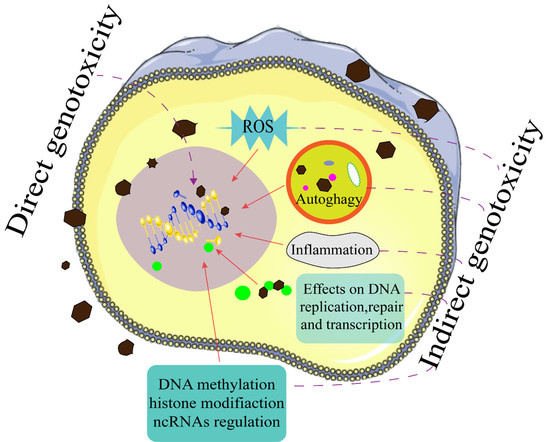
Direct and Indirect Genotoxicity of Graphene Family Nanomaterials on DNA—A Review
Abstract
:1. Introduction
| Products | Supplier or Synthesis Methods | Dose | Animal or Cell Models | Toxicological Mechanisms | Adverse Effects | Ref. |
|---|---|---|---|---|---|---|
| graphene nanoplatelets | cheaptubes.com (Brattleboro, VT, USA) | 0.3, 1 mg/rat | rat | oxidative stress, inflammation | lung inflammation | [55] |
| commercial GO and rGO | Nanjing XFNANO Materials Tech Co., Ltd., (China) | 2.0 mg/kg body weight | rat | transcriptional and epigenetic | liver zonated accumulation | [56] |
| amination GQDs carboxylated GQDs hydroxylated GQDs | Nanjing XFNANO Materials Tech Co., Ltd., (China) | 100, 200 μg/mL | A549 cells | autophagy | cytotoxicity | [57] |
| GO and rGO oxidated from carbon nanofibers | Grupo Antolin (Spain) | 0.1, 1.0, 10, 50 mg/L | erythrocyte cell | oxidative stress | genotoxicity | [58] |
| GO nanosheets | Sigma-Aldrich (St. Louis, MO, USA) | 40, 60, 80 mg/L | Human SH-SY5Y neuroblastoma cell | oxidative stress, autophagy–lysosomal network dysfunction | cytotoxicity | [59] |
| pristine rGO | Chengdu Organic Chemicals Co., Ltd., the Chinese Academy of Sciences | 1–100 mg/L | Earthworm coelomocytes | oxidative stress | immunotoxicity | [60] |
| single layer GO (product no. GNOP10A5) | ACS Materials LLC (Medford, MA, USA) | 1, 10, 50, 150, 250, 500 mg/L | Escherichia coli | physical destruction | toxicity against bacteria | [61] |
| GO | modified Hummers method | 25 mg/L | THP-1 and BEAS-2B cells | lipid peroxidation, membrane adsorption, membrane damage | cytotoxicity | [62] |
| GO | modified Hummers method | 2 mg/kg | rat | lipid peroxidation, membrane adsorption, membrane damage | acute lung inflammation | [62] |
| GO | Nanjing XFNANO Materials Tech Co., Ltd., (China) | 0–100 mg/L | zebrafish embryos | oxidative stress | developmental toxicity | [63] |
| GO | modified Hummers method | 10 mg/L | Caenorhabditis elegans | oxidative stress | toxicity | [64] |
| graphene, GO | modified Hummers method | 3.125–200 mg/L | human erythrocytes and skin fibroblasts | oxidative stress | cytotoxicity | [65] |
| graphene exfoliated form graphite, GO oxidated from carbon fibers | Grupo Antolin Ingeniería (Burgos, Spain) | 1, 10 mg/L | primary neurons | inhibition of synaptic transmission, altered calcium homeostasis | neurotoxicity | [66] |
2. Direct Genotoxicity of GFNs
2.1. Direct Physical Nucleus Damage by GFNs
2.2. Interaction Mechanisms between DNA and GFNs
3. Indirect Genotoxicity of GFNs
3.1. Oxidative Stress
3.2. Epigenetic Toxicity
3.3. The DNA Replication, Repair, and Transcription Affected by GFNs
3.4. Inflammation
3.5. Autophagy
4. Factors Influencing Genotoxicity of GFNs
4.1. Surface Properties
4.2. Size and Structure
4.3. Exposure Dose and Time
4.4. The Resistance of Cell Structures and Biological Barriers
5. Genotoxicity Testing of GFNs
5.1. Detection of GFNs in Cells and Organism Tissues
5.2. Genotoxicity Assay of GFNs
6. Conclusions, Challenges, and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [Green Version]
- Castro Neto, A.H.; Guinea, F.; Peres, N.M.R. Drawing conclusions from graphene. Phys. World 2006, 19, 33–37. [Google Scholar] [CrossRef]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of graphene-family nanoparticles: A general review of the origins and mechanisms. Part. Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Kong, Z.; Kang, Z.; Wang, H.; Zhang, L.; Shen, J.-W. Theoretical evaluation on potential cytotoxicity of graphene quantum dots. ACS Biomater. Sci. Eng. 2016, 2, 1983–1991. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qi, X.Y.; Boey, F.; Zhang, H. Graphene-based composites. Chem. Soc. Rev. 2012, 41, 666–686. [Google Scholar] [CrossRef] [PubMed]
- Lerf, A.; He, H.Y.; Forster, M.; Klinowski, J. Structure of graphite oxide revisited. J. Phys. Chem. B 1998, 102, 4477–4482. [Google Scholar] [CrossRef]
- Mattevi, C.; Eda, G.; Agnoli, S.; Miller, S.; Mkhoyan, K.A.; Celik, O.; Mastrogiovanni, D.; Granozzi, G.; Garfunkel, E.; Chhowalla, M. Evolution of electrical, chemical, and structural properties of transparent and conducting chemically derived graphene thin films. Adv. Funct. Mater. 2009, 19, 2577–2583. [Google Scholar] [CrossRef]
- Singh, V.; Joung, D.; Zhai, L.; Das, S.; Khondaker, S.I.; Seal, S. Graphene based materials: Past, present and future. Prog. Mater. Sci. 2011, 56, 1178–1271. [Google Scholar] [CrossRef]
- Yan, Y.; Gong, J.; Chen, J.; Zeng, Z.; Huang, W.; Pu, K.; Liu, J.; Chen, P. Recent advances on graphene quantum dots: From chemistry and physics to applications. Adv. Mater. 2019, 31, 1808283. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-T.; Song, W.-L.; Fan, L.-Z. Engineering graphene aerogels with porous carbon of large surface area for flexible all-solid-state supercapacitors. Electrochim. Acta 2015, 165, 92–97. [Google Scholar] [CrossRef]
- Kumar, R.; Sahoo, S.; Joanni, E.; Singh, R.K.; Tan, W.K.; Kar, K.K.; Matsuda, A. Recent progress in the synthesis of graphene and derived materials for next generation electrodes of high performance lithium ion batteries. Prog. Energy Combust. Sci. 2019, 75, 100786. [Google Scholar] [CrossRef]
- Li, G.; Huang, B.; Pan, Z.; Su, X.; Shao, Z.; An, L. Advances in three-dimensional graphene-based materials: Configurations, preparation and application in secondary metal (Li, Na, K, Mg, Al)-ion batteries. Energy Environ. Sci. 2019, 12, 2030–2053. [Google Scholar] [CrossRef]
- Li, Z.; Liu, X.; Wang, L.; Bu, F.; Wei, J.; Pan, D.; Wu, M. Hierarchical 3D all-carbon composite structure modified with N-doped graphene quantum dots for high-performance flexible supercapacitors. Small 2018, 14, 1801498. [Google Scholar] [CrossRef]
- Tsai, M.L.; Wei, W.-R.; Tang, L.; Chang, H.-C.; Tai, S.-H.; Yang, P.-K.; Lau, S.P.; Chen, L.-J.; He, J.-H. Si Hybrid Solar Cells with 13% Efficiency via concurrent improvement in optical and electrical properties by employing graphene quantum dots. ACS Nano 2016, 10, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Young, C.; Park, T.; Yi, J.W.; Kim, J.; Hossain, M.S.A.; Kaneti, Y.V.; Yamauchi, Y. Advanced functional carbons and their hybrid nanoarchitectures towards supercapacitor applications. Chemsuschem 2018, 11, 3546–3558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-Q.; Zhao, D.-D.; Tang, P.-Y.; Wang, Y.-M.; Xu, C.-L.; Li, H.-L. MnO2/graphene/nickel foam composite as high performance supercapacitor electrode via a facile electrochemical deposition strategy. Mater. Lett. 2012, 76, 127–130. [Google Scholar] [CrossRef]
- Zhou, S.; Lin, M.; Zhuang, Z.; Liu, P.; Chen, Z. Biosynthetic graphene enhanced extracellular electron transfer for high performance anode in microbial fuel cell. Chemosphere 2019, 232, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Afsahi, S.; Lerner, M.B.; Goldstein, J.M.; Lee, J.; Tang, X.; Bagarozzi, D.A., Jr.; Pan, D.; Locascio, L.; Walker, A.; Barron, F.; et al. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018, 100, 85–88. [Google Scholar] [CrossRef]
- Chen, H.; Wang, Z.; Zong, S.; Chen, P.; Zhu, D.; Wu, L.; Cui, Y. A graphene quantum dot-based FRET system for nuclear-targeted and real-time monitoring of drug delivery. Nanoscale 2015, 7, 15477–15486. [Google Scholar] [CrossRef] [PubMed]
- Iannazzo, D.; Pistone, A.; Ferro, S.; De Luca, L.; Monforte, A.M.; Romeo, R.; Buemi, M.R.; Pannecouque, C. Graphene quantum dots based systems as HIV inhibitors. Bioconjug. Chem. 2018, 29, 3084–3093. [Google Scholar] [CrossRef]
- Li, N.; Than, A.; Chen, J.; Xi, F.; Liu, J.; Chen, P. Graphene quantum dots based fluorescence turn-on nanoprobe for highly sensitive and selective imaging of hydrogen sulfide in living cells. Biomater. Sci. 2018, 6, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Than, A.; Sun, C.; Tian, J.; Chen, J.; Pu, K.; Dong, X.; Chen, P. Monitoring dynamic cellular redox homeostasis using fluorescence-switchable graphene quantum dots. ACS Nano 2016, 10, 11475–11482. [Google Scholar] [CrossRef]
- Li, N.; Than, A.; Wang, X.; Xu, S.; Sun, L.; Duan, H.; Xu, C.; Chen, P. Ultrasensitive profiling of metabolites using tyramine-functionalized graphene quantum dots. ACS Nano 2016, 10, 3622–3629. [Google Scholar] [CrossRef]
- Ma, N.; Liu, J.; He, W.; Li, Z.; Luan, Y.; Song, Y.; Garg, S. lFolic acid-grafted bovine serum albumin decorated graphene oxide: An efficient drug carrier for targeted cancer therapy. J. Colloid Interface Sci. 2017, 490, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Marzo, A.M.L.; Mayorga-Martinez, C.C.; Pumera, M. 3D-printed graphene direct electron transfer enzyme biosensors. Biosens. Bioelectron. 2020, 151, 111980. [Google Scholar] [CrossRef]
- Divyapriya, G.; Vijayakumar, K.K.; Nambi, I. Development of a novel graphene/Co3O4 composite for hybrid capacitive deionization system. Desalination 2019, 451, 102–110. [Google Scholar] [CrossRef]
- Gan, L.; Li, H.; Chen, L.; Xu, L.; Liu, J.; Geng, A.; Mei, C.; Shang, S. Graphene oxide incorporated alginate hydrogel beads for the removal of various organic dyes and bisphenol A in water. Colloid Polym. Sci. 2018, 296, 607–615. [Google Scholar] [CrossRef]
- Liu, S.; Ge, H.; Wang, C.; Zou, Y.; Liu, J. Agricultural waste/graphene oxide 3D bio-adsorbent for highly efficient removal of methylene blue from water pollution. Sci. Total Environ. 2018, 628–629, 959–968. [Google Scholar] [CrossRef]
- Miao, W.; Li, Z.-K.; Yan, X.; Guo, Y.-J.; Lang, W.-Z. Improved ultrafiltration performance and chlorine resistance of PVDF hollow fiber membranes via doping with sulfonated graphene oxide. Chem. Eng. J. 2017, 317, 901–912. [Google Scholar] [CrossRef]
- Zhang, D.; Zhao, Y.; Chen, L. Fabrication and characterization of amino-grafted graphene oxide modified ZnO with high photocatalytic activity. Appl. Surf. Sci. 2018, 458, 638–647. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, J.; Wang, X.; Huo, X.; Xu, Y.; Li, Q.; Wang, L. Improving the hydrophilic and antifouling properties of polyvinylidene fluoride membrane by incorporation of novel nanohybrid GO@SiO2 particles. Chem. Eng. J. 2017, 314, 266–276. [Google Scholar] [CrossRef]
- Afroj, S.; Tan, S.; Abdelkader, A.M.; Novoselov, K.S.; Karim, N. Highly conductive, scalable, and machine washable graphene-based e-textiles for multifunctional wearable electronic applications. Adv. Funct. Mater. 2020, 30, 2000293. [Google Scholar] [CrossRef] [Green Version]
- Kurra, N.; Jiang, Q.; Nayak, P.; Alshareef, H.N. Laser-derived graphene: A three-dimensional printed graphene electrode and its emerging applications. Nano Today 2019, 24, 81–102. [Google Scholar] [CrossRef]
- Liu, C.; Qiu, S.; Du, P.; Zhao, H.; Wang, L. An ionic liquid-graphene oxide hybrid nanomaterial: Synthesis and anticorrosive applications. Nanoscale 2018, 10, 8115–8124. [Google Scholar] [CrossRef]
- Ahmed, F.; Rodrigues, D.F. Investigation of acute effects of graphene oxide on wastewater microbial community: A case study. J. Hazard. Mater. 2013, 256, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Arvidsson, R.; Molander, S.; Sanden, B.A. Review of potential environmental and health risks of the nanomaterial graphene. Hum. Ecol. Risk Assess. 2013, 19, 873–887. [Google Scholar] [CrossRef]
- Lee, J.H.; Han, J.H.; Kim, J.H.; Kim, B.; Bello, D.; Kim, J.K.; Lee, G.H.; Sohn, E.K.; Lee, K.; Ahn, K.; et al. Exposure monitoring of graphene nanoplatelets manufacturing workplaces. Inhal. Toxicol. 2016, 28, 281–291. [Google Scholar] [CrossRef]
- Pelin, M.; Sosa, S.; Prato, M.; Tubaro, A. Occupational exposure to graphene based nanomaterials: Risk assessment. Nanoscale 2018, 10, 15894–15903. [Google Scholar] [CrossRef] [Green Version]
- Seabra, A.B.; Paula, A.J.; De Lima, R.; Alves, O.L.; Duran, N. Nanotoxicity of graphene and graphene oxide. Chem. Res. Toxicol. 2014, 27, 159–168. [Google Scholar] [CrossRef]
- Zhou, J.; Ouedraogo, M.; Qu, F.; Duez, P. Potential genotoxicity of traditional chinese medicinal plants and phytochemicals: An overview. Phytother. Res. 2013, 27, 1745–1755. [Google Scholar] [CrossRef]
- Bohne, J.; Cathomen, T. Genotoxicity in gene therapy: An account of vector integration and designer nucleases. Curr. Opin. Mol. Ther. 2008, 10, 214–223. [Google Scholar]
- Huang, R.; Zhou, Y.; Hu, S.; Zhou, P.-K. Targeting and non-targeting effects of nanomaterials on DNA: Challenges and perspectives. Rev. Environ. Sci. Biotechnol. 2019, 18, 617–634. [Google Scholar] [CrossRef]
- Burgum, M.J.; Clift, M.J.D.; Evans, S.J.; Hondow, N.; Tarat, A.; Jenkins, G.J.; Doak, S.H. Few-layer graphene induces both primary and secondary genotoxicity in epithelial barrier models in vitro. J. Nanobiotechnol. 2021, 19, 24. [Google Scholar] [CrossRef]
- De Souza, T.A.J.; Souza, L.R.R.; Franchi, L.P. Silver nanoparticles: An integrated view of green synthesis methods, transformation in the environment, and toxicity. Ecotoxicol. Environ. Saf. 2019, 171, 691–700. [Google Scholar] [CrossRef]
- Mortezaee, K.; Najafi, M.; Samadian, H.; Barabadi, H.; Azarnezhad, A.; Ahmadi, A. Redox interactions and genotoxicity of metal-based nanoparticles: A comprehensive review. Chem. Biol. Interact. 2019, 312, 108814. [Google Scholar] [CrossRef]
- Lam, P.L.; Wong, R.S.M.; Lam, K.H.; Hung, L.K.; Wong, M.M.; Yung, L.H.; Ho, Y.W.; Wong, W.Y.; Hau, D.K.P.; Gambari, R.; et al. The role of reactive oxygen species in the biological activity of antimicrobial agents: An updated mini review. Chem. Biol. Interact. 2020, 320, 109023. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, Y.; Jia, Y.; Liu, J. Heating Drives DNA to Hydrophobic regions while freezing drives DNA to hydrophilic regions of graphene oxide for highly robust biosensors. J. Am. Chem. Soc. 2020, 142, 14702–14709. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Gou, N.; Gao, C.; He, M.; Gu, A.Z. Comparative and mechanistic genotoxicity assessment of nanomaterials via a quantitative toxicogenomics approach across multiple species. Environ. Sci. Technol. 2014, 48, 12937–12945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavallo, D.; Fanizza, C.; Ursini, C.L.; Casciardi, S.; Paba, E.; Ciervo, A.; Fresegna, A.M.; Maiello, R.; Marcelloni, A.M.; Buresti, G.; et al. Multi-walled carbon nanotubes induce cytotoxicity and genotoxicity in human lung epithelial cells. J. Appl. Toxicol. 2012, 32, 454–464. [Google Scholar] [CrossRef]
- Kim, Y.J.; Rahman, M.M.; Lee, S.M.; Kim, J.M.; Park, K.; Kang, J.-H.; Seo, Y.R. Assessment of in vivo genotoxicity of citrated-coated silver nanoparticles via transcriptomic analysis of rabbit liver tissue. Int. J. Nanomed. 2019, 14, 393–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarosz, A.; Skoda, M.; Dudek, I.; Szukiewicz, D. Oxidative stress and mitochondrial activation as the main mechanisms underlying graphene toxicity against human cancer cells. Oxid. Med. Cell. Longev. 2016, 2016, 5851035. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Hu, X.; Zhou, Q. Envelopment-internalization synergistic effects and metabolic mechanisms of graphene oxide on single-cell chlorella vulgaris are dependent on the nanomaterial particle size. ACS Appl. Mater. Interfaces 2015, 7, 18104–18112. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lin, W.; Lin, B.; Wu, K.; Fan, H.; Yu, Y. Persistent DNA methylation changes in zebrafish following graphene quantum dots exposure in surface chemistry-dependent manner. Ecotoxicol. Environ. Saf. 2019, 169, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, Z.; White, J.C.; Xing, B. Graphene in the aquatic environment: Adsorption, dispersion, toxicity and transformation. Environ. Sci. Technol. 2014, 48, 9995–10009. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, A.Y.; Bae, J.; Seok, J.H.; Yang, J.-Y.; Roh, H.S.; Jeong, J.; Han, Y.; Jeong, J.; Cho, W.-S. The role of surface functionalization on the pulmonary inflammogenicity and translocation into mediastinal lymph nodes of graphene nanoplatelets in rats. Arch. Toxicol. 2017, 91, 667–676. [Google Scholar] [CrossRef]
- Wu, Y.; Feng, W.; Liu, R.; Xia, T.; Liu, S. Graphene oxide causes disordered zonation due to differential intralobular localization in the liver. ACS Nano 2020, 14, 877–890. [Google Scholar] [CrossRef]
- Xie, Y.; Wan, B.; Yang, Y.; Cui, X.; Xin, Y.; Guo, L.-H. Cytotoxicity and autophagy induction by graphene quantum dots with different functional groups. J. Environ. Sci. 2019, 77, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Evariste, L.; Lagier, L.; Gonzalez, P.; Mottier, A.; Mouchet, F.; Cadarsi, S.; Lonchambon, P.; Daffe, G.; Chimowa, G.; Sarrieu, C.; et al. Thermal reduction of graphene oxide mitigates its in vivo genotoxicity toward xenopus laevis tadpoles. Nanomaterials 2019, 9, 584. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.L.; Zhang, Y.Q.; Luo, R.H.; Lai, X.; Chen, A.J.; Zhang, Y.L.; Hu, C.; Chen, L.L.; Shao, L.Q. Graphene oxide disrupted mitochondrial homeostasis through inducing intracellular redox deviation and autophagy-lysosomal network dysfunction in SH-SY5Y cells. J. Hazard. Mater. 2021, 416, 126158. [Google Scholar]
- Xu, K.; Wang, X.; Lu, C.; Liu, Y.; Zhang, D.; Cheng, J. Toxicity of three carbon-based nanomaterials to earthworms: Effect of morphology on biomarkers, cytotoxicity, and metabolomics. Sci. Total Environ. 2021, 777, 146224. [Google Scholar] [CrossRef]
- Barrios, A.C.; Wang, Y.; Gilbertson, L.M.; Perreault, F. Structure-property-toxicity relationships of graphene oxide: Role of surface chemistry on the mechanisms of interaction with bacteria. Environ. Sci. Technol. 2019, 53, 14679–14687. [Google Scholar] [CrossRef]
- Li, R.; Guiney, L.M.; Chang, C.H.; Mansukhani, N.D.; Ji, Z.; Wang, X.; Liao, Y.-P.; Jiang, W.; Sun, B.; Hersam, M.C.; et al. Surface oxidation of graphene oxide determines membrane damage, lipid peroxidation, and cytotoxicity in macrophages in a pulmonary toxicity model. ACS Nano 2018, 12, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ren, C.; Ouyang, S.; Hu, X.; Zhou, Q. Mitigation in multiple effects of graphene oxide toxicity in zebrafish embryogenesis driven by humic acid. Environ. Sci. Technol. 2015, 49, 10147–10154. [Google Scholar] [CrossRef]
- Ding, X.; Rui, Q.; Zhao, Y.; Shao, H.; Yin, Y.; Wu, Q.; Wang, D. Toxicity of graphene oxide in nematodes with a deficit in the epidermal barrier caused by RNA interference knockdown of unc-52. Environ. Sci. Technol. Lett. 2018, 5, 622–628. [Google Scholar] [CrossRef]
- Liao, K.-H.; Lin, Y.-S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl. Mater. Interfaces 2011, 3, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
- Bramini, M.; Sacchetti, S.; Armirotti, A.; Rocchi, A.; Vazquez, E.; Leon Castellanos, V.; Bandiera, T.; Cesca, F.; Benfenati, F. Graphene oxide nanosheets disrupt lipid composition, Ca2+ homeostasis, and synaptic transmission in primary cortical neurons. ACS Nano 2016, 10, 7154–7171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Y.; Lv, M.; Xiu, P.; Huynh, T.; Zhang, M.; Castelli, M.; Liu, Z.; Huang, Q.; Fan, C.; Fang, H.; et al. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Lammel, T.; Boisseaux, P.; Fernandez-Cruz, M.-L.; Navas, J.M. Internalization and cytotoxicity of graphene oxide and carboxyl graphene nanoplatelets in the human hepatocellular carcinoma cell line Hep G2. Part. Fibre Toxicol. 2013, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Dai, Y.; Wang, Z.; Zhao, J.; Li, F.; White, J.C.; Xing, B. Graphene quantum dots in alveolar macrophage: Uptake-exocytosis, accumulation in nuclei, nuclear responses and DNA cleavage. Part. Fibre Toxicol. 2018, 15, 45. [Google Scholar] [CrossRef]
- Mohammed, H.; Kumar, A.; Bekyarova, E.; Al-Hadeethi, Y.; Zhang, X.; Chen, M.; Ansari, M.S.; Cochis, A.; Rimondini, L. Antimicrobial mechanisms and effectiveness of graphene and graphene-functionalized biomaterials. A scope review. Front. Bioeng. Biotechnol. 2020, 8, 465. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Akhavan, A. Size-dependent genotoxicity of graphene nanoplatelets in human stem cells. Biomaterials 2012, 33, 8017–8025. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E.; Emamy, H.; Akhavan, F. Genotoxicity of graphene nanoribbons in human mesenchymal stem cells. Carbon 2013, 54, 419–431. [Google Scholar] [CrossRef]
- Liu, B.; Salgado, S.; Maheshwari, V.; Liu, J. DNA adsorbed on graphene and graphene oxide: Fundamental interactions, desorption and applications. Curr. Opin. Colloid Interface Sci. 2016, 26, 41–49. [Google Scholar] [CrossRef]
- Kong, Z.; Hu, W.; Jiao, F.; Zhang, P.; Shen, J.; Cui, B.; Wang, H.; Liang, L. Theoretical evaluation of DNA genotoxicity of graphene quantum dots: A combination of density functional theory and molecular dynamics simulations. J. Phys. Chem. B 2020, 124, 9335–9342. [Google Scholar] [CrossRef]
- Li, B.; Zhang, Y.; Meng, X.-Y.; Zhou, R. Zipper-Like unfolding of dsDNA caused by graphene wrinkles. J. Phys. Chem. C 2020, 124, 3332–3340. [Google Scholar] [CrossRef]
- Ren, H.; Wang, C.; Zhang, J.; Zhou, X.; Xu, D.; Zheng, J.; Guo, S.; Zhang, J. DNA cleavage system of nanosized graphene oxide sheets and copper ions. ACS Nano 2010, 4, 7169–7174. [Google Scholar] [CrossRef]
- Liu, J. Adsorption of DNA onto gold nanoparticles and graphene oxide: Surface science and applications. Phys. Chem. Chem. Phys. 2012, 14, 10485–10496. [Google Scholar] [CrossRef] [Green Version]
- Flores-Lopez, L.Z.; Espinoza-Gomez, H.; Somanathan, R. Silver nanoparticles: Electron transfer, reactive oxygen species, oxidative stress, beneficial and toxicological effects. mini review. J. Appl. Toxicol. 2019, 39, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Toyokuni, S. Oxidative stress and cancer: The role of redox regulation. Biotherapy 1998, 11, 147–154. [Google Scholar] [CrossRef]
- Singh, N.; Manshian, B.; Jenkins, G.J.S.; Griffiths, S.M.; Williams, P.M.; Maffeis, T.G.G.; Wright, C.J.; Doak, S.H. NanoGenotoxicology: The DNA damaging potential of engineered nanomaterials. Biomaterials 2009, 30, 3891–3914. [Google Scholar] [CrossRef]
- Ou, L.; Lv, X.; Wu, Z.; Xia, W.; Huang, Y.; Chen, L.; Sun, W.; Qi, Y.; Yang, M.; Qi, L. Oxygen content-related DNA damage of graphene oxide on human retinal pigment epithelium cells. J. Mater. Sci. Mater. Med. 2021, 32, 20. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, S.S.; Bengtson, S.; Williams, A.; Jacobsen, N.R.; Troelsen, J.T.; Halappanavar, S.; Vogel, U. A transcriptomic overview of lung and liver changes one day after pulmonary exposure to graphene and graphene oxide. Toxicol. Appl. Pharmacol. 2021, 410, 115343. [Google Scholar] [CrossRef] [PubMed]
- Burgum, M.J.; Clift, M.J.D.; Evans, S.J.; Hondow, N.; Miller, M.; Lopez, S.B.; Williams, A.; Tarat, A.; Jenkins, G.J.; Doak, S.H. In vitro primary-indirect genotoxicity in bronchial epithelial cells promoted by industrially relevant few-layer graphene. Small 2021, 17, 2002551. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, Y.; Duo, L. Biochemical toxicity, lysosomal membrane stability and DNA damage induced by graphene oxide in earthworms. Environ. Pollut. 2021, 269, 116225. [Google Scholar] [CrossRef]
- Wu, T.; Liang, X.; Liu, X.; Li, Y.; Wang, Y.; Kong, L.; Tang, M. Induction of ferroptosis in response to graphene quantum dots through mitochondrial oxidative stress in microglia. Part. Fibre Toxicol. 2020, 17, 30. [Google Scholar] [CrossRef]
- Wang, A.; Pu, K.; Dong, B.; Liu, Y.; Zhang, L.; Zhang, Z.; Duan, W.; Zhu, Y. Role of surface charge and oxidative stress in cytotoxicity and genotoxicity of graphene oxide towards human lung fibroblast cells. J. Appl. Toxicol. 2013, 33, 1156–1164. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhou, Q.; Zeng, H.; Wang, Y.; Hu, X. Natural nanocolloids mediate the phytotoxicity of graphene oxide. Environ. Sci. Technol. 2020, 54, 4865–4875. [Google Scholar] [CrossRef]
- Feng, X.; Chen, Q.; Guo, W.; Zhang, Y.; Hu, C.; Wu, J.; Shao, L. Toxicology data of graphene-family nanomaterials: An update. Arch. Toxicol. 2020, 94, 1915–1939. [Google Scholar]
- Sun, Y.; Dai, H.; Chen, S.; Xu, M.; Wang, X.; Zhang, Y.; Xu, S.; Xu, A.; Weng, J.; Liu, S.; et al. Graphene oxide regulates Cox2 in human embryonic kidney 293T cells via epigenetic mechanisms: Dynamic chromosomal interactions. Nanotoxicology 2018, 12, 117–137. [Google Scholar] [CrossRef] [PubMed]
- Dusinska, M.; Tulinska, J.; El Yamani, N.; Kuricova, M.; Liskova, A.; Rollerova, E.; Runden-Pran, E.; Smolkova, B. Immunotoxicity, genotoxicity and epigenetic toxicity of nanomaterials: New strategies for toxicity testing? Food Chem. Toxicol. 2017, 109, 797–811. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.; Hao, F.; Yang, X.; Rao, Z.; Liu, Q.S.; Sang, N.; Faiola, F.; Zhou, Q.; Jiang, G. Graphene quantum dots disrupt embryonic stem cell differentiation by interfering with the methylation level of Sox2. Environ. Sci. Technol. 2021, 55, 3144–3155. [Google Scholar] [CrossRef]
- Pogribna, M.; Hammons, G. Epigenetic effects of nanomaterials and nanoparticles. J. Nanobiotechnol. 2021, 19, 2. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Cai, H.-Q.; Wang, J.-L.; Mesalam, A.; Reza, A.M.M.T.; Li, L.; Chen, L.; Qian, C. Graphene oxide-silver nanoparticle nanocomposites induce oxidative stress and aberrant methylation in caprine fetal fibroblast cells. Cells 2021, 10, 682. [Google Scholar] [CrossRef]
- Flasz, B.; Dziewiecka, M.; Kedziorski, A.; Tarnawska, M.; Augustyniak, M. Multigenerational graphene oxide intoxication results in reproduction disorders at the molecular level of vitellogenin protein expression in Acheta domesticus. Chemosphere 2021, 280, 130772. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, Q.; Wang, D. An epigenetic signal encoded protection mechanism is activated by graphene oxide to inhibit its induced reproductive toxicity in Caenorhabditis elegans. Biomaterials 2016, 79, 15–24. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Zhao, Y.; Bai, Y.; Chen, P.; Xia, T.; Wang, D. Response of microRNAs to in vitro treatment with graphene oxide. ACS Nano 2014, 8, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, A.; Swaroop, S.; Chandran, P.; Nair, S.; Koyakutty, M. Cellular and molecular mechanistic insight into the DNA-damaging potential of few-layer graphene in human primary endothelial cells. Nanomedicine 2016, 12, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Magdolenova, Z.; Collins, A.; Kumar, A.; Dhawan, A.; Stone, V.; Dusinska, M. Mechanisms of genotoxicity. A review of in vitro and in vivo studies with engineered nanoparticles. Nanotoxicology 2014, 8, 233–278. [Google Scholar] [CrossRef]
- Hashemi, E.; Akhavan, O.; Shamsara, M.; Majd, S.A.; Sanati, M.H.; Joupari, M.D.; Farmany, A. Graphene oxide negatively regulates cell cycle in embryonic fibroblast cells. Int. J. Nanomed. 2020, 15, 6201–6209. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhao, J.; Wang, Z. Genotoxic response and damage recovery of macrophages to graphene quantum dots. Sci. Total Environ. 2019, 664, 536–545. [Google Scholar] [CrossRef]
- Chatterjee, N.; Yang, J.; Choi, J. Differential genotoxic and epigenotoxic effects of graphene family nanomaterials (GFNs) in human bronchial epithelial cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 798, 1–10. [Google Scholar] [CrossRef]
- Keshavan, S.; Calligari, P.; Stella, L.; Fusco, L.; Delogu, L.G.; Fadeel, B. Nano-bio interactions: A neutrophil-centric view. Cell Death Dis. 2019, 10, 569. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, A.; Appleton, I.; Moore, A.R.; Gilroy, D.W.; Willis, D.; Mitchell, J.A.; Willoughby, D.A. Cyclo-oxygenase and nitric oxide synthase isoforms in rat carrageenin-induced pleurisy. Br. J. Pharmacol. 1994, 113, 693–698. [Google Scholar] [CrossRef] [Green Version]
- Khanna, P.; Ong, C.; Bay, B.H.; Baeg, G.H. Nanotoxicity: An interplay of oxidative stress, inflammation and cell death. Nanomaterials 2015, 5, 1163–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, S.J.; Clift, M.J.D.; Singh, N.; De Oliveira Mallia, J.; Burgum, M.; Wills, J.W.; Wilkinson, T.S.; Jenkins, G.J.S.; Doak, S.H. Critical review of the current and future challenges associated with advanced in vitro systems towards the study of nanoparticle (secondary) genotoxicity. Mutagenesis 2017, 32, 233–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domenech, J.; Hernandez, A.; Demir, E.; Marcos, R.; Cortes, C. Interactions of graphene oxide and graphene nanoplatelets with the in vitro Caco-2/HT29 model of intestinal barrier. Sci. Rep. 2020, 10, 2793. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, K.; Li, W.; Yang, N.; Liu, Y.; Chen, C.; Wei, T. The interactions between pristine graphene and macrophages and the production of cytokines/chemokines via TLR- and NF-kappa B-related signaling pathways. Biomaterials 2012, 33, 6933–6942. [Google Scholar] [CrossRef]
- Capasso, L.; Camatini, M.; Gualtieri, M. Nickel oxide nanoparticles induce inflammation and genotoxic effect in lung epithelial cells. Toxicol. Lett. 2014, 226, 28–34. [Google Scholar] [CrossRef]
- Mohammadinejad, R.; Moosavi, M.A.; Tavakol, S.; Vardar, D.O.; Hosseini, A.; Rahmati, M.; Dini, L.; Hussain, S.; Mandegary, A.; Klionsky, D.J. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 2019, 15, 4–33. [Google Scholar] [CrossRef] [Green Version]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science. 2010, 330, 1344–1348. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, G.; Korolchuk, V.I. Repair, reuse, recycle: The expanding role of autophagy in genome maintenance. Trends Cell Biol. 2017, 27, 340–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, Y.; Kanki, T.; Aoki, Y.; Hirota, Y.; Saigusa, T.; Uchiumi, T.; Kang, D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 2012, 287, 3265–3272. [Google Scholar] [CrossRef] [Green Version]
- Wan, B.; Wang, Z.-X.; Lv, Q.-Y.; Dong, P.-X.; Zhao, L.-X.; Yang, Y.; Guo, L.-H. Single-walled carbon nanotubes and graphene oxides induce autophagosome accumulation and lysosome impairment in primarily cultured murine peritoneal macrophages. Toxicol. Lett. 2013, 221, 118–127. [Google Scholar] [CrossRef]
- Feng, X.; Zhang, Y.; Zhang, C.; Lai, X.; Zhang, Y.; Wu, J.; Hu, C.; Shao, L. Nanomaterial-mediated autophagy: Coexisting hazard and health benefits in biomedicine. Part. Fibre Toxicol. 2020, 17, 53. [Google Scholar] [CrossRef]
- Li, X.; Liu, H.; Yu, Y.; Ma, L.; Liu, C.; Miao, L. Graphene oxide quantum dots-induced mineralization via the reactive oxygen species-dependent autophagy pathway in dental pulp stem cells. J. Biomed. Nanotechnol. 2020, 16, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Eapen, V.V.; Waterman, D.P.; Lemos, B.; Haber, J.E. Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Elsevier: Amsterdam, The Netherlands, 2017; pp. 213–236. [Google Scholar]
- Szabo, P.; Zelko, R. Formulation and stability aspects of nanosized solid drug delivery systems. Curr. Pharm. Des. 2015, 21, 3148–3157. [Google Scholar] [CrossRef]
- Ema, M.; Gamo, M.; Honda, K. A review of toxicity studies on graphene-based nanomaterials in laboratory animals. Regul. Toxicol. Pharmacol. 2017, 85, 7–24. [Google Scholar] [CrossRef]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiello, J.; Kozinski, R.; Wierzbicki, M.; Grodzik, M.; Lipinska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar]
- Krasteva, N.; Keremidarska-Markova, M.; Hristova-Panusheva, K.; Andreeva, T.; Speranza, G.; Wang, D.Y.; Draganova-Filipova, M.; Miloshev, G.; Georgieva, M. Aminated graphene oxide as a potential new therapy for colorectal cancer. Oxid. Med. Cell. Longev. 2019, 2019, 3738980. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, E.; Akhavan, O.; Shamsara, M.; Rahighi, R.; Esfandiar, A.; Tayefeh, A.R. Cyto and genotoxicities of graphene oxide and reduced graphene oxide sheets on spermatozoa. RSC Adv. 2014, 4, 27213–27223. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, H.M.; Chen, S.; Yu, H.T.; Quan, X. Salt-controlled assembly of stacked-graphene for capturing fluorescence and its application in chemical genotoxicity screening. J. Mater. Chem. 2011, 21, 15266–15272. [Google Scholar] [CrossRef]
- Chatterjee, N.; Eom, H.-J.; Choi, J. A systems toxicology approach to the surface functionality control of graphene-cell interactions. Biomaterials 2014, 35, 1109–1127. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, A.; Panchakarla, L.S.; Chandran, P.; Menon, D.; Nair, S.; Rao, C.N.R.; Koyakutty, M. Differential nano-bio interactions and toxicity effects of pristine versus functionalized graphene. Nanoscale 2011, 3, 2461–2464. [Google Scholar] [CrossRef]
- Franqui, L.S.; De Farias, M.A.; Portugal, R.V.; Costa, C.A.R.; Domingues, R.R.; Souza, A.G.; Coluci, V.R.; Leme, A.F.P.; Martinez, D.S.T. Interaction of graphene oxide with cell culture medium: Evaluating the fetal bovine serum protein corona formation towards in vitro nanotoxicity assessment and nanobiointeractions. Mater. Sci. Eng. C 2019, 100, 363–377. [Google Scholar] [CrossRef]
- Borandeh, S.; Alimardani, V.; Abolmaali, S.S.; Seppala, J. Graphene family nanomaterials in ocular applications: Physicochemical properties and toxicity. Chem. Res. Toxicol. 2021, 34, 1386–1402. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Mei, K.-C.; Chang, C.H.; Jiang, J.; Liu, X.; Liu, Q.; Guiney, L.M.; Hersam, M.C.; Liao, Y.-P.; et al. Lateral size of graphene oxide determines differential cellular uptake and cell death pathways in Kupffer cells, LSECs, and hepatocytes. Nano Today 2021, 37, 101061. [Google Scholar] [CrossRef]
- Reina, G.; Ngoc Do Quyen, C.; Nishina, Y.; Bianco, A. Graphene oxide size and oxidation degree govern its supramolecular interactions with siRNA. Nanoscale 2018, 10, 5965–5974. [Google Scholar] [CrossRef] [Green Version]
- Syama, S.; Mohanan, P.V. Safety and biocompatibility of graphene: A new generation nanomaterial for biomedical application. Int. J. Biol. Macromol. 2016, 86, 546–555. [Google Scholar] [CrossRef]
- Volkov, Y.; McIntyre, J.; Prina-Mello, A. Graphene toxicity as a double-edged sword of risks and exploitable opportunities: A critical analysis of the most recent trends and developments. 2D Mater. 2017, 4, 022001. [Google Scholar] [CrossRef]
- Yao, J.; Wang, H.; Chen, M.; Yang, M. Recent advances in graphene-based nanomaterials: Properties, toxicity and applications in chemistry, biology and medicine. Microchim. Acta 2019, 186, 395. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef]
- Hu, Q.; Jiao, B.; Shi, X.; Valle, R.P.; Zuo, Y.Y.; Hu, G. Effects of graphene oxide nanosheets on the ultrastructure and biophysical properties of the pulmonary surfactant film. Nanoscale 2015, 7, 18025–18029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, G.A.; Tahiliani, S.; Neu-Baker, N.M.; Brenner, S.A. Hyperspectral microscopy as an analytical tool for nanomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, K.L.; Goreham, R.V.; Nann, T. Graphene quantum dots for theranostics and bioimaging. Pharm. Res. 2016, 33, 2337–2357. [Google Scholar] [CrossRef]
- Wu, C.; Wang, C.; Han, T.; Zhou, X.; Guo, S.; Zhang, J. Insight into the cellular internalization and cytotoxicity of graphene quantum dots. Adv. Healthc. Mater. 2013, 2, 1613–1619. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, J.; Xue, Q.; Yin, X.; Yin, X.; Li, C.; Wang, Y. Acute and subacute toxicity study of graphene-based tumor cell nucleus-targeting fluorescent nanoprobes. Mol. Pharm. 2020, 17, 2682–2690. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xiong, C.; Liu, H.; Wan, Q.; Hou, J.; He, Q.; Badu-Tawiah, A.; Nie, Z. Mass spectrometry imaging reveals the sub-organ distribution of carbon nanomaterials. Nat. Nanotechnol. 2015, 10, 176–182. [Google Scholar] [CrossRef]
- Cardell, C.; Guerra, I. An overview of emerging hyphenated SEM-EDX and Raman spectroscopy systems: Applications in life, environmental and materials sciences. TrAC Trends Anal. Chem. 2016, 77, 156–166. [Google Scholar] [CrossRef]
- Ganguly, P.; Breen, A.; Pillai, S.C. Toxicity of nanomaterials: Exposure, pathways, assessment, and recent advances. ACS Biomater. Sci. Eng. 2018, 4, 2237–2275. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, K.; Zhou, Q.; Ouyang, S. Direct and Indirect Genotoxicity of Graphene Family Nanomaterials on DNA—A Review. Nanomaterials 2021, 11, 2889. https://doi.org/10.3390/nano11112889
Wu K, Zhou Q, Ouyang S. Direct and Indirect Genotoxicity of Graphene Family Nanomaterials on DNA—A Review. Nanomaterials. 2021; 11(11):2889. https://doi.org/10.3390/nano11112889
Chicago/Turabian StyleWu, Kangying, Qixing Zhou, and Shaohu Ouyang. 2021. "Direct and Indirect Genotoxicity of Graphene Family Nanomaterials on DNA—A Review" Nanomaterials 11, no. 11: 2889. https://doi.org/10.3390/nano11112889
APA StyleWu, K., Zhou, Q., & Ouyang, S. (2021). Direct and Indirect Genotoxicity of Graphene Family Nanomaterials on DNA—A Review. Nanomaterials, 11(11), 2889. https://doi.org/10.3390/nano11112889