Zinc Oxide Nanoparticles Damage Tobacco BY-2 Cells by Oxidative Stress Followed by Processes of Autophagy and Programmed Cell Death
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Cultivation of the BY-2 Cell Suspension
2.3. Cell Viability and Growth
2.4. Spectrophotometric Measurements
2.4.1. Dehydrogenase and Oxidoreductase Activity and the Loss of the Plasma Membrane Integrity
2.4.2. Determination of Caspase-Like, Protease and Acid Phosphatase Activities
2.4.3. Quantification of Total Phenolics and PAL Activity
2.5. Extraction and Quantification of Proteins, In-Gel Assays, and Western Blot
2.5.1. Extraction and Quantification of Proteins
2.5.2. In-Gel Assays
2.5.3. Western Blot
2.6. Microscope Observations
2.6.1. Visualization of the Cell Structure—Endoplasmic Reticulum, Golgi Apparatus, and Mitochondria
2.6.2. Visualization of Intracellular ROS and RNS
2.6.3. Visualization of Zinc Ions
2.6.4. Mitotic Index, Nuclear Architecture, and DNA Fragmentation
2.7. Statistical Analyses
3. Results
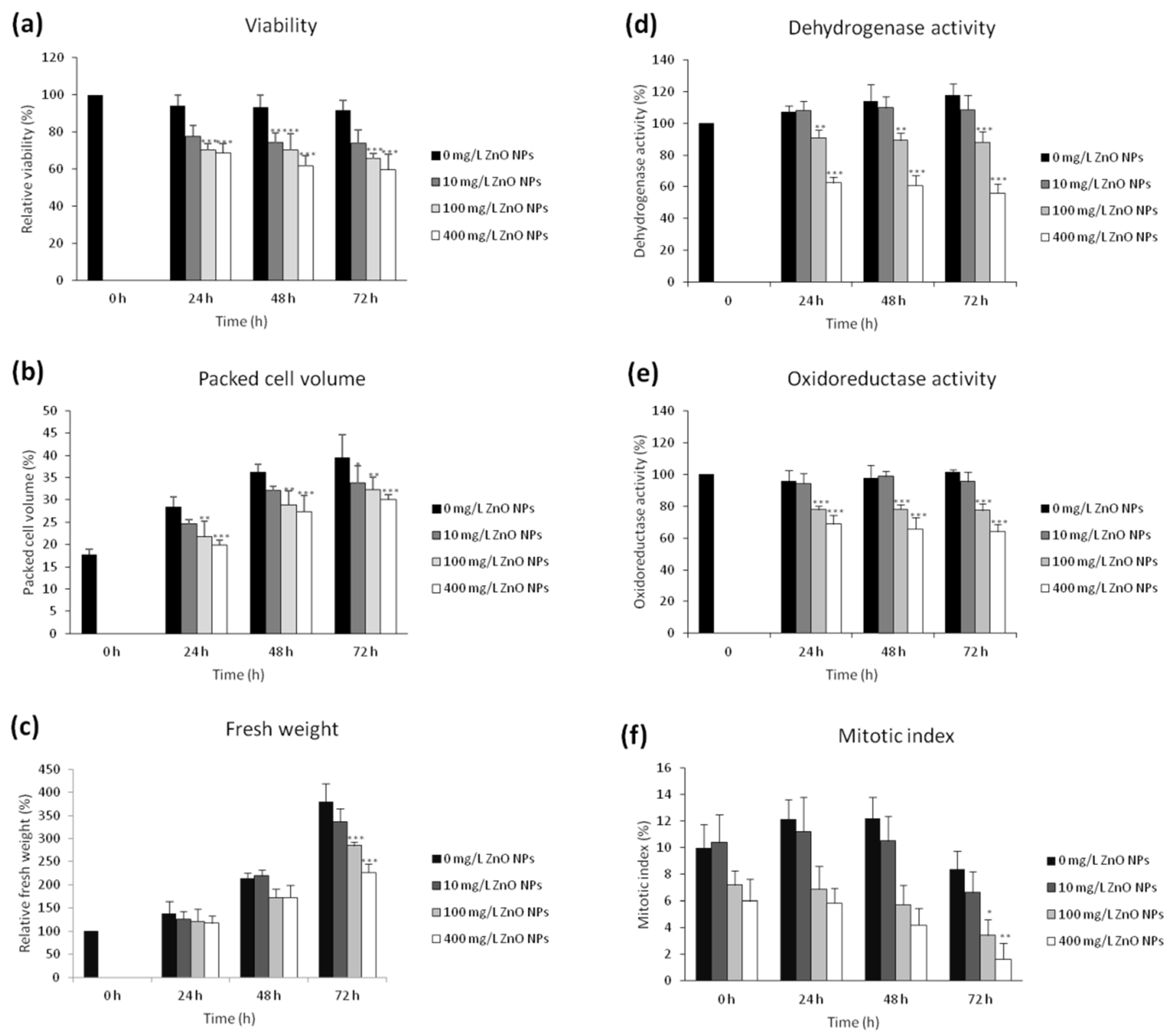
3.1. The Effect of ZnO NPs on the Viability and Cell Growth
3.2. Visualization and Quantification of Intracellular Zinc Ions, ROS and RNS in Connection with Changes in the BY-2 Cell Structure
3.3. Changes in Biochemical Parameters—Antioxidant Enzymes
3.4. Changes in Total Phenolics and PAL
3.5. A Loss of the Plasma Membrane Integrity
3.6. Changes in Nuclear Architecture and Ability of ZnO NPs to Induce Programmed Cell Death and Autophagy
4. Discussion
4.1. ZnO NPs Reduce the Viability and Growth of BY-2 Cells
4.2. The Fate of ZnO NPs and Their Effect on BY-2 Cells and Cellular Structures
4.3. ZnO NPs Increased the Level of ROS and RNS and Activated the ROS Defense System
4.4. ZnO NPs Activate PAL and Enhance Amount of Metabolites with Scavenging Activity
4.5. ZnO NPs Induce Processes of Autophagy
4.6. ZnO NPs Induce Processes of PCD
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nel, A.; Xia, T.; Madler, L.; Li, N. Toxic potential of materials at the nanolevel. Science 2006, 311, 622–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberdorster, G.; Oberdorster, E.; Oberdorster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Borm, P.J.A.; Robbins, D.; Haubold, S.; Kuhlbusch, T.; Fissan, H.; Donaldson, K.; Schins, R.; Stone, V.; Kreyling, W.; Lademann, J.; et al. The potential risks of nanomaterials: A review carried out for ECETOC. Part. Fibre Toxicol. 2006, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Cook, S.; Wang, P.; Hwang, H.-M. In vitro evaluation of cytotoxicity of engineered metal oxide nanoparticles. Sci. Total Environ. 2009, 407, 3070–3072. [Google Scholar] [CrossRef]
- Vance, M.E.; Kuiken, T.; Vejerano, E.P.; McGinnis, S.P.; Hochella, M.F.; Rejeski, D.; Hull, M.S. Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory. Beilstein J. Nanotechnol. 2015, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rico, C.M.; Majumdar, S.; Duarte-Gardea, M.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Interaction of nanoparticles with edible plants and their possible implications in the food chain. J. Agric. Food Chem. 2011, 59, 3485–3498. [Google Scholar] [CrossRef] [Green Version]
- Judy, J.D.; Unrine, J.M.; Bertsch, P.M. Evidence for biomagnification of gold nanoparticles within a terrestrial food chain. Environ. Sci. Technol. 2011, 45, 776–781. [Google Scholar] [CrossRef]
- Gottschalk, F.; Nowack, B. The release of engineered nanomaterials to the environment. J. Environ. Monit. 2011, 13, 1145–1155. [Google Scholar] [CrossRef]
- Lee, J.; Mahendra, S.; Alvarez, P.J.J. Nanomaterials in the construction industry: A review of their applications and environmental health and safety considerations. ACS Nano 2010, 4, 3580–3590. [Google Scholar] [CrossRef]
- Seil, J.T.; Webster, T.J. Antimicrobial applications of nanotechnology: Methods and literature. Int. J. Nanomed. 2012, 7, 2767–2781. [Google Scholar]
- Shaw, B.J.; Handy, R.D. Physiological effects of nanoparticles on fish: A comparison of nanometals versus metal ions. Environ. Int. 2011, 37, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.N. Do nanoparticles present ecotoxicological risks for the health of the aquatic environment? Environ. Int. 2006, 32, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Besha, A.T.; Liu, Y.J.; Bekele, D.N.; Dong, Z.M.; Naidu, R.; Gebremariam, G.N. Sustainability and environmental ethics for the application of engineered nanoparticles. Environ. Sci. Policy 2020, 103, 85–98. [Google Scholar] [CrossRef]
- Sharma, V.; Anderson, D.; Dhawan, A. Zinc oxide nanoparticles induce oxidative DNA damage and ROS-triggered mitochondria mediated apoptosis in human liver cells (HepG2). Apoptosis 2012, 17, 852–870. [Google Scholar] [CrossRef] [PubMed]
- Saquib, Q.; Al-Khedhairy, A.A.; Siddiqui, M.A.; Abou-Tarboush, F.M.; Azam, A.; Musarrat, J. Titanium dioxide nanoparticles induced cytotoxicity, oxidative stress and DNA damage in human amnion epithelial (WISH) cells. Toxicol. In Vitro 2012, 26, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Aronstam, R.S.; Chen, D.R.; Huang, Y.W. Oxidative stress, calcium homeostasis, and altered gene expression in human lung epithelial cells exposed to ZnO nanoparticles. Toxicol. In Vitro 2010, 24, 45–55. [Google Scholar] [CrossRef]
- Asare, N.; Duale, N.; Slagsvold, H.H.; Lindeman, B.; Olsen, A.K.; Gromadzka-Ostrowska, J.; Meczynska-Wielgosz, S.; Kruszewski, M.; Brunborg, G.; Instanes, C. Genotoxicity and gene expression modulation of silver and titanium dioxide nanoparticles in mice. Nanotoxicology 2016, 10, 312–321. [Google Scholar] [CrossRef]
- Kohan-Baghkheirati, E.; Geisler-Lee, J. Gene expression, protein function and pathways of arabidopsis thaliana responding to silver nanoparticles in comparison to silver ions, cold, salt, drought, and heat. Nanomaterials 2015, 5, 436–467. [Google Scholar] [CrossRef]
- Kumaran, R.S.; Choi, Y.K.; Singh, V.; Kim, K.J.; Kim, H.J. Cytotoxic effects of ZnO nanoparticles on the expression of ROS-responsive genes in the human cell lines. J. Nanosci. Nanotechnol. 2016, 16, 210–218. [Google Scholar] [CrossRef]
- Ebbs, S.D.; Bradfield, S.J.; Kumar, P.; White, J.C.; Ma, X.M. Projected dietary intake of zinc, copper, and cerium from consumption of carrot (daucus carota) exposed to metal oxide nanoparticles or metal ions. Front. Plant Sci. 2016, 7, 188. [Google Scholar] [CrossRef] [Green Version]
- Ebbs, S.D.; Bradfield, S.J.; Kumar, P.; White, J.C.; Musante, C.; Ma, X.M. Accumulation of zinc, copper, or cerium in carrot (Daucus carota) exposed to metal oxide nanoparticles and metal ions. Environ. Sci.-Nano 2016, 3, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Triboulet, S.; Aude-Garcia, C.; Armand, L.; Gerdil, A.; Diemer, H.; Proamer, F.; Collin-Faure, V.; Habert, A.; Strub, J.M.; Hanau, D.; et al. Analysis of cellular responses of macrophages to zinc ions and zinc oxide nanoparticles: A combined targeted and proteomic approach. Nanoscale 2014, 6, 6102–6114. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Zhang, Y.Y.; Mao, Z.W.; Yu, D.H.; Gao, C.Y. Toxicity of ZnO nanoparticles to macrophages due to cell uptake and intracellular release of zinc ions. J. Nanosci. Nanotechnol. 2014, 14, 5688–5696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, N.M.; Rogers, N.J.; Apte, S.C.; Batley, G.E.; Gadd, G.E.; Casey, P.S. Comparative toxicity of nanoparticulate ZnO, bulk ZnO, and ZnCl2 to a freshwater microalga (Pseudokirchneriella subcapitata): The importance of particle solubility. Environ. Sci. Technol. 2007, 41, 8484–8490. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, M.; Kim, D.H. In vitro toxicity of zinc oxide nanoparticles: A review. J. Nanopart. Res. 2015, 17, 158. [Google Scholar] [CrossRef]
- Xiao, L.; Liu, C.H.; Chen, X.N.; Yang, Z. Zinc oxide nanoparticles induce renal toxicity through reactive oxygen species. Food Chem. Toxicol. 2016, 90, 76–83. [Google Scholar] [CrossRef]
- Wu, D.M.; Ma, Y.; Cao, Y.N.; Zhang, T. Mitochondrial toxicity of nanomaterials. Sci. Total Environ. 2020, 702, 134994. [Google Scholar] [CrossRef]
- Sun, Z.Q.; Xiong, T.T.; Zhang, T.; Wang, N.F.; Chen, D.; Li, S.S. Influences of zinc oxide nanoparticles on Allium cepa root cells and the primary cause of phytotoxicity. Ecotoxicology 2019, 28, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Burman, U.; Saini, M.; Praveen, K. Effect of zinc oxide nanoparticles on growth and antioxidant system of chickpea seedlings. Toxicol. Environ. Chem. 2013, 95, 605–612. [Google Scholar] [CrossRef]
- Pokhrel, L.R.; Dubey, B. Evaluation of developmental responses of two crop plants exposed to silver and zinc oxide nanoparticles. Sci. Total Environ. 2013, 452, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.N.V.K.V.; Sudhakar, P.; Sreenivasulu, Y.; Latha, P.; Munaswamy, V.; Reddy, K.R.; Sreeprasad, T.S.; Sajanlal, P.R.; Pradeep, T. Effect of nanoscale zinc oxide particles on the germination, growth and yield of peanut. J. Plant Nutr. 2012, 35, 905–927. [Google Scholar] [CrossRef]
- Yoon, S.-J.; Kwak, J.I.; Lee, W.-M.; Holden, P.A.; An, Y.-J. Zinc oxide nanoparticles delay soybean development: A standard soil microcosm study. Ecotoxicol. Environ. Saf. 2014, 100, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Balazova, L.; Babula, P.; Balaz, M.; Backorova, M.; Bujnakova, Z.; Briancin, J.; Kurmanbayeva, A.; Sagi, M. Zinc oxide nanoparticles phytotoxicity on halophyte from genus Salicornia. Plant Physiol. Biochem. 2018, 130, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.P.; Wang, R.T.; Wang, R.L.; Ju, Q.; Wang, Y.B.; Xu, J. Comparative physiological and transcriptomic analyses reveal the toxic effects of ZnO nanoparticles on plant growth. Environ. Sci. Technol. 2019, 53, 4235–4244. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.; Kim, S.; Lee, I. Assessment of phytotoxicity of ZnO NPs on a medicinal plant, Fagopyrum esculentum. Environ. Sci. Pollut. Res. 2013, 20, 848–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youssef, M.S.; Elamawi, R.M. Evaluation of phytotoxicity, cytotoxicity, and genotoxicity of ZnO nanoparticles in Vicia faba. Environ. Sci. Pollut. Res. Int. 2018. [Google Scholar] [CrossRef]
- Zhu, J.; Zou, Z.; Shen, Y.; Li, J.; Shi, S.; Han, S.; Zhan, X. Increased ZnO nanoparticle toxicity to wheat upon co-exposure to phenanthrene. Environ. Pollut. 2019, 247, 108–117. [Google Scholar] [CrossRef]
- Zakharova, O.V.; Gusev, A.A. Photocatalytically active zinc oxide and titanium dioxide nanoparticles in clonal micropropagation of plants: Prospects. Nanotechnologies Russia 2019, 14, 311–324. [Google Scholar] [CrossRef]
- Brunner, T.J.; Wick, P.; Manser, P.; Spohn, P.; Grass, R.N.; Limbach, L.K.; Bruinink, A.; Stark, W.J. In vitro cytotoxicity of oxide nanoparticles: Comparison to asbestos, silica, and the effect of particle solubility. Environ. Sci. Technol. 2006, 40, 4374–4381. [Google Scholar] [CrossRef]
- Xia, T.; Kovochich, M.; Brant, J.; Hotze, M.; Sempf, J.; Oberley, T.; Sioutas, C.; Yeh, J.I.; Wiesner, M.R.; Nel, A.E. Comparison of the abilities of ambient and manufactured nanoparticles to induce cellular toxicity according to an oxidative stress paradigm. Nano Lett. 2006, 6, 1794–1807. [Google Scholar] [CrossRef]
- Lin, D.; Xing, B. Root uptake and phytotoxicity of ZnO nanoparticles. Environ. Sci. Technol. 2008, 42, 5580–5585. [Google Scholar] [CrossRef] [PubMed]
- Asli, S.; Neumann, P.M. Colloidal suspensions of clay or titanium dioxide nanoparticles can inhibit leaf growth and transpiration via physical effects on root water transport. Plant Cell Environ. 2009, 32, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Corredor, E.; Testillano, P.S.; Coronado, M.-J.; Gonzalez-Melendi, P.; Fernandez-Pacheco, R.; Marquina, C.; Ricardo Ibarra, M.; de la Fuente, J.M.; Rubiales, D.; Perez-de-Luque, A.; et al. Nanoparticle penetration and transport in living pumpkin plants: In situ subcellular identification. BMC Plant Biol. 2009, 9, 45. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Williams, P.L.; Diamond, S.A. Ecotoxicity of manufactured ZnO nanoparticles—A review. Environ. Pollut. 2013, 172, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Krystofova, O.; Sochor, J.; Zitka, O.; Babula, P.; Kudrle, V.; Adam, V.; Kizek, R. Effect of magnetic nanoparticles on tobacco BY-2 cell suspension culture. Int. J. Environ. Res. Public Health 2013, 10, 47–71. [Google Scholar]
- Poborilova, Z.; Opatrilova, R.; Babula, P. Toxicity of aluminium oxide nanoparticles demonstrated using a BY-2 plant cell suspension culture model. Environ. Exp. Bot. 2013, 91, 1–11. [Google Scholar]
- Trojan, V.; Chomoucka, J.; Krystofova, O.; Hubalek, J.; Babula, P.; Kizek, R. Quantum dots (CdSe) modified by glutathione and their localization of tobacco BY-2 cells. J. Biotechnol. 2010, 150, S479. [Google Scholar] [CrossRef]
- Svobodnikova, L.; Kummerova, M.; Zezulka, S.; Babula, P. Possible use of a Nicotiana tabacum ‘Bright Yellow 2’ cell suspension as a model to assess phytotoxicity of pharmaceuticals (diclofenac). Ecotoxicol. Environ. Saf. 2019, 182, 109369. [Google Scholar] [CrossRef]
- Poborilova, Z.; Ohlsson, A.B.; Berglund, T.; Vildova, A.; Provaznik, I.; Babula, P. DNA hypomethylation concomitant with the overproduction of ROS induced by naphthoquinone juglone on tobacco BY-2 suspension cells. Environ. Exp. Bot. 2015, 113, 28–39. [Google Scholar] [CrossRef]
- Nagata, T.; Nemoto, Y.; Hasezawa, S. Tobacco BY-2 cell line as the HeLa cell in the cell biology of higher plants. Int. Rev. Cytol. 1992, 132, 1–30. [Google Scholar]
- Babula, P.; Vodicka, O.; Adam, V.; Kummerova, M.; Havel, L.; Hosek, J.; Provaznik, I.; Skutkova, H.; Beklova, M.; Kizek, R. Effect of fluoranthene on plant cell model: Tobacco BY-2 suspension culture. Environ. Exp. Bot. 2012, 78, 117–126. [Google Scholar] [CrossRef]
- Vagnerova, K.; Macura, J. Determination of protease activity of plant roots. Folia Microbiol. 1974, 19, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Charney, J.; Tomarelli, R.M. A colorimetric method for the determination of the proteolytic activity of duodenal juice. J. Biol. Chem. 1947, 171, 501–505. [Google Scholar]
- Ferrarese, M.L.L.; Rodrigues, J.D.; Ferrarese, O. Phenylalanine ammonia-lyase activity in soybean roots extract measured by reverse-phase high performance liquid chromatography. Plant Biol. 2000, 2, 152–153. [Google Scholar] [CrossRef]
- dos Santos, W.D.; Ferrarese, M.D.L.; Finger, A.; Teixeira, A.C.N.; Ferrarese, O. Lignification and related enzymes in Glycine max root growth-inhibition by ferulic acid. J. Chem. Ecol. 2004, 30, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Kovacik, J.; Klejdus, B.; Backor, M.; Repcak, M. Phenylalanine ammonia-lyase activity and phenolic compounds accumulation in nitrogen-deficient Matricaria chamomilla leaf rosettes. Plant Sci. 2007, 172, 393–399. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for quantification of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brychkova, G.; Yarmolinsky, D.; Ventura, Y.; Sagi, M. A novel in-gel assay and an improved kinetic assay for determining In vitro sulfite reductase activity in plants. Plant Cell Physiol. 2012, 53, 1507–1516. [Google Scholar] [CrossRef]
- Brewer, G.J. Achromatic regions of tetrazolium stained starch gels: Inherited electrophoretic variation. Am. J. Hum. Genet. 1967, 19, 674. [Google Scholar]
- Mittler, R.; Zilinskas, B.A. Detection of ascorbate peroxidase activity in native gels by inhibition of the ascorbate-dependent reduction of nitroblue tetrazolium. Anal. Biochem. 1993, 212, 540–546. [Google Scholar]
- Gregory, E.M.; Fridovic, I. Visualization of catalase on acrylamide gels. Anal. Biochem. 1974, 58, 57–62. [Google Scholar]
- Sagi, M.; Davydov, O.; Orazova, S.; Yesbergenova, Z.; Ophir, R.; Stratmann, J.W.; Fluhr, R. Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 2004, 16, 616–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brychkova, G.; Xia, Z.; Yang, G.; Yesbergenova, Z.; Zhang, Z.; Davydov, O.; Fluhr, R.; Sagi, M. Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 2007, 50, 696–709. [Google Scholar] [CrossRef]
- Helaly, M.S.; El-Metwally, M.A.; El-Hoseiny, H.; Omar, S.A.; El-Sheery, N.I. Effect of nanoparticles on biological contamination of in vitro cultures and organogenic regeneration of banana. Aust. J. Crop Sci. 2014, 8, 612–624. [Google Scholar]
- Czyzowska, A.; Barbasz, A. Effect of ZnO, TiO2, Al2O3, and ZrO2 nanoparticles on wheat callus cells. Acta Biochim. Pol. 2019, 66, 365–370. [Google Scholar] [CrossRef] [Green Version]
- Peralta-Videa, J.R.; Hernandez-Viezcas, J.A.; Zhao, L.; Diaz, B.C.; Ge, Y.; Priester, J.H.; Holden, P.A.; Gardea-Torresdey, J.L. Cerium dioxide and zinc oxide nanoparticles alter the nutritional value of soil cultivated soybean plants. Plant Physiol. Biochem. 2014, 80, 128–135. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Chen, S.; Li, Q.; Wang, W.; Hou, C.; Gao, X.; Wang, L.; Wang, S. Zinc oxide nanoparticles affect biomass accumulation and photosynthesis in Arabidopsis. Front. Plant Sci. 2016, 6, 1243. [Google Scholar] [CrossRef] [Green Version]
- Thunugunta, T.; Reddy, A.C.; Seetharamaiah, S.K.; Hunashikatti, L.R.; Chandrappa, S.G.; Kalathil, N.C.; Reddy, L. Impact of Zinc oxide nanoparticles on eggplant (S-melongena): Studies on growth and the accumulation of nanoparticles. IET Nanobiotechnol. 2018, 12, 706–713. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; McLean, J.E.; Latta, D.E.; Manangon, E.; Britt, D.W.; Johnson, W.P.; Boyanov, M.I.; Anderson, A.J. CuO and ZnO nanoparticles: Phytotoxicity, metal speciation, and induction of oxidative stress in sand-grown wheat. J. Nanopart. Res. 2012, 14, 1125. [Google Scholar] [CrossRef]
- Kim, S.; Lee, S.; Lee, I. Alteration of phytotoxicity and oxidant stress potential by metal oxide nanoparticles in cucumis sativus. Water Air Soil Pollut. 2012, 223, 2799–2806. [Google Scholar] [CrossRef]
- Lin, D.; Xing, B. Phytotoxicity of nanoparticles: Inhibition of seed germination and root growth. Environ. Pollut. 2007, 150, 243–250. [Google Scholar] [CrossRef]
- Khudsar, T.; Mahmooduzzafar; Iqbal, M.; Sairam, R.K. Zinc-induced changes in morpho-physiological and biochemical parameters in Artemisia annua. Biol. Plant. 2004, 48, 255–260. [Google Scholar] [CrossRef]
- Brown, P.H.; Cakmak, I.; Zhang, Q.L. Form and function of zinc plants. In Zinc in Soils and Plants; Springer: Dordrecht, The Netherlands, 1993; Volume 55, pp. 93–106. [Google Scholar]
- Cakmak, I. Tansley review No. 111—Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 2000, 146, 185–205. [Google Scholar] [CrossRef]
- Hernandez-Viezcas, J.A.; Castillo-Michel, H.; Servin, A.D.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Spectroscopic verification of zinc absorption and distribution in the desert plant Prosopis juliflora-velutina (velvet mesquite) treated with ZnO nanoparticles. Chem. Eng. J. 2011, 170, 346–352. [Google Scholar] [CrossRef] [Green Version]
- Pullagurala, V.L.R.; Adisa, I.O.; Rawat, S.; Kim, B.; Barrios, A.C.; Medina-Velo, I.A.; Hernandez-Viezcas, J.A.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Finding the conditions for the beneficial use of ZnO nanoparticles towards plants-A review. Environ. Pollut. 2018, 241, 1175–1181. [Google Scholar] [CrossRef]
- Hernandez-Viezcas, J.A.; Castillo-Michel, H.; Andrews, J.C.; Cotte, M.; Rico, C.; Peralta-Videa, J.R.; Ge, Y.; Priester, J.H.; Holden, P.A.; Gardea-Torresdey, J.L. In situ synchrotron X-ray fluorescence mapping and speciation of CeO2 and ZnO nanoparticles in soil cultivated soybean (Glycine max). ACS Nano 2013, 7, 1415–1423. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; McLean, J.E.; Britt, D.W.; Anderson, A.J. Antifungal activity of ZnO nanoparticles and their interactive effect with a biocontrol bacterium on growth antagonism of the plant pathogen Fusarium graminearum. BioMetals 2013, 26, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Khan, S.S.; Pakrashi, S.; Mukherjee, A.; Chandrasekaran, N. Cytogenetic and genotoxic effects of zinc oxide nanoparticles on root cells of Allium cepa. J. Hazard. Mater. 2011, 190, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.M.G.; Chung, I.M. Regulation of morphological, molecular and nutrient status in Arabidopsis thaliana seedlings in response to ZnO nanoparticles and Zn ion exposure. Sci. Total Environ. 2017, 575, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Muschitz, A.; Riou, C.; Mollet, J.C.; Gloaguen, V.; Faugeron, C. Modifications of cell wall pectin in tomato cell suspension in response to cadmium and zinc. Acta Physiol. Plant. 2015, 37, 245. [Google Scholar] [CrossRef]
- Cifuentes, Z.; Custardoy, L.; de la Fuente, J.M.; Marquina, C.; Ibarra, M.R.; Rubiales, D.; Perez-De-Luque, A. Absorption and translocation to the aerial part of magnetic carbon-coated nanoparticles through the root of different crop plants. J. Nanobiotechnol. 2010, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hischemoller, A.; Nordmann, J.; Ptacek, P.; Mummenhoff, K.; Haase, M. In-vivo imaging of the uptake of upconversion nanoparticles by plant roots. J. Biomed. Nanotechnol. 2009, 5, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.T.; Zhang, S.Z.; Luo, L.; Zhang, J.; Yang, K.; Christie, P. Accumulation, speciation and uptake pathway of ZnO nanoparticles in maize. Environ. Sci.-Nano 2015, 2, 68–77. [Google Scholar] [CrossRef]
- Qin, Y.; Dittmer, P.J.; Park, J.G.; Jansen, K.B.; Palmer, A.E. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc. Natl. Acad. Sci. USA 2011, 108, 7351–7356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Huo, L.L.; Shi, X.F.; Bai, R.; Zhang, Z.J.; Zhao, Y.L.; Chang, Y.Z.; Chen, C.Y. Endoplasmic reticulum stress induced by zinc oxide nanoparticles is an earlier biomarker for nanotoxicological evaluation. ACS Nano 2014, 8, 2562–2574. [Google Scholar] [CrossRef]
- Tuncay, E.; Cicek, F.A.; Toy, A.; Turan, B. Intracellular free zinc ion increase triggers hyperglycemia-induced cardiomyocyte dysfunction through endoplasmic reticulum stress. Biophys. J. 2014, 106, 113A. [Google Scholar] [CrossRef] [Green Version]
- Singh, H.; Lee, H.W.; Heo, C.H.; Byun, J.W.; Sarkar, A.R.; Kim, H.M. A Golgi-localized two-photon probe for imaging zinc ions. Chem. Commun. 2015, 51, 12099–12102. [Google Scholar] [CrossRef]
- Maret, W. Molecular aspects of human cellular zinc homeostasis: Redox control of zinc potentials and zinc signals. BioMetals 2009, 22, 149–157. [Google Scholar] [CrossRef]
- Ma, H.; Kabengi, N.J.; Bertsch, P.M.; Unrine, J.M.; Glenn, T.C.; Williams, P.L. Comparative phototoxicity of nanoparticulate and bulk ZnO to a free-living nematode Caenorhabditis elegans: The importance of illumination mode and primary particle size. Environ. Pollut. 2011, 159, 1473–1480. [Google Scholar] [CrossRef]
- Navarro, E.; Baun, A.; Behra, R.; Hartmann, N.B.; Filser, J.; Miao, A.-J.; Quigg, A.; Santschi, P.H.; Sigg, L. Environmental behavior and ecotoxicity of engineered nanoparticles to algae, plants, and fungi. Ecotoxicology 2008, 17, 372–386. [Google Scholar] [CrossRef] [Green Version]
- Chevion, M. A site-specific mechanism for free-radical induced biological damage: The essential role of redox-active transition-metals. Free Radical Biol. Med. 1988, 5, 27–37. [Google Scholar] [CrossRef]
- Hou, J.; Wu, Y.Z.; Li, X.; Wei, B.B.; Li, S.G.; Wang, X.K. Toxic effects of different types of zinc oxide nanoparticles on algae, plants, invertebrates, vertebrates and microorganisms. Chemosphere 2018, 193, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Kovacik, J.; Babula, P. Fluorescence microscopy as a tool for visualization of metal-induced oxidative stress in plants. Acta Physiol. Plant. 2017, 39, 157. [Google Scholar] [CrossRef]
- Feigl, G.; Lehotai, N.; Molnar, A.; Ordog, A.; Rodriguez-Ruiz, M.; Palma, J.M.; Corpas, F.J.; Erdei, L.; Kolbert, Z. Zinc induces distinct changes in the metabolism of reactive oxygen and nitrogen species (ROS and RNS) in the roots of two Brassica species with different sensitivity to zinc stress. Ann. Bot. 2015, 116, 613–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, A.; Vangronsveld, J.; Clijsters, H. The redox status of plant cells (AsA and GSH) is sensitive to zinc imposed oxidative stress in roots and primary leaves of Phaseolus vulgaris. Plant Physiol. Biochem. 2001, 39, 657–664. [Google Scholar] [CrossRef]
- Panda, S.K.; Chaudhury, I.; Khan, M.H. Heavy metals induce lipid peroxidation and affect antioxidants in wheat leaves. Biol. Plant. 2003, 46, 289–294. [Google Scholar] [CrossRef]
- Aarti, P.D.; Tanaka, R.; Tanaka, A. Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant. 2006, 128, 186–197. [Google Scholar] [CrossRef]
- Thwala, M.; Musee, N.; Sikhwivhilu, L.; Wepener, V. The oxidative toxicity of Ag and ZnO nanoparticles towards the aquatic plant Spirodela punctuta and the role of testing media parameters. Environ. Sci.-Proc. Imp. 2013, 15, 1830–1843. [Google Scholar] [CrossRef]
- Hafizi, Z.; Nasr, N. The effect of zinc oxide nanoparticles on safflower plant growth and physiology. Eng. Technol. Appl. Sci. Res. 2018, 8, 2508–2513. [Google Scholar]
- Parlak, K.U.; Yilmaz, D.D. Response of antioxidant defences to Zn stress in three duckweed species. Ecotoxicol. Environ. Saf. 2012, 85, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Marichali, A.; Dallali, S.; Ouerghemmi, S.; Sebei, H.; Casabianca, H.; Hosni, K. Responses of Nigella sativa L. to zinc excess: Focus on germination, growth, yield and yield components, lipid and terpene metabolism, and total phenolics and antioxidant activities. J. Agric. Food Chem. 2016, 64, 1664–1675. [Google Scholar] [CrossRef]
- Parrotta, L.; Guerriero, G.; Sergeant, K.; Cal, G.; Hausman, J.F. Target or barrier? The cell wall of early- and later-diverging plants vs cadmium toxicity: Differences in the response mechanisms. Front. Plant Sci. 2015, 6, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zafar, H.; Ali, A.; Ali, J.S.; Haq, I.U.; Zia, M. Effect of ZnO nanoparticles on brassica nigra seedlings and stem explants: Growth dynamics and antioxidative response. Front. Plant Sci. 2016, 7, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, N.; Arizumi, T.; Takita, M.; Kitai, S.; Yada, N.; Hagiwara, S.; Inoue, T.; Minami, Y.; Ueshima, K.; Sakurai, T.; et al. Reactive oxygen species induce epigenetic instability through the formation of 8-hydroxydeoxyguanosine in human hepatocarcinogenesis. Dig. Dis. 2013, 31, 459–466. [Google Scholar] [CrossRef]
- Solis, M.-T.; Chakrabarti, N.; Corredor, E.; Cortes-Eslava, J.; Rodriguez-Serrano, M.; Biggiogera, M.; Risueno, M.C.; Testillano, P.S. Epigenetic changes accompany developmental programmed cell death in tapetum cells. Plant Cell Physiol. 2014, 55, 16–29. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhao, L.; Hou, H.; Zhang, H.; Huang, Y.; Wang, Y.; Li, H.; Gao, F.; Yan, S.; Li, L. Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol. 2015, 56, 965–976. [Google Scholar] [CrossRef] [Green Version]
- Michaeli, S.; Galili, G.; Genschik, P.; Fernie, A.R.; Avin-Wittenberg, T. Autophagy in plants—What’s new on the menu? Trends Plant Sci. 2016, 21, 134–144. [Google Scholar] [CrossRef]
- Kaufman, R.J. Protein folding, oxidative stress, and the unfolded protein response. Int. J. Clin. Pharmacol. Ther. 2010, 48, S4–S5. [Google Scholar] [CrossRef]
- Hetz, C. Protein folding stress in neurodegenerative disease: An interplay between ER stress and autophagy. FEBS J. 2012, 279, 15. [Google Scholar]
- Esther Perez-Perez, M.; Lemaire, S.D.; Crespo, J.L. Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 2012, 160, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denton, D.; Shravage, B.; Simin, R.; Mills, K.; Berry, D.L.; Baehrecke, E.H.; Kumar, S. Autophagy, not apoptosis, is essential for midgut cell death in drosophila. Curr. Biol. 2009, 19, 1741–1746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyasu, Y.; Ohsumi, Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996, 111, 1233–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hailey, D.W.; Rambold, A.S.; Satpute-Krishnan, P.; Mitra, K.; Sougrat, R.; Kim, P.K.; Lippincott-Schwartz, J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010, 141, 656–667. [Google Scholar] [CrossRef] [Green Version]
- Hayashi-Nishino, M.; Fujita, N.; Noda, T.; Yamaguchi, A.; Yoshimori, T.; Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009, 11, 1433–1437. [Google Scholar] [CrossRef]
- Yen, W.-L.; Shintani, T.; Nair, U.; Cao, Y.; Richardson, B.C.; Li, Z.; Hughson, F.M.; Baba, M.; Klionsky, D.J. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 2010, 188, 101–114. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, M.; Jana, A.; Sinha, S.; Jothiramajayam, M.; Nag, A.; Chakraborty, A.; Mukherjee, A.; Mukherjee, A. Effects of ZnO nanoparticles in plants: Cytotoxicity, genotoxicity, deregulation of antioxidant defenses, and cell-cycle arrest. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 807, 25–32. [Google Scholar] [CrossRef]
- Minibayeva, F.; Dmitrieva, S.; Ponomareva, A.; Ryabovol, V. Oxidative stress-induced autophagy in plants: The role of mitochondria. Plant Physiol. Biochem 2012, 59, 11–19. [Google Scholar] [CrossRef]
- Minibayeva, F.V.; Ryabovol, V.; Ponomareva, A. Oxidative stress-induced autophagy in wheat seedlings. S. Afr. J. Bot. 2013, 86, 165. [Google Scholar] [CrossRef] [Green Version]
- Perez-Martin, M.; Perez-Perez, M.E.; Lemaire, S.D.; Crespo, J.L. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 2014, 166, 997–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liuzzi, J.P.; Guo, L.; Yoo, C.; Stewart, T.S. Zinc and autophagy. BioMetals 2014, 27, 1087–1096. [Google Scholar] [CrossRef]
- Wei, L.M.; Wang, J.F.; Chen, A.J.; Liu, J.; Feng, X.L.; Shao, L.Q. Involvement of PINK1/parkin-mediated mitophagy in ZnO nanoparticle-induced toxicity in BV-2 cells. Int. J. Nanomed. 2017, 12, 1891–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Xu, Q.; Xing, D.; Gao, C.; Xiong, H. Real-time detection of caspase-3-like protease activation in vivo using fluorescence resonance energy transfer during plant programmed cell death induced by ultraviolet C overexposure. Plant Physiol. 2009, 150, 1773–1783. [Google Scholar] [CrossRef] [Green Version]
- Siczek, L.; Mostowska, A. Characteristics and function of plant caspases during programmed cell death in plants. Postepy Biol. Komorki 2012, 39, 159–172. [Google Scholar]
- Debnath, P.; Mondal, A.; Sen, K.; Mishra, D.; Mondal, N.K. Genotoxicity study of nano Al2O3, TiO2 and ZnO along with UV-B exposure: An Allium cepa root tip assay. Sci. Total Environ. 2020, 713, 136592. [Google Scholar] [CrossRef]
- Takada, S.; Shirakata, Y.; Kaneniwa, N.; Koike, K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 1999, 18, 6965–6973. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.S.; Ren, X.; Zhu, R.; Luo, Z.Y.; Ren, B.X. Zinc oxide nanoparticles induce oxidative DNA damage and ROS-triggered mitochondria-mediated apoptosis in zebrafish embryos. Aquat. Toxicol. 2016, 180, 56–70. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Haga, N.; Fujita, N.; Tsuruo, T. Mitochondrial aggregation precedes cytochrome c 1release from mitochondria during apoptosis. Oncogene 2003, 22, 5579–5585. [Google Scholar] [CrossRef] [Green Version]
- Panda, K.K.; Golari, D.; Venugopal, A.; Achary, V.M.M.; Phaomei, G.; Parinandi, N.L.; Sahu, H.K.; Panda, B.B. Green synthesized zinc oxide (ZnO) nanoparticles induce oxidative stress and DNA damage in Lathyrus sativus L. root bioassay system. Antioxidants 2017, 6, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balážová, Ľ.; Baláž, M.; Babula, P. Zinc Oxide Nanoparticles Damage Tobacco BY-2 Cells by Oxidative Stress Followed by Processes of Autophagy and Programmed Cell Death. Nanomaterials 2020, 10, 1066. https://doi.org/10.3390/nano10061066
Balážová Ľ, Baláž M, Babula P. Zinc Oxide Nanoparticles Damage Tobacco BY-2 Cells by Oxidative Stress Followed by Processes of Autophagy and Programmed Cell Death. Nanomaterials. 2020; 10(6):1066. https://doi.org/10.3390/nano10061066
Chicago/Turabian StyleBalážová, Ľudmila, Matej Baláž, and Petr Babula. 2020. "Zinc Oxide Nanoparticles Damage Tobacco BY-2 Cells by Oxidative Stress Followed by Processes of Autophagy and Programmed Cell Death" Nanomaterials 10, no. 6: 1066. https://doi.org/10.3390/nano10061066
APA StyleBalážová, Ľ., Baláž, M., & Babula, P. (2020). Zinc Oxide Nanoparticles Damage Tobacco BY-2 Cells by Oxidative Stress Followed by Processes of Autophagy and Programmed Cell Death. Nanomaterials, 10(6), 1066. https://doi.org/10.3390/nano10061066





