Sci. USA 2016, 113, 4164–4169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Q.; Wang, S.-B.; Hu, J.-J.; Lin, Y.-X.; Zhu, C.-H.; Rong, L.; Zhang, X.-Z. Stimuli-Responsive “Cluster Bomb” for programmed tumor therapy. ACS Nano 2017, 11, 7201–7214. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-J.; Du, J.-Z.; Liu, J.; Du, X.-J.; Shen, S.; Zhu, Y.-H.; Wang, X.; Ye, X.; Nie, S.; Wang, J. Smart superstructures with ultrahigh ph-sensitivity for targeting acidic tumor microenvironment: Instantaneous size switching and improved tumor penetration. ACS Nano 2016, 10, 6753–6761. [Google Scholar] [CrossRef]
- Xue, X.; Huang, Y.; Bo, R.; Jia, B.; Wu, H.; Yuan, Y.; Wang, Z.; Ma, Z.; Jing, D.; Xu, X.; et al. Trojan horse nanotheranostics with dual transformability and multifunctionality for highly effective cancer treatment. Nat. Commun. 2018, 9, 3653. [Google Scholar] [CrossRef] [PubMed]
- Ruan, S.; Cao, X.; Cun, X.; Hu, G.; Zhou, Y.; Zhang, Y.; Lu, L.; He, Q.; Gao, H. Matrix metalloproteinase-sensitive size-shrinkable nanoparticles for deep tumor penetration and pH triggered doxorubicin release. Biomaterials 2015, 60, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Huang-Fu, M.-Y.; Guo, W.-W.; Guo, N.-N.; Chen, J.; Liu, H.-N.; Xie, Z.-Q.; Lin, M.-T.; Wei, Q.-C.; Gao, J.-Q. MMP-2-Sensitive HA End-Conjugated Poly (amidoamine) dendrimers via click reaction to enhance drug penetration into solid tumor. ACS Appl. Mater. Interfaces 2017, 9, 42459–42470. [Google Scholar] [CrossRef]
- Gao, H.; Xiong, Y.; Zhang, S.; Yang, Z.; Cao, S.; Jiang, X. RGD and interleukin-13 peptide functionalized nanoparticles for enhanced glioblastoma cells and neovasculature dual targeting delivery and elevated tumor penetration. Mol. Pharm. 2014, 11, 1042–1052. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Y.; Li, J.; Peng, Z.-H.; Sheinin, Y.; Zhou, J.; Oupický, D. Tumor-penetrating nanoparticles for enhanced anticancer activity of combined photodynamic and hypoxia-activated therapy. ACS Nano 2017, 11, 2227–2238. [Google Scholar] [CrossRef]
- Wang, R.; Han, Y.; Sun, B.; Zhao, Z.; Opoku-Damoah, Y.; Cheng, H.; Zhang, H.; Zhou, J.; Ding, Y. Deep tumor penetrating bioparticulates inspired burst intracellular drug release for precision chemo-phototherapy. Small 2018, 14, 1703110. [Google Scholar] [CrossRef]
- Kang, T.; Jiang, M.; Jiang, D.; Feng, X.; Yao, J.; Song, Q.; Chen, H.; Gao, X.; Chen, J. Enhancing glioblastoma-specific penetration by functionalization of nanoparticles with an iron-mimic peptide targeting transferrin/transferrin receptor complex. Mol. Pharm. 2015, 12, 2947–2961. [Google Scholar] [CrossRef] [PubMed]
- van Oppen, L.M.P.E.; Pille, J.; Stuut, C.; van Stevendaal, M.; van der Vorm, L.N.; Smeitink, J.A.M.; Koopman, W.J.H.; Willems, P.H.G.M.; van Hest, J.C.M.; Brock, R. Octa-arginine boosts the penetration of elastin-like polypeptide nanoparticles in 3D cancer models. Eur. J. Pharm. Biopharm. 2019, 137, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, V.M.; Costa, E.C.; Queiroz, J.A.; Pichon, C.; Sousa, F.; Correia, I.J. Folate-targeted multifunctional amino acid-chitosan nanoparticles for improved cancer therapy. Pharm. Res. 2015, 32, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.-L.; Yu, T.-W.; Chiang, W.-H.; Chiu, H.-C.; Chang, C.-H.; Chiang, C.-S.; Hu, S.-H. Hierarchically targeted and penetrated delivery of drugs to tumors by size-changeable graphene quantum dot nanoaircrafts for photolytic therapy. Adv. Funct. Mater. 2017, 27, 1700056. [Google Scholar] [CrossRef]
- Kulkarni, P.; Haldar, M.K.; Katti, P.; Dawes, C.; You, S.; Choi, Y.; Mallik, S. Hypoxia responsive, tumor penetrating lipid nanoparticles for delivery of chemotherapeutics to pancreatic cancer cell spheroids. Bioconjugate Chem. 2016, 27, 1830–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Lu, H.; Wong, S.; Lu, M.; Xiao, P.; Stenzel, M.H. Influence of nanoparticle shapes on cellular uptake of paclitaxel loaded nanoparticles in 2D and 3D cancer models. Polym. Chem. 2017, 8, 3317–3326. [Google Scholar] [CrossRef]
- Zhao, J.; Lu, H.; Yao, Y.; Ganda, S.; Stenzel, M.H. Length vs. stiffness: Which plays a dominant role in the cellular uptake of fructose-based rod-like micelles by breast cancer cells in 2D and 3D cell culture models? J. Mater. Chem. B 2018, 6, 4223–4231. [Google Scholar] [CrossRef]
- Stern, T.; Kaner, I.; Laser Zer, N.; Shoval, H.; Dror, D.; Manevitch, Z.; Chai, L.; Brill-Karniely, Y.; Benny, O. Rigidity of polymer micelles affects interactions with tumor cells. J. Control. Release 2017, 257, 40–50. [Google Scholar] [CrossRef]
- Solomon, M.A.; Lemera, J.; D’Souza, G.G.M. Development of an in vitro tumor spheroid culture model amenable to high-throughput testing of potential anticancer nanotherapeutics. J. Liposome Res. 2016, 26, 246–260. [Google Scholar] [CrossRef]
- Tang, L.; Gabrielson, N.P.; Uckun, F.M.; Fan, T.M.; Cheng, J. Size-dependent tumor penetration and in vivo efficacy of monodisperse drug–silica nanoconjugates. Mol. Pharm. 2013, 10, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Zheng, J. Clearance pathways and tumor targeting of imaging nanoparticles. ACS Nano 2015, 9, 6655–6674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Quan, L.; Zhou, C.; Zhan, Q. Factors relating to the biodistribution & clearance of nanoparticles & their effects on in vivo application. Nanomedicine 2018, 13, 1495–1512. [Google Scholar] [PubMed]
- Ernsting, M.J.; Murakami, M.; Roy, A.; Li, S.-D. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. J. Control Release 2013, 172, 782–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Yuan, S.; Huo, M.; Chaudhuri, A.S.; Zhao, M.; Wu, Z.; Qi, X. Spatial controlled multistage nanocarriers through hybridization of dendrimers and gelatin nanoparticles for deep penetration and therapy into tumor tissue. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1399–1410. [Google Scholar] [CrossRef]
- Fam, S.Y.; Chee, C.F.; Yong, C.Y.; Ho, K.L.; Mariatulqabtiah, A.R.; Tan, W.S. Stealth coating of nanoparticles in drug-delivery systems. Nanomaterials 2020, 10, 787. [Google Scholar] [CrossRef] [Green Version]
- De la Rosa, V.R. Poly (2-oxazoline) s as materials for biomedical applications. J. Mater. Sci Mater. Med. 2014, 25, 1211–1225. [Google Scholar] [CrossRef]
- Cao, Z.; Jiang, S. Super-hydrophilic zwitterionic poly (carboxybetaine) and amphiphilic non-ionic poly (ethylene glycol) for stealth nanoparticles. Nano Today 2012, 7, 404–413. [Google Scholar] [CrossRef]
- Cao, J.; Chen, Y.-W.; Wang, X.; Luo, X.-L. Enhancing blood compatibility of biodegradable polymers by introducing sulfobetaine. J. Biomed. Mater. Res. Part A 2011, 97A, 472–479. [Google Scholar] [CrossRef]
- Fortuni, B.; Inose, T.; Ricci, M.; Fujita, Y.; Zundert, I.V.; Masuhara, A.; Fron, E.; Mizuno, H.; Latterini, L.; Rocha, S.; et al. Polymeric engineering of nanoparticles for highly efficient multifunctional drug delivery systems. Sci. Rep. 2019, 9, 2666. [Google Scholar] [CrossRef]
- Yoo, M.-K.; Park, I.-K.; Lim, H.-T.; Lee, S.-J.; Jiang, H.-L.; Kim, Y.-K.; Choi, Y.-J.; Cho, M.-H.; Cho, C.-S. Folate–PEG–superparamagnetic iron oxide nanoparticles for lung cancer imaging. Acta Biomater. 2012, 8, 3005–3013. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science 2010, 328, 1031–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessner, I.; Neundorf, I. Nanoparticles modified with cell-penetrating peptides: Conjugation mechanisms, physicochemical properties and application in cancer diagnosis and therapy. Int. J. Mol. Sci. 2020, 21, 2536. [Google Scholar] [CrossRef] [Green Version]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef]
- Salomone, F.; Cardarelli, F.; Di Luca, M.; Boccardi, C.; Nifosì, R.; Bardi, G.; di Bari, L.; Serresi, M.; Beltram, F. A novel chimeric cell-penetrating peptide with membrane-disruptive properties for efficient endosomal escape. J. Control Release 2012, 163, 293–303. [Google Scholar] [CrossRef]
- Zhou, H.; Fan, Z.; Deng, J.; Lemons, P.K.; Arhontoulis, D.C.; Bowne, W.B.; Cheng, H. Hyaluronidase embedded in nanocarrier peg shell for enhanced tumor penetration and highly efficient antitumor efficacy. Nano Lett. 2016, 16, 3268–3277. [Google Scholar] [CrossRef]
- Bremnes, R.M.; Dønnem, T.; Al-Saad, S.; Al-Shibli, K.; Andersen, S.; Sirera, R.; Camps, C.; Marinez, I.; Busund, L.-T. The role of tumor stroma in cancer progression and prognosis: Emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Werb, Z.; Lu, P. The role of stroma in tumor development. Cancer J. 2015, 21, 250–253. [Google Scholar] [CrossRef] [Green Version]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Leong, D.T.; Ng, K.W. Probing the relevance of 3D cancer models in nanomedicine research. Adv. Drug Deliv. Rev. 2014, 79–80, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Kouwer, P.H.J.; Koepf, M.; le Sage, V.A.A.; Jaspers, M.; van Buul, A.M.; Eksteen-Akeroyd, Z.H.; Woltinge, T.; Schwartz, E.; Kitto, H.J.; Hoogenboom, R.; et al. Responsive biomimetic networks from polyisocyanopeptide hydrogels. Nature 2013, 493, 651–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandaele, J.; Louis, B.; Liu, K.; Camacho, R.; Kouwer, P.H.J.; Rocha, S. Structural characterization of fibrous synthetic hydrogels using fluorescence microscopy. Soft Matter 2020, 16, 4210–4219. [Google Scholar] [CrossRef]
- Liu, K.; Mihaila, S.M.; Rowan, A.; Oosterwijk, E.; Kouwer, P.H.J. Synthetic extracellular matrices with nonlinear elasticity regulate cellular organization. Biomacromolecules 2019, 20, 826–834. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, C.; Span, P.N.; Rowan, A.E.; Aalders, T.W.; Schalken, J.A.; Adema, G.J.; Kouwer, P.H.J.; Zegers, M.M.P.; Ansems, M. Polyisocyanide hydrogels as a tunable platform for mammary gland organoid formation. Adv. Sci. 2020. [Google Scholar] [CrossRef]
- Belli, V.; Guarnieri, D.; Biondi, M.; Sala, F.D.; Netti, P.A. Dynamics of nanoparticle diffusion and uptake in three-dimensional cell cultures. Colloids Surf. B Biointerfaces 2017, 149, 7–15. [Google Scholar] [CrossRef]
- Ramanujan, S.; Pluen, A.; McKee, T.D.; Brown, E.B.; Boucher, Y.; Jain, R.K. Diffusion and convection in collagen gels: Implications for transport in the tumor interstitium. Biophys. J. 2002, 83, 1650–1660. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Lee, M.-Y.; Hogg, M.G.; Dordick, J.S.; Sharfstein, S.T. Gene delivery in three-dimensional cell cultures by superparamagnetic nanoparticles. ACS Nano 2010, 4, 4733–4743. [Google Scholar] [CrossRef]
- Dai, Q.; Wilhelm, S.; Ding, D.; Syed, A.M.; Sindhwani, S.; Zhang, Y.; Chen, Y.Y.; MacMillan, P.; Chan, W.C.W. Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors. ACS Nano 2018, 12, 8423–8435. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Pitek, A.S.; Lynch, I.; Dawson, K.A. Formation and characterization of the nanoparticle-protein corona. Methods Mol. Biol. 2013, 1025, 137–155. [Google Scholar] [PubMed]
- Ke, P.C.; Lin, S.; Parak, W.J.; Davis, T.P.; Caruso, F. A decade of the protein corona. ACS Nano 2017, 11, 11773–11776. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.A.; Addison, C.L. β1 integrin: An emerging player in the modulation of tumorigenesis and response to therapy. Cell Adh. Migr. 2012, 6, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, V.C.; Higuita-Castro, N.; Nana-Sinkam, P.; Ghadiali, S.N. Substrate stiffness modulates lung cancer cell migration but not epithelial to mesenchymal transition. J. Biomed. Mater. Res. Part A 2016, 104, 1182–1193. [Google Scholar] [CrossRef]
- Prina-Mello, A.; Jain, N.; Liu, B.; Kilpatrick, J.I.; Tutty, M.A.; Bell, A.P.; Jarvis, S.P.; Volkov, Y.; Movia, D. Culturing substrates influence the morphological, mechanical and biochemical features of lung adenocarcinoma cells cultured in 2D or 3D. Tissue Cell 2018, 50, 15–30. [Google Scholar] [CrossRef]
- Hutmacher, D.W. Biomaterials offer cancer research the third dimension. Nat. Mater. 2010, 9, 90–93. [Google Scholar] [CrossRef]
- Friedrich, J.; Ebner, R.; Kunz-Schughart, L.A. Experimental anti-tumor therapy in 3-D: Spheroids–old hat or new challenge? Int. J. Radiat. Biol. 2007, 83, 849–871. [Google Scholar] [CrossRef]
- Jeng, R.L.; Welch, M.D. Cytoskeleton: Actin and endocytosis—No longer the weakest link. Curr. Biol. 2001, 11, R691–R694. [Google Scholar] [CrossRef] [Green Version]
- Qualmann, B.; Kessels, M.M. Endocytosis and the cytoskeleton. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 2002; Volume 220, pp. 93–144. [Google Scholar]
- Wei, X.; Wei, R.; Jiang, G.; Jia, Y.; Lou, H.; Yang, Z.; Luo, D.; Huang, Q.; Xu, S.; Yang, X.; et al. Mechanical cues modulate cellular uptake of nanoparticles in cancer via clathrin-mediated and caveolae-mediated endocytosis pathways. Nanomedicine 2019, 14, 613–626. [Google Scholar] [CrossRef]
- Lutolf, M.P.; Gilbert, P.M.; Blau, H.M. Designing materials to direct stem-cell fate. Nature 2009, 462, 433–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assoian, R.K.; Klein, E.A. Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol. 2008, 18, 347–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT–mTOR–S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longati, P.; Jia, X.; Eimer, J.; Wagman, A.; Witt, M.-R.; Rehnmark, S.; Verbeke, C.; Toftgård, R.; Löhr, M.; Heuchel, R.L. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 2013, 13, 95. [Google Scholar] [CrossRef] [Green Version]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Boretto, M.; Maenhoudt, N.; Luo, X.; Hennes, A.; Boeckx, B.; Bui, B.; Heremans, R.; Perneel, L.; Kobayashi, H.; Van Zundert, I.; et al. Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 2019, 21, 1041–1051. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Huch, M. Disease modelling in human organoids. Dis. Model Mech. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Busslinger, G.A.; Lissendorp, F.; Franken, I.A.; van Hillegersberg, R.; Ruurda, J.P.; Clevers, H.; de Maat, M.F.G. The potential and challenges of patient-derived organoids in guiding the multimodality treatment of upper gastrointestinal malignancies. Open Biol. 2020, 10, 190274. [Google Scholar] [CrossRef] [Green Version]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing organoids from ovarian cancer as experimental and preclinical models. Stem. Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colella, G.; Fazioli, F.; Gallo, M.; De Chiara, A.; Apice, G.; Ruosi, C.; Cimmino, A.; de Nigris, F. Sarcoma spheroids and organoids—Promising tools in the era of personalized medicine. Int. J. Mol. Sci. 2018, 19, 615. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; Kolders, S.; Spelier, S.; Lõhmussaar, K.; Willems, S.M.; Devriese, L.A.; de Bree, R.; de Ruiter, E.J.; Korving, J.; Begthel, H.; et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 2019. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef]
- Park, S.B.; Jung, W.H.; Kim, K.Y.; Koh, B. Toxicity assessment of SiO2 and TiO2 in normal colon cells, in vivo and in human colon organoids. Molecules 2020, 25, 3594. [Google Scholar] [CrossRef]
- Bergenheim, F.; Seidelin, J.B.; Pedersen, M.T.; Mead, B.E.; Jensen, K.B.; Karp, J.M.; Nielsen, O.H. Fluorescence-based tracing of transplanted intestinal epithelial cells using confocal laser endomicroscopy. Stem. Cell Res. Ther. 2019, 10, 148. [Google Scholar] [CrossRef]
- Pastuła, A.; Middelhoff, M.; Brandtner, A.; Tobiasch, M.; Höhl, B.; Nuber, A.H.; Demir, I.E.; Neupert, S.; Kollmann, P.; Mazzuoli-Weber, G.; et al. Three-Dimensional Gastrointestinal Organoid Culture in Combination with Nerves or Fibroblasts: A Method to Characterize the Gastrointestinal Stem Cell Niche. Available online: https://www.hindawi.com/journals/sci/2016/3710836/ (accessed on 1 August 2020).
- Fede, C.; Fortunati, I.; Weber, V.; Rossetto, N.; Bertasi, F.; Petrelli, L.; Guidolin, D.; Signorini, R.; de Caro, R.; Albertin, G.; et al. Evaluation of gold nanoparticles toxicity towards human endothelial cells under static and flow conditions. Microvasc. Res. 2015, 97, 147–155. [Google Scholar] [CrossRef]
- Ozcelikkale, A.; Moon, H.; Linnes, M.; Han, B. In vitro microfluidic models of tumor microenvironment to screen transport of drugs and nanoparticles. WIREs Nanomed. Nanobiotechnol. 2017, 9, e1460. [Google Scholar] [CrossRef]
- Ruppen, J.; Cortes-Dericks, L.; Marconi, E.; Karoubi, G.; Schmid, R.A.; Peng, R.; Marti, T.M.; Guenat, O.T. A microfluidic platform for chemoresistive testing of multicellular pleural cancer spheroids. Lab Chip 2014, 14, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; Boerhan, R.; Liu, C.; Jiang, G. Nanoparticles penetrate into the multicellular spheroid-on-chip: Effect of surface charge, protein corona and exterior flow. Mol. Pharm. 2017, 14, 4618–4627. [Google Scholar] [CrossRef] [PubMed]
- Patra, B.; Peng, C.-C.; Liao, W.-H.; Lee, C.-H.; Tung, Y.-C. Drug testing and flow cytometry analysis on a large number of uniform sized tumor spheroids using a microfluidic device. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astolfi, M.; Péant, B.; Lateef, M.A.; Rousset, N.; Kendall-Dupont, J.; Carmona, E.; Monet, F.; Saad, F.; Provencher, D.; Mes-Masson, A.-M.; et al. Micro-dissected tumor tissues on chip: An ex vivo method for drug testing and personalized therapy. Lab Chip 2016, 16, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Toley, B.J.; Lovatt, Z.T.; Harrington, J.L.; Forbes, N.S. Microfluidic technique to measure intratumoral transport and calculate drug efficacy shows that binding is essential for doxorubicin and release hampers doxil. Integr. Biol. 2013, 5, 1184–1196. [Google Scholar] [CrossRef]
- Albanese, A.; Lam, A.K.; Sykes, E.A.; Rocheleau, J.V.; Chan, W.C.W. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 2013, 4, 2718. [Google Scholar] [CrossRef] [Green Version]
- Farahat, W.A.; Wood, L.B.; Zervantonakis, I.K.; Schor, A.; Ong, S.; Neal, D.; Kamm, R.D.; Asada, H.H. Ensemble analysis of angiogenic growth in three-dimensional microfluidic cell cultures. PLoS ONE 2012, 7, e37333. [Google Scholar] [CrossRef] [Green Version]
- Schuerle, S.; Soleimany, A.P.; Yeh, T.; Anand, G.M.; Häberli, M.; Fleming, H.E.; Mirkhani, N.; Qiu, F.; Hauert, S.; Wang, X.; et al. Synthetic and living micropropellers for convection-enhanced nanoparticle transport. Sci. Adv. 2019, 5, eaav4803. [Google Scholar] [CrossRef]
- Feiner-Gracia, N.; Mares, A.G.; Buzhor, M.; Rodriguez-Trujillo, R.; Samitier, J.; Amir, R.J.; Pujals, S.; Albertazzi, L. Real-time ratiometric imaging of micelles assembly state in a microfluidic cancer-on-a-chip. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kwak, B.; Ozcelikkale, A.; Shin, C.S.; Park, K.; Han, B. Simulation of complex transport of nanoparticles around a tumor using tumor-microenvironment-on-chip. J. Control Release 2014, 194, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Greish, K. Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. Methods Mol. Biol. 2010, 624, 25–37. [Google Scholar] [PubMed]
- Wang, A.Z. EPR or no EPR? The billion-dollar question. Sci. Transl. Med. 2015, 7, ec112–ec294. [Google Scholar] [CrossRef]
- Nel, A.; Ruoslahti, E.; Meng, H. New insights into “permeability” as in the enhanced permeability and retention effect of cancer nanotherapeutics. ACS Nano 2017, 11, 9567–9569. [Google Scholar] [CrossRef]
- Sindhwani, S.; Syed, A.M.; Ngai, J.; Kingston, B.R.; Maiorino, L.; Rothschild, J.; MacMillan, P.; Zhang, Y.; Rajesh, N.U.; Hoang, T.; et al. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. [Google Scholar] [CrossRef]
- Agarwal, P.; Wang, H.; Sun, M.; Xu, J.; Zhao, S.; Liu, Z.; Gooch, K.J.; Zhao, Y.; Lu, X.; He, X. Microfluidics enabled bottom-up engineering of 3D Vascularized tumor for drug discovery. ACS Nano 2017, 11, 6691–6702. [Google Scholar] [CrossRef]
- Ozcelikkale, A.; Shin, K.; Noe-Kim, V.; Elzey, B.D.; Dong, Z.; Zhang, J.-T.; Kim, K.; Kwon, I.C.; Park, K.; Han, B. Differential response to doxorubicin in breast cancer subtypes simulated by a microfluidic tumor model. J. Control Release 2017, 266, 129–139. [Google Scholar] [CrossRef]
- Choi, Y.; Hyun, E.; Seo, J.; Blundell, C.; Kim, H.C.; Lee, E.; Lee, S.H.; Moon, A.; Moon, W.K.; Huh, D. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip 2015, 15, 3350–3357. [Google Scholar] [CrossRef]
- Ghaemmaghami, A.M.; Hancock, M.J.; Harrington, H.; Kaji, H.; Khademhosseini, A. Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 2012, 17, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Sung, J.H.; Kam, C.; Shuler, M.L. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip 2010, 10, 446–455. [Google Scholar] [CrossRef]
- Esch, M.B.; Mahler, G.J.; Stokol, T.; Shuler, M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip 2014, 14, 3081–3092. [Google Scholar] [CrossRef] [PubMed]
- Booij, T.H.; Price, L.S.; Danen, E.H.J. 3D Cell-based assays for drug screens: Challenges in imaging, image analysis and high-content analysis. SLAS Discov. Adv. Life Sci. R D 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Rourke, K.P.; Dow, L.E.; Lowe, S.W. Immunofluorescent staining of mouse intestinal stem cells. Bio. Protoc. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiswald, L.-B.; Guinebretière, J.-M.; Richon, S.; Bellet, D.; Saubaméa, B.; Dangles-Marie, V. In situ protein expression in tumour spheres: Development of an immunostaining protocol for confocal microscopy. BMC Cancer 2010, 10, 106. [Google Scholar] [CrossRef] [Green Version]
- Dekkers, J.F.; Alieva, M.; Wellens, L.M.; Ariese, H.C.R.; Jamieson, P.R.; Vonk, A.M.; Amatngalim, G.D.; Hu, H.; Oost, K.C.; Snippert, H.J.G.; et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019, 14, 1756–1771. [Google Scholar] [CrossRef]
- Cora, V.; Haderspeck, J.; Antkowiak, L.; Mattheus, U.; Neckel, P.H.; Mack, A.F.; Bolz, S.; Ueffing, M.; Pashkovskaia, N.; Achberger, K.; et al. A cleared view on retinal organoids. Cells 2019, 8, 391. [Google Scholar] [CrossRef] [Green Version]
- Boutin, M.E.; Voss, T.C.; Titus, S.A.; Cruz-Gutierrez, K.; Michael, S.; Ferrer, M. A high-throughput imaging and nuclear segmentation analysis protocol for cleared 3D culture models. Sci. Rep. 2018, 8, 11135. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Tillberg, P.W.; Boyden, E.S. Expansion microscopy. Science 2015, 347, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Edwards, S.J.; Carannante, V.; Kuhnigk, K.; Ring, H.; Tararuk, T.; Hallböök, F.; Blom, H.; Önfelt, B.; Brismar, H. High-Resolution Imaging of Tumor Spheroids and Organoids Enabled by Expansion Microscopy. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Santi, P.A. Light Sheet Fluorescence Microscopy. J. Histochem. Cytochem. 2011, 59, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Sapoznik, E.; Chang, B.-J.; Ju, R.J.; Welf, E.S.; Broadbent, D.; Carisey, A.F.; Stehbens, S.J.; Lee, K.; Marín, A.; Hanker, A.B.; et al. A Single-Objective Light-Sheet Microscope with 200 nm-Scale Resolution. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Alladin, A.; Chaible, L.; Reither, S.; Löschinger, M.; Wachsmuth, M.; Hériché, J.K.; Tischer, C.; Jechlinger, M. Tracking the cells of tumor origin in breast organoids by light sheet microscopy. bioRxiv 2019, 617837. [Google Scholar] [CrossRef]
- Ingber, D.E. Is it time for reviewer 3 to request human organ chip experiments instead of animal validation studies? Adv. Sci. 2020. [Google Scholar] [CrossRef]




| Nanoparticle | Diameter (nm) | Cell Type 1 | Cancer Type | Strategy | Parameter(s) Studied | Ref. |
|---|---|---|---|---|---|---|
| Lipid nanoparticle (Lipidots) | 50, 120 | CALL-33 | Oral tongue cancer | n/a | Penetration Viability | [36] |
| Polystyrene | 30, 50, 100 | HCT116 | Colorectal cancer | n/a | Penetration Surface charge dependence | [37] |
| Dendrimer | 2 and 8 | KB | Epidermal | n/a | Penetration Surface charge dependence | [38] |
| AuNPs | 50 and 100 | MCF-7 | Breast cancer | n/a | Penetration | [39] |
| AuNPs | 2, 6 and 15 | MCF-7 | Breast cancer | n/a | Penetration | [40] |
| Silica | 30, 100 | 4T1 and 3T3 co-culture | Breast cancer | n/a | Penetration influenced by tumor stroma | [41] |
| PGLA | 200 | 4T1 and 3T3 co-culture | Breast cancer | n/a | Penetration influenced by tumor stroma | [41] |
| Au nanorod | 55 × 15 | MCF-7 | Breast cancer | n/a | Penetration Surface charge dependence | [42] |
| ECM destabilization | ||||||
| Polystyrene | 20, 40, 100 and 200 | SiHa | Cervix cancer | Collagenase treatment | Penetration after ECM degradation | [34] |
| Size-switching | ||||||
| Dendrimeric iCluster | 100 → 5 | BxPC-3 | Pancreatic cancer | Size-switching, trigger = pH | Penetration Therapeutic activity | [43] |
| MSN WS2-HP Cluster Bomb | 50 → 5 | 4T1 | Breast cancer | Size-switching, trigger = pH | Penetration Therapeutic activity | [44] |
| Dendrimeric nanobomb | 80 → 10 | BxPC-3 | Pancreatic cancer | Size-switching, trigger = pH | Penetration Therapeutic effect | [45] |
| PEG conjugated Multi-Micelles | 80 → 4 | BxPC-3 | Pancreatic cancer | Size-switching, trigger = pH | Penetration Therapeutic effect | [46] |
| Gelatin | 186.5 → 59.5 | 4T1 and B16F10 | Breast cancer | Size-switching, trigger = matrix metalloproteinase-2 | Penetration | [47] |
| Hyaluronic acid modified dendrimer | 200 → 10 | A549 | Lung cancer | Size-switching, trigger = matrix metalloproteinase-2 | Penetration | [48] |
| Ligand functionalization | ||||||
| PEG-PCL nanoparticle | 120 | C6 | Brain cancer | iRGD functionalizationInterleukin-13 functionalization | Penetration | [49] |
| PLGA | 112 | 4T1 | Breast cancer | iRGD functionalization | Penetration Viability | [50] |
| HDL (lipoprotein) nanoparticle | 136 | A549 | Lung cancer | iRGD functionalization | Penetration Viability | [51] |
| PLGA-b-PEG nanoparticle | 107 | C6 | Brain cancer | CRT peptide functionalization, Tf receptor targeting | Penetration Viability | [52] |
| Elastin-like polypeptide nanoparticles | 60 | U-87 | Brain cancer | Cell-penetrating peptide functionalization | Penetration | [53] |
| Folic acid-CM-PFA/pDNA | 126–176 | HeLa | Cervix cancer | Folic acid | Penetration pDNA expression Viability | [54] |
| Ligand functionalization (L) and size-Switching (SS) | ||||||
| Graphene quantum dot-loaded nanoparticle | 150 → 5 | RG2 | Brain cancer | L: pH sensitive compound functionalization SS: trigger disassembly = irradiation with NIR light | Penetration | [55] |
| Lipid nanoparticle | 180 | BxPC-3 | Pancreatic cancer | L: iRGD functionalization SS: trigger = hypoxia | Viability | [56] |
| Nanoparticle shape | ||||||
| Glycopolymer nanoparticle | sphere: 30 rod: 122 vesicle: 165 | MCF-7 | Breast cancer | Sphere/rod/vesicles | Penetration Viability | [57] |
| Nanoparticle stiffness | ||||||
| Fructose-based micelle nanorod | varies | MCF-7 | Breast cancer | Stiff/soft | Penetration | [58] |
| polymer micelles | varies | BxPC-3 | Pancreatic cancer | Stiff/soft | Penetration | [59] |
| Model | Advantages | Disadvantages | What Can we Learn? |
|---|---|---|---|
| Scaffold free spheroids |
|
|
|
| Scaffold-embedded cells 1 |
|
|
|
| Scaffold-embedded organoids 1 |
|
|
|
| Micro-fluidics |
|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Zundert, I.; Fortuni, B.; Rocha, S. From 2D to 3D Cancer Cell Models—The Enigmas of Drug Delivery Research. Nanomaterials 2020, 10, 2236. https://doi.org/10.3390/nano10112236
Van Zundert I, Fortuni B, Rocha S. From 2D to 3D Cancer Cell Models—The Enigmas of Drug Delivery Research. Nanomaterials. 2020; 10(11):2236. https://doi.org/10.3390/nano10112236
Chicago/Turabian StyleVan Zundert, Indra, Beatrice Fortuni, and Susana Rocha. 2020. "From 2D to 3D Cancer Cell Models—The Enigmas of Drug Delivery Research" Nanomaterials 10, no. 11: 2236. https://doi.org/10.3390/nano10112236
APA StyleVan Zundert, I., Fortuni, B., & Rocha, S. (2020). From 2D to 3D Cancer Cell Models—The Enigmas of Drug Delivery Research. Nanomaterials, 10(11), 2236. https://doi.org/10.3390/nano10112236







