Transcriptome Profiling and Toxicity Following Long-Term, Low Dose Exposure of Human Lung Cells to Ni and NiO Nanoparticles—Comparison with NiCl2
Abstract
1. Introduction
2. Materials and Methods
2.1. Nanoparticles and Characteristics
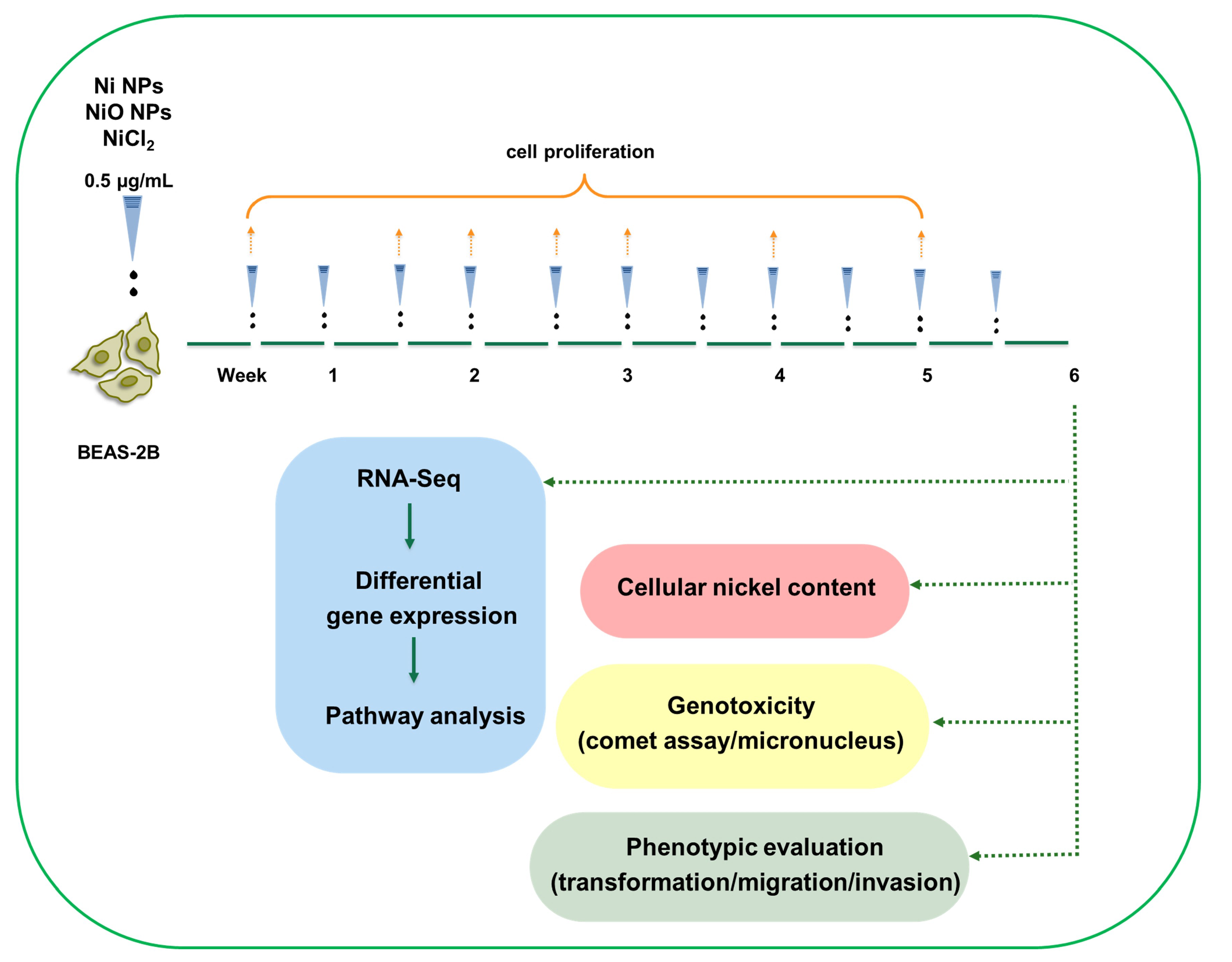
2.2. Cell Culture and Exposures
2.3. Alamar Blue
2.4. Inductively Coupled Plasma Mass Spectrometry (ICP-MS)
2.5. Comet Assay
2.6. Micronucleus Flow Cytometric Assay
2.7. Invasion-Migration Assay
2.8. Soft Agar Cell Transformation and Colony-Forming Efficiency Assays
2.9. RNA Extraction
2.10. RNA Sequencing and Data Analysis
2.11. Pathway, Network and Gene Enrichment Analysis
2.12. Statistical Analysis
3. Results
3.1. Ni and NiO NPs, but Not Soluble Ni, Are Readily Taken up by Human Lung Cells
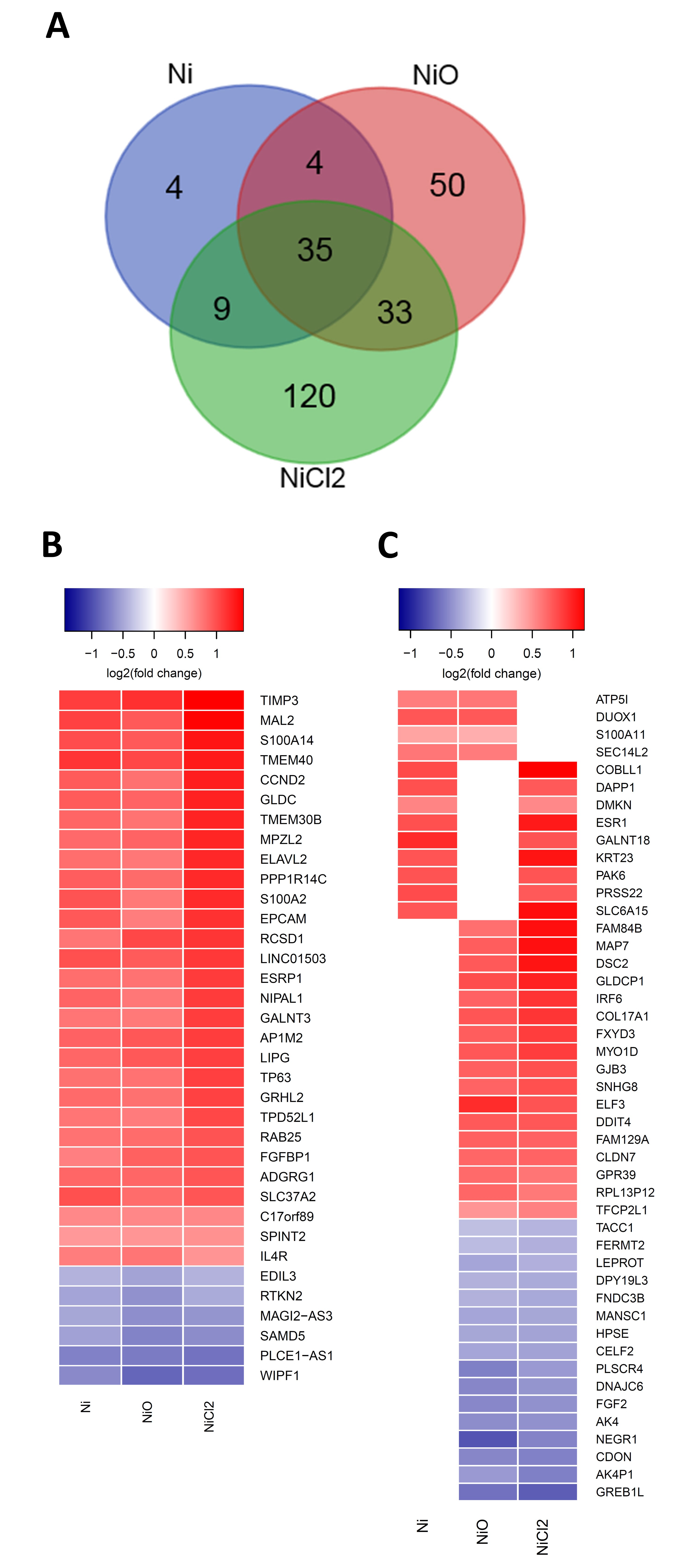
3.2. RNA Sequencing Reveals Gene Expression Changes Following Six-Week Exposure of BEAS-2B Cells to Ni
3.3. Pathway Enrichment and Upstream Regulator Analysis
3.4. Nickel-Containing NPs Induce DNA Strand Breaks and Alter Cell Cycle after Six Weeks of Repeated Exposure in BEAS-2B Cells
3.5. No Clear Changes in Cell Transformation or Cell Motility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Journeay, W.S.; Goldman, R.H. Occupational handling of nickel nanoparticles: A case report. Am. J. Ind. Med. 2014, 57, 1073–1076. [Google Scholar] [CrossRef]
- Phillips, J.; Green, F.Y.; Davies, J.C.; Murray, J. Pulmonary and systemic toxicity following exposure to nickel nanoparticles. Am. J. Ind. Med. 2010, 53, 763–767. [Google Scholar] [CrossRef]
- IARC. Arsenic, metals, fibres and dusts. In IARC Monographs Volume 100C; IARC: Lyon, France, 2012. [Google Scholar]
- Oller, A.R. Respiratory carcinogenicity assessment of soluble nickel compounds. Environ. Health Perspect. 2002, 110, 841–844. [Google Scholar] [CrossRef]
- Goodman, J.E.; Prueitt, R.L.; Thakali, S.; Oller, A.R. The nickel ion bioavailability model of the carcinogenic potential of nickel-containing substances in the lung. Crit. Rev. Toxicol. 2010, 41, 142–174. [Google Scholar] [CrossRef] [PubMed]
- Åkerlund, E.; Cappellini, F.; Di Bucchianico, S.; Islam, S.; Skoglund, S.; Derr, R.; Wallinder, I.O.; Hendriks, G.; Karlsson, H.L.; Johnson, G. Genotoxic and mutagenic properties of Ni and NiO nanoparticles investigated by comet assay, γ-H2AX staining, Hprt mutation assay and ToxTracker reporter cell lines. Environ. Mol. Mutagen. 2017, 59, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Di Bucchianico, S.; Gliga, A.R.; Åkerlund, E.; Skoglund, S.; Wallinder, I.O.; Fadeel, B.; Karlsson, H.L. Calcium-dependent cyto- and genotoxicity of nickel metal and nickel oxide nanoparticles in human lung cells. Part. Fibre Toxicol. 2018, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Beyersmann, D.; Hartwig, A. Carcinogenic metal compounds: Recent insight into molecular and cellular mechanisms. Arch. Toxicol. 2008, 82, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Davidson, G.; Huang, Y.; Jiang, B.-H.; Shi, X.; Costa, M.; Huang, C. Nickel compounds act through phosphatidylinositol-3-kinase/Akt-dependent, p70(S6k)-independent pathway to induce hypoxia inducible factor transactivation and Cap43 expression in mouse epidermal Cl41 cells. Cancer Res. 2004, 64, 94–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pietruska, J.R.; Liu, X.; Smith, A.; McNeil, K.; Weston, P.; Zhitkovich, A.; Hurt, R.; Kane, A.B. Bioavailability, Intracellular Mobilization of Nickel, and HIF-1α Activation in Human Lung Epithelial Cells Exposed to Metallic Nickel and Nickel Oxide Nanoparticles. Toxicol. Sci. 2011, 124, 138–148. [Google Scholar] [CrossRef]
- Salnikow, K.; Blagosklonny, M.V.; Ryan, H.; Johnson, R.; Costa, M. Carcinogenic nickel induces genes involved with hypoxic stress. Cancer Res. 2000, 60, 38–41. [Google Scholar]
- Efremenko, A.Y.; Campbell, J.; Dodd, D.; Oller, A.; Clewell, H. Time-and concentration-dependent genomic responses of the rat airway to inhaled nickel subsulfide. Toxicol. Appl. Pharmacol. 2014, 279, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Efremenko, A.Y.; Campbell, J.L.; Dodd, D.E.; Oller, A.R.; Clewell, H.J. Time-and concentration-dependent genomic responses of the rat airway to inhaled nickel sulfate. Environ. Mol. Mutagen. 2017, 58, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Saquib, Q.; Siddiqui, M.A.; Ahmad, J.; Ansari, S.M.; Faisal, M.; Wahab, R.; Alatar, A.A.; Al-Khedhairy, A.; Musarrat, J. Nickel Oxide Nanoparticles Induced Transcriptomic Alterations in HEPG2 Cells. Adv. Exp. Med. Biol. 2018, 1048, 163–174. [Google Scholar]
- Di Bucchianico, S.; Cappellini, F.; Le Bihanic, F.; Zhang, Y.; Dreij, K.; Karlsson, H.L. Genotoxicity of TiO2 nanoparticles assessed by mini-gel comet assay and micronucleus scoring with flow cytometry. Mutagenesis 2016, 32, 127–137. [Google Scholar] [CrossRef]
- Gliga, A.R.; Di Bucchianico, S.; Lindvall, J.; Fadeel, B.; Karlsson, H.L. RNA-sequencing reveals long-term effects of silver nanoparticles on human lung cells. Sci. Rep. 2018, 8, 6668. [Google Scholar] [CrossRef]
- Lampa, S.; Dahlö, M.; Olason, P.I.; Hagberg, J.; Spjuth, O. Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data. GigaScience 2013, 2, 9. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Boil. 2014, 15, 31. [Google Scholar] [CrossRef]
- Su, C.-W.; Lin, C.-W.; Yang, W.-E.; Yang, S.-F. TIMP-3 as a therapeutic target for cancer. Ther. Adv. Med Oncol. 2019, 11, 1758835919864247. [Google Scholar] [CrossRef]
- Wang, T.; Huo, X.; Chong, Z.; Khan, H.; Liu, R. A review of S100 protein family in lung cancer. Clin. Chim. Acta 2018, 476, 54–59. [Google Scholar] [CrossRef]
- Meyyappan, M.; Wong, H.; Hull, C.; Riabowol, K.T. Increased Expression of Cyclin D2 during Multiple States of Growth Arrest in Primary and Established Cells. Mol. Cell. Boil. 1998, 18, 3163–3172. [Google Scholar] [CrossRef][Green Version]
- Huang, L.; Yang, Y.; Yang, F.; Liu, S.; Zhu, Z.; Lei, Z.; Guo, J. Functions of EpCAM in physiological processes and diseases (Review). Int. J. Mol. Med. 2018, 42, 1771–1785. [Google Scholar] [CrossRef]
- Melino, G.; Memmi, E.M.; Pelicci, P.G.; Bernassola, F. Maintaining epithelial stemness with p63. Sci. Signal. 2015, 8, re9. [Google Scholar] [CrossRef]
- Bankaitis, K.V.; Fingleton, B. Targeting IL4/IL4R for the treatment of epithelial cancer metastasis. Clin. Exp. Metastasis 2015, 32, 847–856. [Google Scholar] [CrossRef]
- Lin, C.-H.; Chung, C.; Wong, J.-H.; Chen, B.-K.; Chiu, S.-J.; Klahan, S.; Lee, Y.-C.; Chang, W.-C. Involvement of L-type Ca2+channel and toll-like receptor-4 in nickel-induced interleukin-8 gene expression. Environ. Toxicol. 2014, 31, 5–12. [Google Scholar] [CrossRef]
- Ouyang, W.; Li, J.; Shi, X.; Costa, M.; Huang, C. Essential role of PI-3K, ERKs and calcium signal pathways in nickel-induced VEGF expression. Mol. Cell. Biochem. 2005, 279, 35–43. [Google Scholar] [CrossRef]
- Wang, J.; Chen, S.; Wang, M.; Ko, J.-L.; Wu, C.; Chen, C.; Lin, H.; Chang, Y.-Y. Nickel-induced VEGF expression via regulation of Akt, ERK1/2, NFκB, and AMPK pathways in H460 cells. Environ. Toxicol. 2019, 34, 652–658. [Google Scholar] [CrossRef]
- Åkerlund, E.; Islam, S.; McCarrick, S.; Alfaro-Moreno, E.; Karlsson, H.L. Inflammation and (secondary) genotoxicity of Ni and NiO nanoparticles. Nanotoxicology 2019, 13, 1060–1072. [Google Scholar] [CrossRef]
- Nissinen, L.; Kähäri, V. Matrix metalloproteinases in inflammation. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2571–2580. [Google Scholar] [CrossRef]
- Mo, Y.; Jiang, M.; Zhang, Y.; Wan, R.; Li, J.; Zhong, C.-J.; Li, H.; Tang, S.; Zhang, Q. Comparative mouse lung injury by nickel nanoparticles with differential surface modification. J. Nanobiotechnol. 2019, 17, 2. [Google Scholar] [CrossRef]
- Wan, R.; Mo, Y.; Chien, S.; Li, Y.; Li, Y.; Tollerud, D.J.; Zhang, Q. The role of hypoxia inducible factor-1α in the increased MMP-2 and MMP-9 production by human monocytes exposed to nickel nanoparticles. Nanotoxicology 2011, 5, 568–582. [Google Scholar] [CrossRef]
- Raulf, M.; Weiss, T.; Lotz, A.; Lehnert, M.; Hoffmeyer, F.; Liebers, V.; Van Gelder, R.; Käfferlein, H.; Hartwig, A.; Pesch, B.; et al. Analysis of inflammatory markers and metals in nasal lavage fluid of welders. J. Toxicol. Environ. Health Part A 2016, 79, 1144–1157. [Google Scholar] [CrossRef]
- Arita, A.; Munoz, A.; Chervona, Y.; Niu, J.; Qu, Q.; Zhao, N.; Ruan, Y.; Kiok, K.; Kluz, T.; Sun, H.; et al. Gene expression profiles in peripheral blood mononuclear cells of Chinese nickel refinery workers with high exposures to nickel and control subjects. Cancer Epidemiol. Biomark. Prev. 2012, 22, 261–269. [Google Scholar] [CrossRef]
- Ellisen, L.W.; Ramsayer, K.D.; Johannessen, C.M.; Yang, A.; Beppu, H.; Minda, K.; Oliner, J.D.; McKeon, F.; A Haber, D. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 2002, 10, 995–1005. [Google Scholar] [CrossRef]
- Sofer, A.; Lei, K.; Johannessen, C.M.; Ellisen, L.W. Regulation of mTOR and Cell Growth in Response to Energy Stress by REDD1. Mol. Cell. Boil. 2005, 25, 5834–5845. [Google Scholar] [CrossRef]
- Xu, Z.; Ren, T.; Xiao, C.; Li, H.; Wu, T. Nickel promotes the invasive potential of human lung cancer cells via TLR4/MyD88 signaling. Toxicology 2011, 285, 25–30. [Google Scholar] [CrossRef]
- Wang, L.; Fan, J.; Hitron, J.A.; Son, Y.; Wise, J.T.; Roy, R.V.; Kim, D.; Dai, J.; Pratheeshkumar, P.; Zhang, Z.; et al. Cancer Stem-Like Cells Accumulated in Nickel-Induced Malignant Transformation. Toxicol. Sci. 2016, 151, 376–387. [Google Scholar] [CrossRef][Green Version]
- Shi, X.; Pan, J.-J.; Chang, Q.; Wang, X.; Son, Y.; Liu, J.; Zhang, Z.; Bi, Y.-Y. Activation of Akt/GSK3beta and Akt/Bcl-2 signaling pathways in nickel-transformed BEAS-2B cells. Int. J. Oncol. 2011, 39, 1285–1294. [Google Scholar]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gliga, A.R.; Di Bucchianico, S.; Åkerlund, E.; Karlsson, H.L. Transcriptome Profiling and Toxicity Following Long-Term, Low Dose Exposure of Human Lung Cells to Ni and NiO Nanoparticles—Comparison with NiCl2. Nanomaterials 2020, 10, 649. https://doi.org/10.3390/nano10040649
Gliga AR, Di Bucchianico S, Åkerlund E, Karlsson HL. Transcriptome Profiling and Toxicity Following Long-Term, Low Dose Exposure of Human Lung Cells to Ni and NiO Nanoparticles—Comparison with NiCl2. Nanomaterials. 2020; 10(4):649. https://doi.org/10.3390/nano10040649
Chicago/Turabian StyleGliga, Anda R., Sebastiano Di Bucchianico, Emma Åkerlund, and Hanna L. Karlsson. 2020. "Transcriptome Profiling and Toxicity Following Long-Term, Low Dose Exposure of Human Lung Cells to Ni and NiO Nanoparticles—Comparison with NiCl2" Nanomaterials 10, no. 4: 649. https://doi.org/10.3390/nano10040649
APA StyleGliga, A. R., Di Bucchianico, S., Åkerlund, E., & Karlsson, H. L. (2020). Transcriptome Profiling and Toxicity Following Long-Term, Low Dose Exposure of Human Lung Cells to Ni and NiO Nanoparticles—Comparison with NiCl2. Nanomaterials, 10(4), 649. https://doi.org/10.3390/nano10040649




