Single-Particle Tracking with Scanning Non-Linear Microscopy
Abstract
:1. Introduction
2. Material and Methods
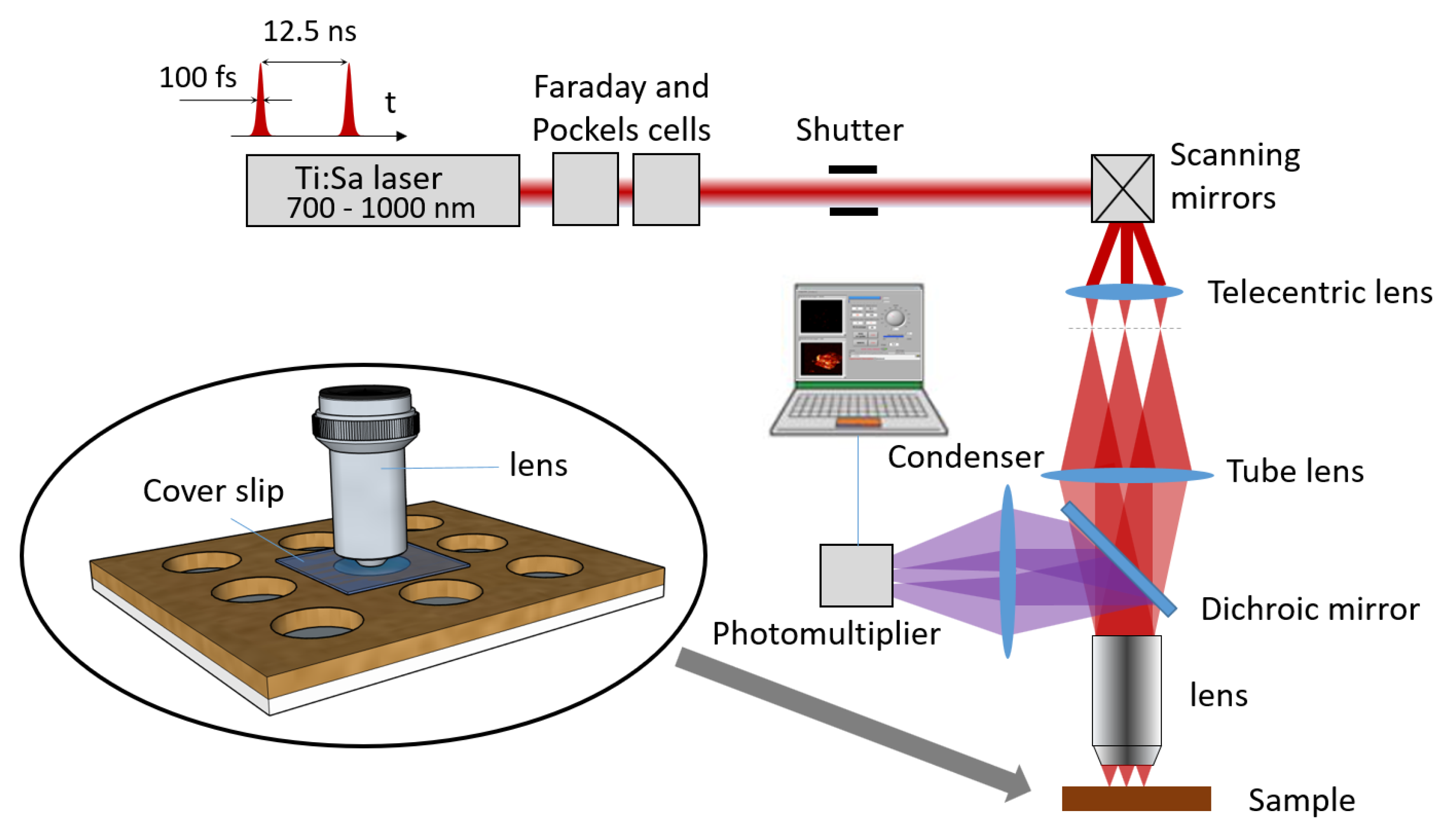
2.1. Equipment
2.2. Consumables
2.3. Software
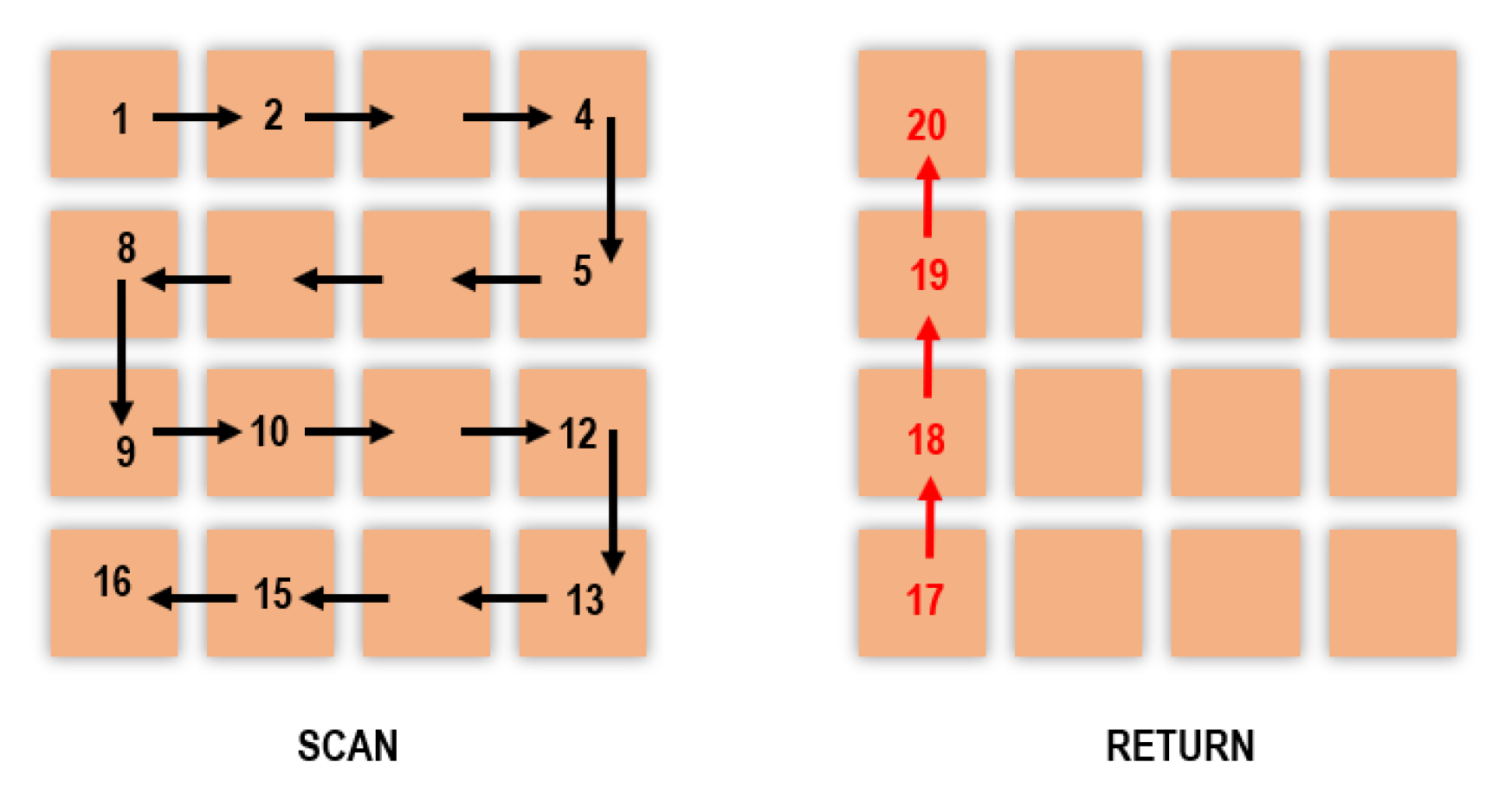
2.4. Scanning Methods
2.5. Track Analysis Methods
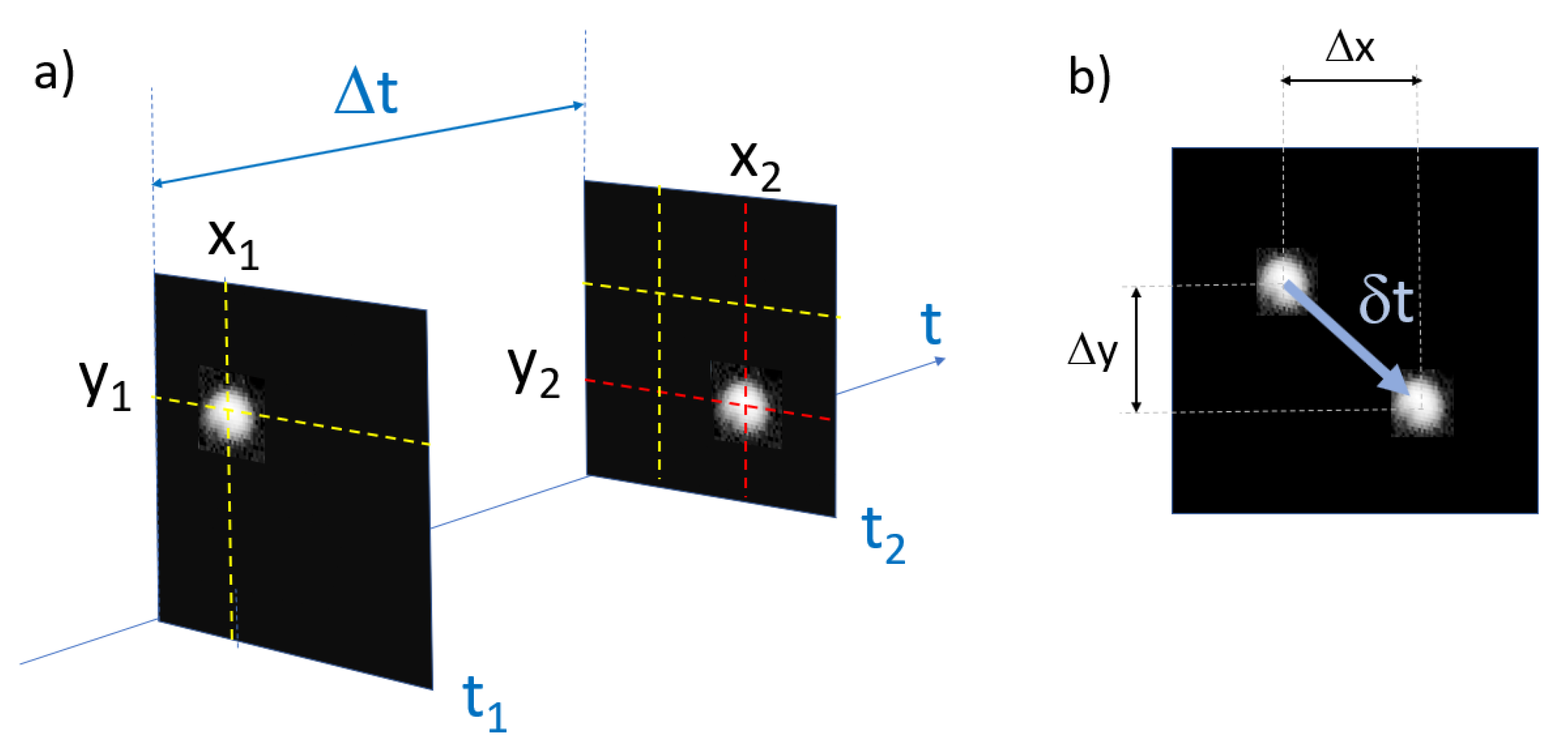
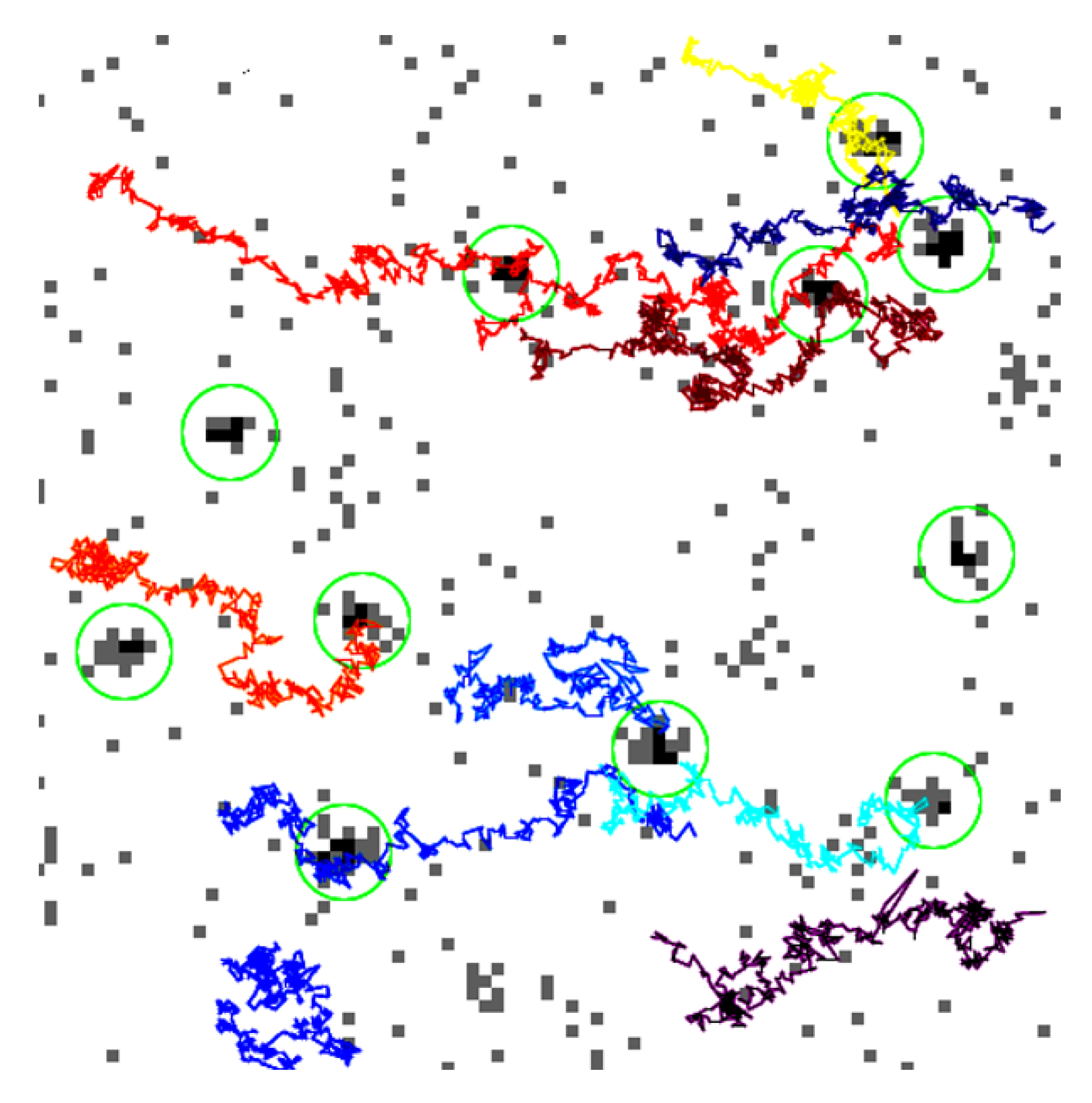
2.6. Tracking
3. Estimator
3.1. Theory
3.2. Experimental Estimation
4. Overview of Improvements and Limits with NLO Microscopy
4.1. Photobleaching
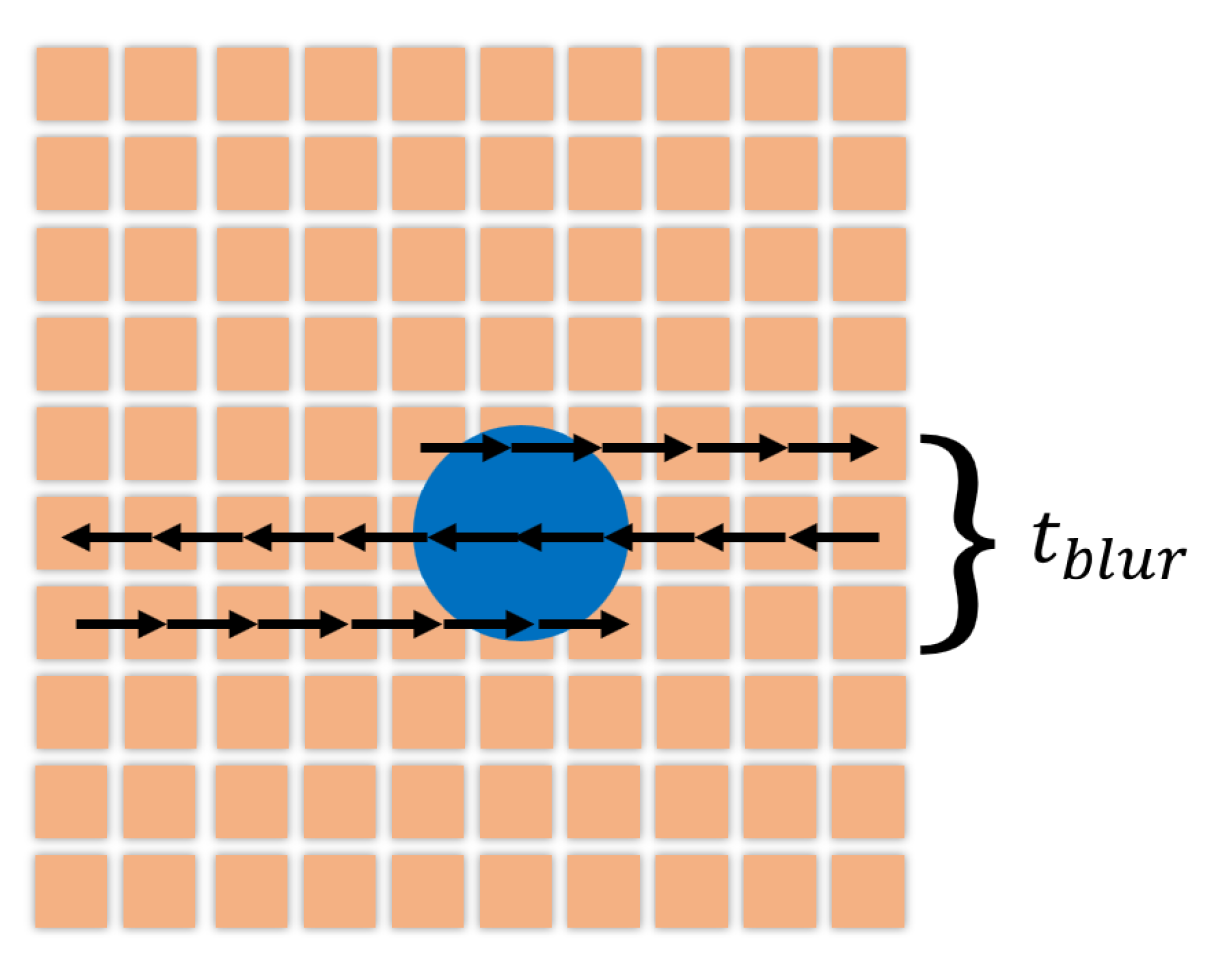
4.2. Motion Blur
4.3. Brilliance and Numerical Aperture
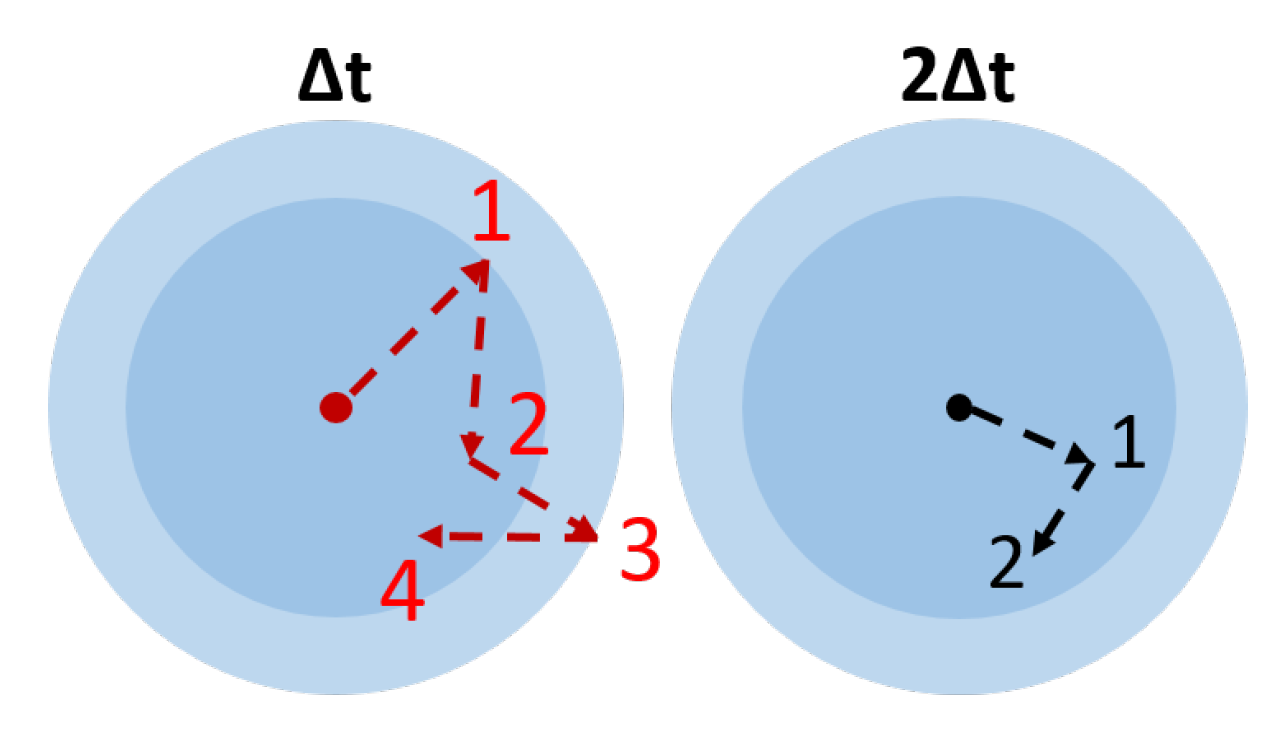
4.4. Image Resolution and Frame Rate
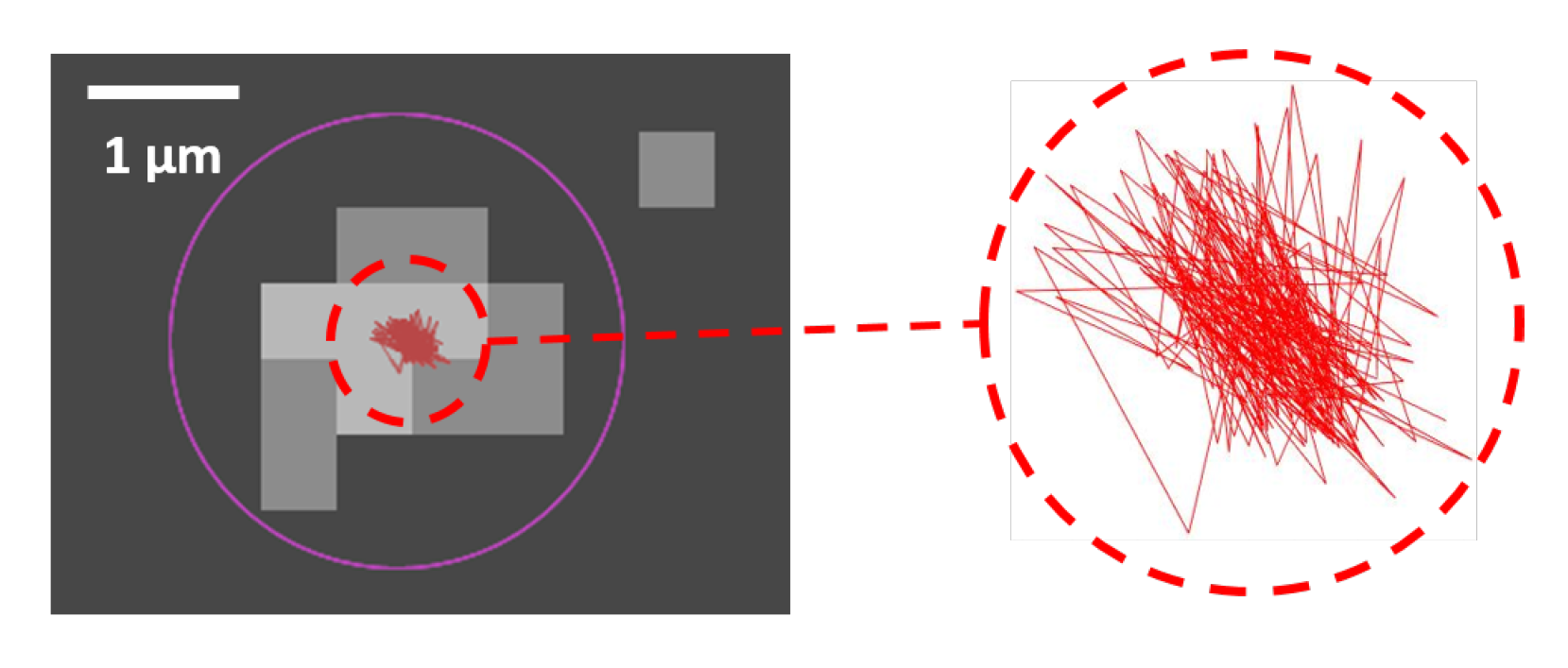
4.5. Image Resolution and Localization Error
4.6. Scanned Area and Image Resolution
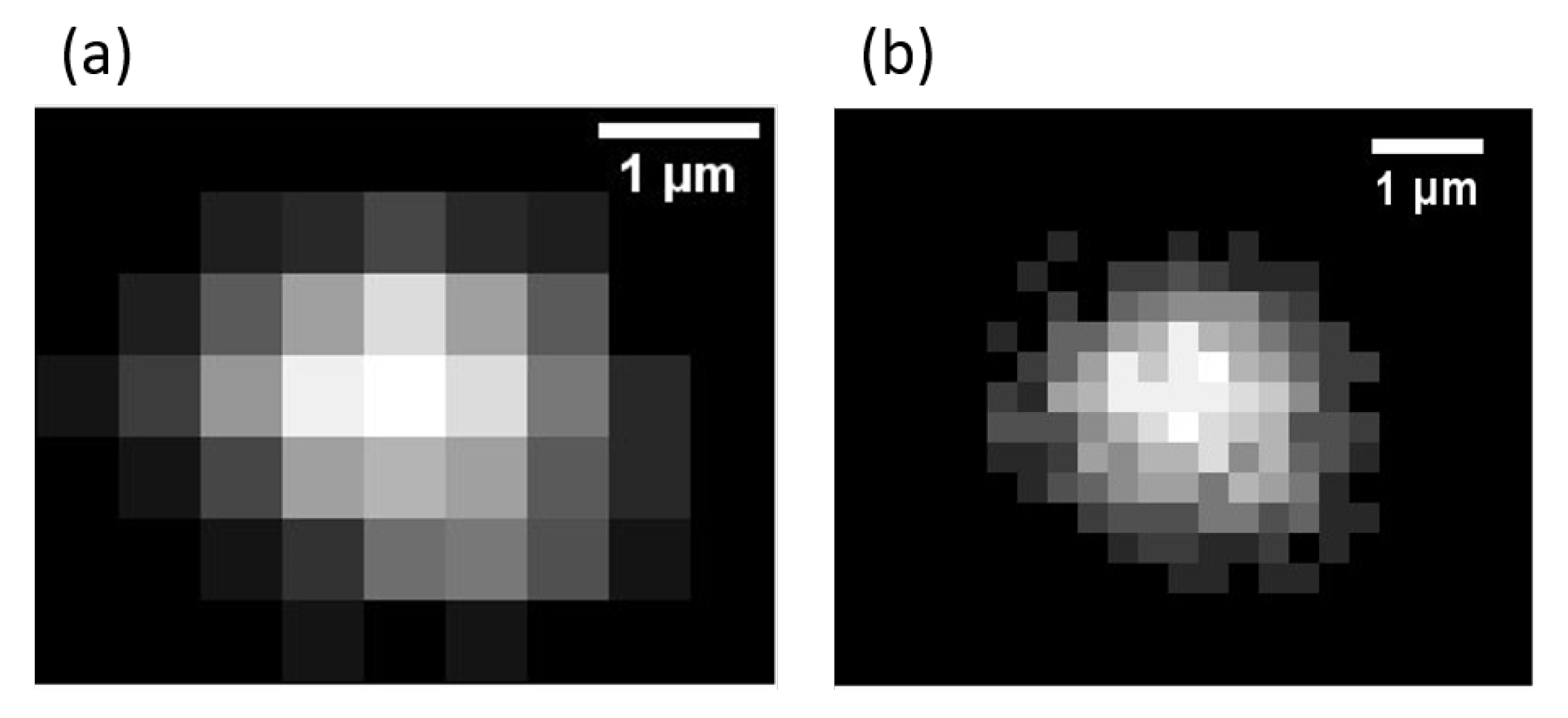
4.7. Signal-To-Noise Ratio Localization and Particle Size
5. Results and Discussion
5.1. Errors
5.2. Size Estimation
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fick, A. Ueber Diffusion. Ann. Phys. 1855, 170, 59–86. [Google Scholar] [CrossRef]
- Einstein, A. Über die von der molekularkinetischen Theorie der Wärme geforderte Bewegung von in ruhenden Flüssigkeiten suspendierten Teilchen. Ann. Phys. 1905, 322, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Schütz, G.J.; Baumgartner, W.; Gruber, H.J.; Schindler, H. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA 1996, 93, 2926–2929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, M. Single-particle tracking: The distribution of diffusion coefficients. Biophys. J. 1997, 72, 1744–1753. [Google Scholar] [CrossRef] [Green Version]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical Evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the Measurement of Nanoparticles and Protein Aggregates. Pharm. Res. 2010, 27, 796–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchetti, L.; Bonsignore, F.; Gobbo, F.; Amodeo, R.; Calvello, M.; Jacob, A.; Signore, G.; Schirripa Spagnolo, C.; Porciani, D.; Mainardi, M.; et al. Fast-diffusing p75NTR monomers support apoptosis and growth cone collapse by neurotrophin ligands. Proc. Natl. Acad. Sci. USA 2019, 116, 21563–21572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amodeo, R.; Nifosa, R.; Giacomelli, C.; Ravelli, C.; La Rosa, L.; Callegari, A.; Trincavelli, L.; Mitola, S.; Luin, S.; Marchetti, L. Molecular insight on the altered membrane trafficking of TrkA kinase dead mutants. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2019, 1867, 118614. [Google Scholar] [CrossRef]
- Savin, T.; Doyle, P.S. Static and Dynamic Errors in Particle Tracking Microrheology. Biophys. J. 2005, 88, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Vestergaard, C.L.; Blainey, P.C.; Flyvbjerg, H. Optimal estimation of diffusion coefficients from single-particle trajectories. Phys. Rev. E 2014, 89, 022726. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; Tauzin, L.J.; Baiyasi, R.; Wang, W.; Moringo, N.; Shuang, B.; Landes, C.F. Single Particle Tracking: From Theory to Biophysical Applications. Chem. Rev. 2017, 117, 7331–7376. [Google Scholar] [CrossRef]
- Walker, J.G. Improved nano-particle tracking analysis. Meas. Sci. Technol. 2012, 23, 065605. [Google Scholar] [CrossRef]
- Chenouard, N.; Smal, I.; De Chaumont, F.; Maska, M.; Sbalzarini, I.F.; Gon, Y.; Cardinale, J.; Carthel, C.; Coraluppi, S.; Winter, M.; et al. Objective comparison of particle tracking methods. Nat. Methods 2014, 11, 281–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oron, D.; Tal, E.; Silberberg, Y. Scanningless depth-resolved microscopy. Opt. Express 2005, 13, 1468–1476. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, C. Dual-color multiple-particle tracking at 50-nm localization and over 100-nm range in 3D with temporal focusing two-photon microscopy. Biomed. Opt. Express 2016, 7, 4187–4197. [Google Scholar] [CrossRef] [Green Version]
- Durso, W.; Martins, M.; Marchetti, L.; Cremisi, F.; Luin, S.; Cardarelli, F. Lysosome Dynamic Properties during Neuronal Stem Cell Differentiation Studied by Spatiotemporal Fluctuation Spectroscopy and Organelle Tracking. Int. J. Mol. Sci. 2020, 21, 3397. [Google Scholar] [CrossRef]
- Gregor, I.; Spiecker, M.; Petrovsky, R.; Großhans, J.; Ros, R.; Enderlein, J. Rapid nonlinear image scanning microscopy. Nat. Methods 2017, 14, 1087–1089. [Google Scholar] [CrossRef]
- Levi, V.; Ruan, Q.; Gratton, E. 3-D Particle Tracking in a Two-Photon Microscope: Application to the Study of Molecular Dynamics in Cells. Biophys. J. 2005, 88, 2919–2928. [Google Scholar] [CrossRef] [Green Version]
- Ferrand, P. GPScan.VI: A general-purpose LabVIEW program for scanning imaging or any application requiring synchronous analog voltage generation and data acquisition. Comput. Phys. Commun. 2015, 192, 342–347. [Google Scholar] [CrossRef] [Green Version]
- Squier, J.; Müller, M. High resolution nonlinear microscopy: A review of sources and methods for achieving optimal imaging. Rev. Sci. Instrum. 2001, 72, 2855–2867. [Google Scholar] [CrossRef]
- Freund, I.; Deutsch, M. Second-harmonic microscopy of biological tissue. Opt. Lett. 1986, 11, 94–96. [Google Scholar] [CrossRef]
- Franken, P.A.; Hill, A.E.; Peters, C.W.; Weinreich, G. Generation of Optical Harmonics. Phys. Rev. Lett. 1961, 7, 118–119. [Google Scholar] [CrossRef] [Green Version]
- Geissbuehler, M.; Bonacina, L.; Shcheslavskiy, V.; Bocchio, N.L.; Geissbuehler, S.; Leutenegger, M.; Märki, I.; Wolf, J.P.; Lasser, T. Nonlinear Correlation Spectroscopy (NLCS). Nano Lett. 2012, 12, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Mertz, J. Nonlinear microscopy: New techniques and applications. Curr. Opin. Neurobiol. 2004, 14, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Andersson, S.B. Simultaneous Localization and Parameter Estimation for Single Particle Tracking via Sigma Points based EM. In Proceedings of the 2019 IEEE 58th Conference on Decision and Control (CDC), Nice, France, 11–13 December 2019; pp. 6467–6472. [Google Scholar]
- Michalet, X.; Berglund, A.J. Optimal diffusion coefficient estimation in single-particle tracking. Phys. Rev. E 2012, 85, 061916. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef]
- Ernst, D.; Köhler, J. Measuring a diffusion coefficient by single-particle tracking: Statistical analysis of experimental mean squared displacement curves. Phys. Chem. Chem. Phys. 2013, 15, 845–849. [Google Scholar] [CrossRef]
- Kepten, E.; Bronshtein, I.; Garini, Y. Improved estimation of anomalous diffusion exponents in single-particle tracking experiments. Phys. Rev. E 2013, 87, 052713. [Google Scholar] [CrossRef] [Green Version]
- Berglund, A.J. Statistics of camera-based single-particle tracking. Phys. Rev. E 2010, 82, 011917. [Google Scholar] [CrossRef] [Green Version]
- Giloh, H.; Sedat, J. Fluorescence microscopy: Reduced photobleaching of rhodamine and fluorescein protein conjugates by n-propyl gallate. Science 1982, 217, 1252–1255. [Google Scholar] [CrossRef]
- Zipfel, W.R.; Williams, R.M.; Webb, W.W. Nonlinear magic: Multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.R. An Introduction to Error Analysis: The Study of Uncertainties in Physical Measurements. Meas. Sci. Technol. 1998, 9. [Google Scholar] [CrossRef]
- Michalet, X. Mean square displacement analysis of single-particle trajectories with localization error: Brownian motion in an isotropic medium. Phys. Rev. E 2010, 82, 041914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, T.; Lipinski, H.G.; Wiemann, M. Dark field nanoparticle tracking analysis for size characterization of plasmonic and non-plasmonic particles. J. Nanoparticle Res. 2014, 16, 2419. [Google Scholar] [CrossRef] [Green Version]
| Particle | Image Size | Dwell Time | Frame Rate | Scale |
|---|---|---|---|---|
| Radius (nm) | (pixels) | (s) | (fps) | (pix/m) |
| 250 | 100 × 100 | 5 | 20 | 2.53 |
| 110 | 100 × 100 | 3 | 33 | 3.81 |
| (A) r = 250 nm | MSD | <> | <> | |||||
|---|---|---|---|---|---|---|---|---|
| Mean radius (nm) | 411 | 252 | 308 | 253 | 308 | 269 | 270 | 418 |
| (nm) | 115 | 46 | 58 | 46 | 58 | 58 | 58 | 127 |
| Mean radius (nm) | 725 | 221 | 325 | 221 | 325 | 230 | 231 | 746 |
| (nm) | 439 | 41 | 69 | 42 | 68 | 51 | 51 | 507 |
| = (+)/2 (nm) | 568 | 237 | 316 | 237 | 316 | 250 | 250 | 582 |
| (B) r = 110 nm | MSD | <> | <> | |||||
| Mean radius (nm) | 143 | 101 | 116 | 101 | 116 | 105 | 106 | 160 |
| (nm) | 31 | 15 | 13 | 15 | 13 | 19 | 19 | 43 |
| Mean radius (nm) | 286 | 90 | 131 | 90 | 131 | 92 | 92 | 90 |
| (nm) | 220 | 12 | 25 | 12 | 25 | 14 | 15 | 12 |
| = (+)/2 (nm) | 214 | 96 | 124 | 96 | 123 | 99 | 99 | 125 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Travers, T.; Colin, V.G.; Loumaigne, M.; Barillé, R.; Gindre, D. Single-Particle Tracking with Scanning Non-Linear Microscopy. Nanomaterials 2020, 10, 1519. https://doi.org/10.3390/nano10081519
Travers T, Colin VG, Loumaigne M, Barillé R, Gindre D. Single-Particle Tracking with Scanning Non-Linear Microscopy. Nanomaterials. 2020; 10(8):1519. https://doi.org/10.3390/nano10081519
Chicago/Turabian StyleTravers, Théo, Vincent G. Colin, Matthieu Loumaigne, Régis Barillé, and Denis Gindre. 2020. "Single-Particle Tracking with Scanning Non-Linear Microscopy" Nanomaterials 10, no. 8: 1519. https://doi.org/10.3390/nano10081519





