Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. CuO NPs Generation, Exposure of Mice, Analysis of Cu and CuO NPs in the Lungs
2.2. DNA, RNA, and miRNA Extraction
2.3. Whole Genome Transcriptome Analysis by Next Generation Sequencing
2.4. miRNA Expression Analysis
2.5. Global DNA Methylation Analysis
2.6. Data Analysis
3. Results
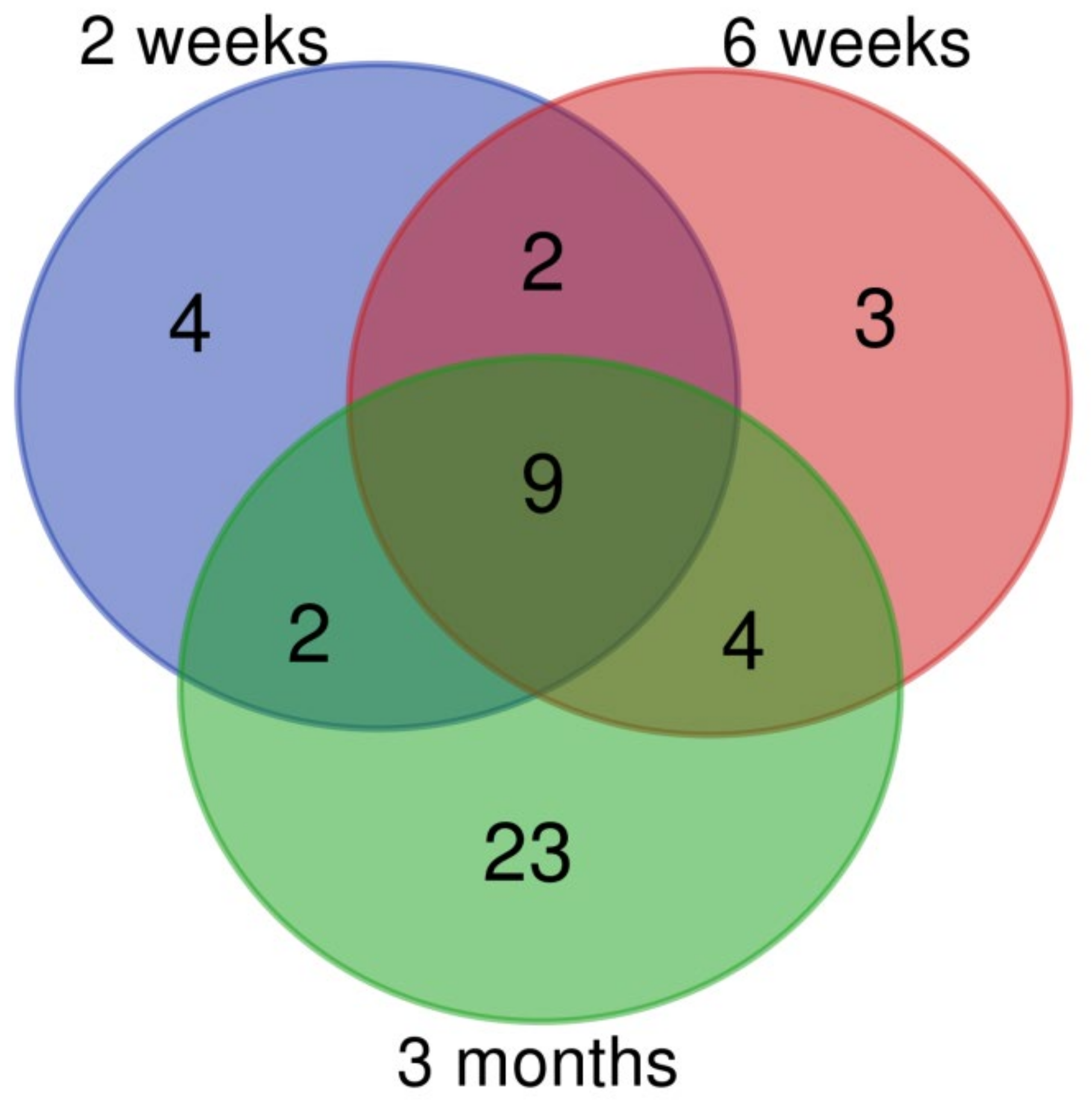
3.1. mRNA Expression Changes Induced by CuO NPs Inhalation
3.2. Changes of miRNA Expression Affected by CuO NPs
3.3. Global DNA Methylation after CuO NPs Inhalation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Magaye, R.; Zhao, J.; Bowman, L.; Ding, M. Genotoxicity and carcinogenicity of cobalt-, nickel- and copper-based nanoparticles. Exp. Ther. Med. 2012, 4, 551–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sengupta, J.; Ghosh, S.; Datta, P.; Gomes, A.; Gomes, A. Physiologically important metal nanoparticles and their toxicity. J. Nanosci. Nanotechnol. 2014, 14, 990–1006. [Google Scholar] [CrossRef] [PubMed]
- Bondarenko, O.; Juganson, K.; Ivask, A.; Kasemets, K.; Mortimer, M.; Kahru, A. Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: A critical review. Arch. Toxicol. 2013, 87, 1181–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubilar, O.; Rai, M.; Tortella, G.; Diez, M.C.; Seabra, A.B.; Durán, N. Biogenic nanoparticles: Copper, copper oxides, copper sulphides, complex copper nanostructures and their applications. Biotechnol. Lett. 2013, 35, 1365–1375. [Google Scholar] [CrossRef]
- Su, Y.; Zheng, X.; Chen, Y.; Li, M.; Liu, K. Alteration of intracellular protein expressions as a key mechanism of the deterioration of bacterial denitrification caused by copper oxide nanoparticles. Sci. Rep. 2015, 5, 15824. [Google Scholar] [CrossRef] [Green Version]
- Ahamed, M.; Akhtar, M.J.; Alhadlaq, H.A.; Alrokayan, S.A. Assessment of the lung toxicity of copper oxide nanoparticles: Current status. Nanomedicine 2015, 10, 2365–2377. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Y.; Zhang, L.; Wang, T.; Wang, J.; Jiao, F.; Li, W.; Liu, Y.; Li, Y.; Li, B.; et al. Potential health impact on mice after nasal instillation of nano-sized copper particles and their translocation in mice. J. Nanosci. Nanotechnol. 2009, 9, 6335–6343. [Google Scholar] [CrossRef]
- Holan, V.; Javorkova, E.; Vrbova, K.; Vecera, Z.; Mikuska, P.; Coufalik, P.; Kulich, P.; Skoupy, R.; Machala, M.; Zajicova, A.; et al. A murine model of the effects of inhaled CuO nanoparticles on cells of innate and adaptive immunity—a kinetic study of a continuous three-month exposure. Nanotoxicology 2019, 13, 952–963. [Google Scholar] [CrossRef]
- Yokohira, M.; Hashimoto, N.; Yamakawa, K.; Suzuki, S.; Saoo, K.; Kuno, T.; Imaida, K. Lung Carcinogenic Bioassay of CuO and TiO2 Nanoparticles with Intratracheal Instillation Using F344 Male Rats. J. Toxicol. Pathol. 2009, 22, 71–78. [Google Scholar] [CrossRef] [Green Version]
- Costa, P.M.; Gosens, I.; Williams, A.; Farcal, L.; Pantano, D.; Brown, D.M.; Stone, V.; Cassee, F.R.; Halappanavar, S.; Fadeel, B. Transcriptional profiling reveals gene expression changes associated with inflammation and cell proliferation following short-term inhalation exposure to copper oxide nanoparticles. J. Appl. Toxicol. 2018, 38, 385–397. [Google Scholar] [CrossRef]
- Vecera, Z.; Mikuska, P.; Moravec, P.; Smolik, J. Unique Exposure System for the Whole Body Inhalation Experiments with Small Animals; Tanger Ltd: Brno, Czech Republic, 2011. [Google Scholar]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Vila-Casadesús, M.; Gironella, M.; Lozano, J.J. MiRComb: An R Package to Analyse miRNA–mRNA Interactions. Examples across Five Digestive Cancers. PLoS ONE 2016, 11, e0151127. [Google Scholar] [CrossRef] [PubMed]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chibber, S.; Shanker, R. Can CuO nanoparticles lead to epigenetic regulation of antioxidant enzyme system? CuO NPs can result in epigenetic regulation: A hypothetical view. J. Appl. Toxicol. 2017, 37, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Kung, M.-L.; Hsieh, S.-L.; Wu, C.-C.; Chu, T.-H.; Lin, Y.-C.; Yeh, B.-W.; Hsieh, S. Enhanced reactive oxygen species overexpression by CuO nanoparticles in poorly differentiated hepatocellular carcinoma cells. Nanoscale 2015, 7, 1820–1829. [Google Scholar] [CrossRef]
- Rabbani, G.; Khan, M.J.; Ahmad, A.; Maskat, M.Y.; Khan, R.H. Effect of copper oxide nanoparticles on the conformation and activity of β-galactosidase. Colloids Surf. B Biointerfaces 2014, 123, 96–105. [Google Scholar] [CrossRef]
- Cheng, T.F.; Choudhuri, S.; Muldoon-Jacobs, K. Epigenetic targets of some toxicologically relevant metals: A review of the literature. J. Appl. Toxicol. 2012, 32, 643–653. [Google Scholar] [CrossRef]
- Lai, X.; Zhao, H.; Zhang, Y.; Guo, K.; Xu, Y.; Chen, S.; Zhang, J. Intranasal Delivery of Copper Oxide Nanoparticles Induces Pulmonary Toxicity and Fibrosis in C57BL/6 mice. Sci. Rep. 2018, 8, 4499. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Li, Y.; Yang, L.; Long, J.; Zheng, Y.; Liu, J.; Xiao, S.; Jia, J. Activation of Erk and p53 regulates copper oxide nanoparticle-induced cytotoxicity in keratinocytes and fibroblasts. Int. J. Nanomed. 2014, 9, 4763–4772. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Wang, W.; Zhao, P.; Qi, Z.; Zhao, S. Cuprous oxide nanoparticles inhibit angiogenesis via down regulation of VEGFR2 expression. Nanoscale 2014, 6, 3206. [Google Scholar] [CrossRef] [PubMed]
- Andl, C.D. The Misregulation of Cell Adhesion Components during Tumorigenesis: Overview and Commentary. J. Oncol. 2010, 2010, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dymacek, J.; Snyder-Talkington, B.N.; Porter, D.W.; Mercer, R.R.; Wolfarth, M.G.; Castranova, V.; Qian, Y.; Guo, N.L. mRNA and miRNA Regulatory Networks Reflective of Multi-Walled Carbon Nanotube-Induced Lung Inflammatory and Fibrotic Pathologies in Mice. Toxicol. Sci. 2015, 144, 51–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halappanavar, S.; Jackson, P.; Williams, A.; Jensen, K.A.; Hougaard, K.S.; Vogel, U.; Yauk, C.L.; Wallin, H. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: A toxicogenomic study. Environ. Mol. Mutagen. 2011, 52, 425–439. [Google Scholar] [CrossRef] [Green Version]
- Conickx, G.; Avila Cobos, F.; Van den Berge, M.; Faiz, A.; Timens, W.; Hiemstra, P.S.; Joos, G.F.; Brusselle, G.G.; Mestdagh, P.; Bracke, K.R. microRNA profiling in lung tissue and bronchoalveolar lavage of cigarette smoke-exposed mice and in COPD patients: A translational approach. Sci. Rep. 2017, 7, 12871. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, Y.; Mise, N.; Suzuki, Y.; Tada-Oikawa, S.; Izuoka, K.; Zhang, L.; Zong, C.; Takai, A.; Yamada, Y.; Ichihara, S. MicroRNAs as Potential Mediators for Cigarette Smoking Induced Atherosclerosis. Int. J. Mol. Sci. 2018, 19, 1097. [Google Scholar] [CrossRef] [Green Version]
- Pei, W.; Tao, L.; Zhang, L.W.; Zhang, S.; Cao, J.; Jiao, Y.; Tong, J.; Nie, J. Circular RNA profiles in mouse lung tissue induced by radon. Environ. Health Prev. Med. 2017, 22, 36. [Google Scholar] [CrossRef] [Green Version]
- Luettich, K.; Xiang, Y.; Iskandar, A.; Sewer, A.; Martin, F.; Talikka, M.; Vanscheeuwijck, P.; Berges, A.; Veljkovic, E.; Gonzalez-Suarez, I.; et al. Systems toxicology approaches enable mechanistic comparison of spontaneous and cigarette smoke-related lung tumor development in the A/J mouse model. Interdiscip. Toxicol. 2014, 7, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, Q.; Du, M.; Xiong, D.; Wang, Y.; Mohammed, A.; Lubet, R.; Wang, L.; You, M. Exosomal miRNAs as Novel Pharmacodynamic Biomarkers for Cancer Chemopreventive Agent Early Stage Treatments in Chemically Induced Mouse Model of Lung Squamous Cell Carcinoma. Cancers 2019, 11, 477. [Google Scholar] [CrossRef] [Green Version]
- Xiong, D.; Pan, J.; Zhang, Q.; Szabo, E.; Miller, M.S.; Lubet, R.A.; You, M.; Wang, Y. Bronchial airway gene expression signatures in mouse lung squamous cell carcinoma and their modulation by cancer chemopreventive agents. Oncotarget 2017, 8, 18885–18900. [Google Scholar] [CrossRef] [Green Version]
- Plank, M.W.; Maltby, S.; Tay, H.L.; Stewart, J.; Eyers, F.; Hansbro, P.M.; Foster, P.S. MicroRNA Expression Is Altered in an Ovalbumin-Induced Asthma Model and Targeting miR-155 with Antagomirs Reveals Cellular Specificity. PLoS ONE 2015, 10, e0144810. [Google Scholar] [CrossRef]
- Tan, K.; Choi, H.; Jiang, X.; Yin, L.; Seet, J.; Patzel, V.; Engelward, B.P.; Chow, V.T. Micro-RNAs in regenerating lungs: An integrative systems biology analysis of murine influenza pneumonia. BMC Genom. 2014, 15, 587. [Google Scholar] [CrossRef] [Green Version]
- Gomez, J.C.; Dang, H.; Kanke, M.; Hagan, R.S.; Mock, J.R.; Kelada, S.N.P.; Sethupathy, P.; Doerschuk, C.M. Predicted effects of observed changes in the mRNA and microRNA transcriptome of lung neutrophils during S. pneumoniae pneumonia in mice. Sci. Rep. 2017, 7, 11258. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-J.; Kim, H.B.; Baek, Y.H.; Kim, E.-H.; Pascua, P.N.Q.; Park, S.-J.; Kwon, H.; Lim, G.-J.; Kim, S.; Kim, Y.-I.; et al. Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 2014, 14, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasarre, P.; Potiron, V.; Drabkin, H.; Roche, J. Guidance molecules in lung cancer. Cell Adhes. Migr. 2010, 4, 130–145. [Google Scholar] [CrossRef] [Green Version]
- Aasen, T.; Sansano, I.; Montero, M.Á.; Romagosa, C.; Temprana-Salvador, J.; Martínez-Marti, A.; Moliné, T.; Hernández-Losa, J.Y.; Cajal, S.R. Insight into the Role and Regulation of Gap Junction Genes in Lung Cancer and Identification of Nuclear Cx43 as a Putative Biomarker of Poor Prognosis. Cancers 2019, 11, 320. [Google Scholar] [CrossRef] [Green Version]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Q.; He, J.; Lu, W. Regulation of calcium signaling in lung cancer. J. Thorac. Dis. 2010, 2, 52–56. [Google Scholar]
- Sanghera, C.; Wong, L.M.; Panahi, M.; Sintou, A.; Hasham, M.; Sattler, S. Cardiac phenotype in mouse models of systemic autoimmunity. Dis. Model. Mech. 2019, 12, 036947. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.; Tay, H.L.; Plank, M.; Essilfie, A.-T.; Hansbro, P.M.; Foster, P.S.; Yang, M. Activation of Olfactory Receptors on Mouse Pulmonary Macrophages Promotes Monocyte Chemotactic Protein-1 Production. PLoS ONE 2013, 8, e80148. [Google Scholar] [CrossRef] [PubMed]
- Tsugita, M.; Morimoto, N.; Nakayama, M. SiO2 and TiO2 nanoparticles synergistically trigger macrophage inflammatory responses. Part. Fibre Toxicol. 2017, 14, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Miousse, I.R.; Pirela, S.V.; Moore, J.K.; Melnyk, S.; Koturbash, I.; Demokritou, P. In vivo epigenetic effects induced by engineered nanomaterials: A case study of copper oxide and laser printer-emitted engineered nanoparticles. Nanotoxicology 2016, 10, 629–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| ID | Deregulated Biological Process/Pathway | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 3 Days | ||||
| GO:0030574 | collagen catabolic process | 0.003 | 7 | 67 |
| GO:0043062 | extracellular structure organization | 0.005 | 13 | 355 |
| GO:0030198 | extracellular matrix organization | 0.005 | 13 | 354 |
| GO:0032963 | collagen metabolic process | 0.028 | 7 | 122 |
| GO:0061077 | chaperone-mediated protein folding | 0.028 | 5 | 52 |
| 1269985 | Chondroitin sulfate biosynthesis | 0.009 | 4 | 22 |
| 82982 | Glycosaminoglycan biosynthesis - chondroitin sulfate/dermatan sulfate | 0.009 | 4 | 20 |
| 545276 | chondroitin sulfate biosynthesis (late stages) | 0.009 | 3 | 8 |
| 1270245 | Collagen formation | 0.019 | 6 | 93 |
| 1270247 | Assembly of collagen fibrils and other multimeric structures | 0.019 | 5 | 60 |
| 2 Weeks | ||||
| GO:0006955 | immune response | < 0.001 | 165 | 1572 |
| GO:0002682 | regulation of immune system process | < 0.001 | 140 | 1506 |
| GO:0006952 | defense response | < 0.001 | 142 | 1651 |
| GO:0002684 | positive regulation of immune system process | < 0.001 | 102 | 976 |
| GO:0001775 | cell activation | < 0.001 | 96 | 1001 |
| 1269203 | Innate Immune System | < 0.001 | 101 | 1312 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 21 | 49 |
| 83051 | Cytokine-cytokine receptor interaction | < 0.001 | 41 | 270 |
| 1457780 | Neutrophil degranulation | < 0.001 | 50 | 492 |
| 1269310 | Cytokine Signaling in Immune system | < 0.001 | 64 | 763 |
| 6 Weeks | ||||
| GO:0006955 | immune response | < 0.001 | 122 | 1572 |
| GO:0002682 | regulation of immune system process | < 0.001 | 113 | 1506 |
| GO:0006954 | inflammatory response | < 0.001 | 76 | 711 |
| GO:0006952 | defense response | < 0.001 | 115 | 1651 |
| GO:0001816 | cytokine production | < 0.001 | 70 | 700 |
| 1269203 | Innate Immune System | < 0.001 | 97 | 1312 |
| 1457780 | Neutrophil degranulation | < 0.001 | 56 | 492 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 17 | 49 |
| M5889 | Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins | < 0.001 | 70 | 1028 |
| M5885 | Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors | < 0.001 | 57 | 753 |
| 3 Months | ||||
| GO:0006955 | immune response | < 0.001 | 282 | 1572 |
| GO:0006952 | defense response | < 0.001 | 277 | 1651 |
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 260 | 1610 |
| GO:0002682 | regulation of immune system process | < 0.001 | 249 | 1506 |
| GO:0001816 | cytokine production | < 0.001 | 154 | 700 |
| 1269203 | Innate Immune System | < 0.001 | 210 | 1312 |
| 1457780 | Neutrophil degranulation | < 0.001 | 105 | 492 |
| 1269310 | Cytokine Signaling in Immune system | < 0.001 | 125 | 763 |
| 1470924 | Interleukin-10 signaling | < 0.001 | 25 | 49 |
| 83051 | Cytokine-cytokine receptor interaction | < 0.001 | 62 | 270 |
| ID | Deregulated Biological Process | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 2 Weeks | ||||
| GO:0006955 | immune response | 0.019 | 33 | 1572 |
| GO:0045776 | negative regulation of blood pressure | 0.019 | 6 | 56 |
| GO:0010941 | regulation of cell death | 0.019 | 33 | 1650 |
| GO:0002682 | regulation of immune system process | 0.019 | 31 | 1506 |
| GO:0043067 | regulation of programmed cell death | 0.022 | 31 | 1536 |
| GO:0002683 | negative regulation of immune system process | 0.022 | 14 | 420 |
| GO:0012501 | programmed cell death | 0.022 | 36 | 1952 |
| GO:0001775 | cell activation | 0.025 | 23 | 1001 |
| GO:0042981 | regulation of apoptotic process | 0.025 | 30 | 1519 |
| GO:0006915 | apoptotic process | 0.027 | 35 | 1923 |
| GO:0006955 | immune response | 0.019 | 33 | 1572 |
| 6 Weeks | ||||
| GO:0000280 | nuclear division | < 0.001 | 24 | 599 |
| GO:0048285 | organelle fission | < 0.001 | 24 | 636 |
| GO:1903047 | mitotic cell cycle process | < 0.001 | 26 | 931 |
| GO:0000278 | mitotic cell cycle | < 0.001 | 27 | 1016 |
| GO:0022402 | cell cycle process | < 0.001 | 32 | 1385 |
| GO:0000070 | mitotic sister chromatid segregation | < 0.001 | 10 | 136 |
| GO:0051241 | negative regulation of multicellular organismal process | 0.001 | 27 | 1127 |
| GO:0051301 | cell division | 0.001 | 20 | 668 |
| GO:0007049 | cell cycle | 0.001 | 35 | 1766 |
| GO:0055118 | negative regulation of cardiac muscle contraction | 0.001 | 3 | 4 |
| 3 Months | ||||
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 171 | 1610 |
| GO:0022610 | biological adhesion | < 0.001 | 164 | 1542 |
| GO:0007155 | cell adhesion | < 0.001 | 161 | 1530 |
| GO:0040011 | locomotion | < 0.001 | 170 | 1735 |
| GO:0016477 | cell migration | < 0.001 | 137 | 1300 |
| GO:0001816 | cytokine production | < 0.001 | 90 | 700 |
| GO:0051674 | localization of cell | < 0.001 | 141 | 1428 |
| GO:0048870 | cell motility | < 0.001 | 141 | 1428 |
| GO:0006928 | movement of cell or subcellular component | < 0.001 | 170 | 1882 |
| GO:0009611 | response to wounding | < 0.001 | 106 | 967 |
| GO:0051240 | positive regulation of multicellular organismal process | < 0.001 | 171 | 1610 |
| miRNA | Target mRNA (N) |
|---|---|
| 2 Weeks | |
| mmu-miR-135b-5p | 50 |
| mmu-miR-147-3p | 11 |
| mmu-miR-449a-5p | 87 |
| 6 Weeks | |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:17:27580702:27580732:Minus | 1 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:18:60298547:60298577:Plus | 1 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:3:46956408:46956438:Minus | 2 |
| mmu_piR_017289/gb/DQ696831/Mus_musculus:6:128725729:128725759:Minus | 4 |
| mmu-miR-1264-3p | 134 |
| mmu-miR-1298-5p | 183 |
| mmu-miR-135b-3p | 22 |
| mmu-miR-135b-5p | 68 |
| mmu-miR-146b-5p | 11 |
| mmu-miR-147-3p | 13 |
| mmu-miR-155-5p | 15 |
| mmu-miR-21a-5p | 5 |
| mmu-miR-448-3p | 104 |
| mmu-miR-489-3p | 1 |
| mmu-miR-672-5p | 2 |
| mmu-miR-6972-5p | 4 |
| mmu-miR-703 | 1 |
| mmu-miR-7048-5p | 4 |
| 3 Months | |
| mmu-miR-1264-3p | 515 |
| mmu-miR-1298-5p | 461 |
| mmu-miR-135b-5p | 29 |
| mmu-miR-144-5p | 249 |
| mmu-miR-146a-5p | 23 |
| mmu-miR-146b-5p | 15 |
| mmu-miR-147-3p | 26 |
| mmu-miR-155-5p | 34 |
| mmu-miR-21a-3p | 43 |
| mmu-miR-21a-5p | 11 |
| mmu-miR-21b | 11 |
| mmu-miR-223-3p | 8 |
| mmu-miR-342-3p | 12 |
| mmu-miR-3967 | 14 |
| mmu-miR-451a | 91 |
| ID | Deregulated Biological Process/Pathway | q-Value | Deregulated Genes (N) | Genes in Pathway (N) |
|---|---|---|---|---|
| 3 Days | ||||
| mmu04510 | Focal adhesion | <0.001 | 19 | 199 |
| mmu04512 | ECM receptor interaction | <0.001 | 11 | 88 |
| mmu05200 | Pathways in cancer | 0.003 | 21 | 539 |
| 2 Weeks | ||||
| mmu04520 | Adherens junction | <0.001 | 27 | 71 |
| mmu04510 | Focal adhesion | <0.001 | 44 | 199 |
| mmu04270 | Vascular smooth muscle contraction | <0.001 | 29 | 140 |
| mmu04360 | Axon guidance | <0.001 | 30 | 180 |
| mmu04916 | Melanogenesis | <0.001 | 24 | 100 |
| mmu05414 | Dilated cardiomyopathy | <0.001 | 23 | 94 |
| mmu05200 | Pathways in cancer | <0.001 | 51 | 539 |
| mmu04330 | Notch signaling pathway | <0.001 | 16 | 54 |
| mmu04540 | Gap junction | <0.001 | 21 | 86 |
| mmu04810 | Regulation of actin cytoskeleton | <0.001 | 36 | 216 |
| mmu04740 | Olfactory transduction | <0.001 | 5 | 1134 |
| mmu04912 | GnRH signaling pathway | <0.001 | 21 | 90 |
| mmu04310 | Wnt signaling pathway | <0.001 | 26 | 162 |
| mmu05217 | Basal cell carcinoma | <0.001 | 14 | 63 |
| mmu04020 | Calcium signaling pathway | 0.001 | 27 | 192 |
| mmu05410 | Hypertrophic cardiomyopathy (HCM) | 0.001 | 17 | 91 |
| mmu04512 | ECM receptor interaction | 0.002 | 16 | 88 |
| mmu05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.002 | 15 | 77 |
| mmu05213 | Endometrial cancer | 0.003 | 11 | 58 |
| mmu04144 | Endocytosis | 0.003 | 27 | 269 |
| mmu11190 | Dorso-ventral axis formation | 0.005 | 7 | 22 |
| mmu05211 | Renal cell carcinoma | 0.005 | 13 | 68 |
| mmu05016 | Huntingtons disease | 0.005 | 25 | 199 |
| mmu04720 | Long term potentiation | 0.006 | 13 | 67 |
| mmu04260 | Cardiac muscle contraction | 0.010 | 14 | 86 |
| 6 Weeks | ||||
| mmu04142 | Lysosome | <0.001 | 23 | 124 |
| 3 Months | ||||
| mmu04142 | Lysosome | <0.001 | 43 | 124 |
| mmu04740 | Olfactory transduction | <0.001 | 4 | 1134 |
| mmu04060 | Cytokine-cytokine receptor interaction | <0.001 | 60 | 296 |
| mmu05140 | Leishmania infection | <0.001 | 27 | 69 |
| mmu04062 | Chemokine signaling pathway | <0.001 | 45 | 198 |
| mmu04650 | Natural killer cell mediated cytotoxicity | <0.001 | 34 | 117 |
| mmu04612 | Antigen processing and presentation | <0.001 | 23 | 91 |
| mmu05332 | Graft versus host disease | <0.001 | 15 | 65 |
| mmu04662 | B cell receptor signaling pathway | <0.001 | 22 | 82 |
| mmu04666 | Fc gamma R-mediated phagocytosis | <0.001 | 26 | 90 |
| mmu04620 | Toll like receptor signaling pathway | <0.001 | 25 | 99 |
| mmu05330 | Allograft rejection | <0.001 | 14 | 63 |
| mmu16848 | Epithelial cell signaling in helicobacter pylori infection | <0.001 | 21 | 69 |
| mmu04940 | Type I diabetes mellitus | <0.001 | 15 | 70 |
| mmu04664 | Fc epsilon RI signaling pathway | 0.001 | 20 | 67 |
| mmu05320 | Autoimmune thyroid disease | 0.002 | 14 | 78 |
| mmu05416 | Viral myocarditis | 0.006 | 17 | 88 |
| mmu04660 | T cell receptor signaling pathway | 0.006 | 23 | 103 |
| mmu04640 | Hematopoietic cell lineage | 0.008 | 18 | 95 |
| mmu04672 | Intestinal immune network for IgA production | 0.009 | 12 | 42 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossner, P., Jr.; Vrbova, K.; Rossnerova, A.; Zavodna, T.; Milcova, A.; Klema, J.; Vecera, Z.; Mikuska, P.; Coufalik, P.; Capka, L.; et al. Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles. Nanomaterials 2020, 10, 550. https://doi.org/10.3390/nano10030550
Rossner P Jr., Vrbova K, Rossnerova A, Zavodna T, Milcova A, Klema J, Vecera Z, Mikuska P, Coufalik P, Capka L, et al. Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles. Nanomaterials. 2020; 10(3):550. https://doi.org/10.3390/nano10030550
Chicago/Turabian StyleRossner, Pavel, Jr., Kristyna Vrbova, Andrea Rossnerova, Tana Zavodna, Alena Milcova, Jiri Klema, Zbynek Vecera, Pavel Mikuska, Pavel Coufalik, Lukas Capka, and et al. 2020. "Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles" Nanomaterials 10, no. 3: 550. https://doi.org/10.3390/nano10030550
APA StyleRossner, P., Jr., Vrbova, K., Rossnerova, A., Zavodna, T., Milcova, A., Klema, J., Vecera, Z., Mikuska, P., Coufalik, P., Capka, L., Krumal, K., Docekal, B., Holan, V., Machala, M., & Topinka, J. (2020). Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles. Nanomaterials, 10(3), 550. https://doi.org/10.3390/nano10030550







