Development and Characterization of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers
Abstract
:1. Introduction
2. Experimental Section
2.1. Reagents and Materials
2.2. Instrumentation
2.3. Computational Methods
2.4. Synthesis of MNPs
2.5. Synthesis of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers
2.6. Binding Studies
2.7. Selectivity Studies
3. Results and Discussion
3.1. Computational Screening
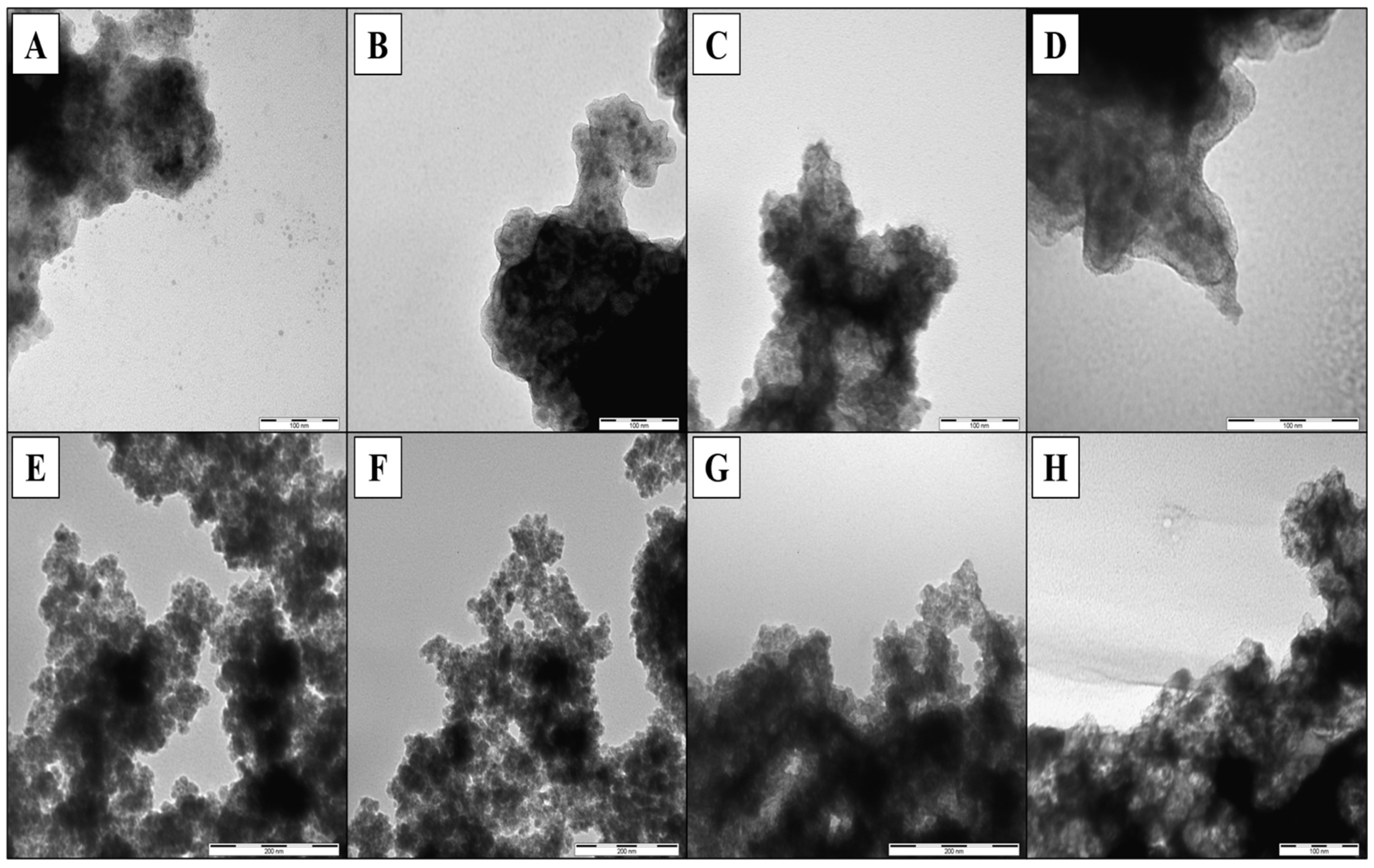
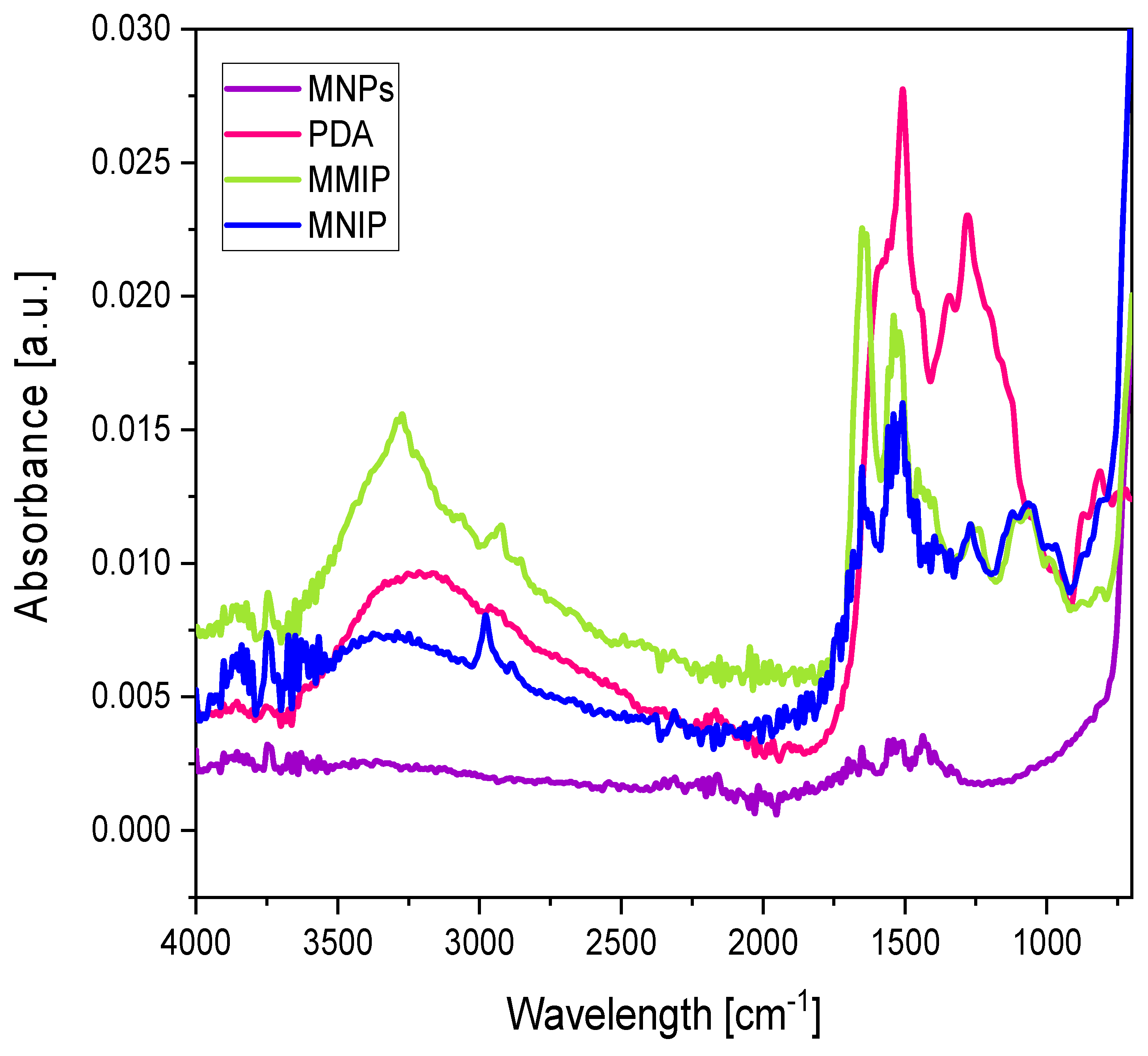
3.2. Synthesis and Characterization of Magnetic Epitope-Imprinted Polymers
3.3. Selectivity Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bedford, J.; Enria, D.; Giesecke, J.; Heymann, D.L.; Ihekweazu, C.; Kobinger, G.; Lane, H.C.; Memish, Z.; Oh, M.D.; Schuchat, A. COVID-19: Towards controlling of a pandemic. Lancet 2020, 395, 1015–1018. [Google Scholar] [CrossRef]
- Kumar, S.U.; Priya, N.M.; Nithya, S.R.; Kannan, P.; Jain, N.; Kumar, D.T.; Magesh, R.; Younes, S.; Zayed, H.; Doss, C.G.P. A review of novel coronavirus disease (COVID-19): Based on genomic structure, phylogeny, current shreds of evidence, candidate vaccines, and drug repurposing. 3 Biotech 2021, 11, 1–22. [Google Scholar] [CrossRef]
- Halpin, S.; O’Connor, R.; Sivan, M. Long COVID and chronic COVID syndromes. J. Med Virol. 2021, 93, 1242–1243. [Google Scholar] [CrossRef]
- Zhao, D.; Yao, F.; Wang, L.; Zheng, L.; Gao, Y.; Ye, J.; Guo, F.; Zhao, H.; Gao, R. A Comparative Study on the Clinical Features of Coronavirus 2019 (COVID-19) Pneumonia with Other Pneumonias. Clin. Infect. Dis. 2020, 71, 756–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadian, E.; Khatibi, S.M.H.; Soofiyani, S.R.; Abediazar, S.; Shoja, M.M.; Ardalan, M.; Vahed, S.Z. COVID-19 and kidney injury: Pathophysiology and molecular mechanisms. Rev. Med Virol. 2021, 31, e2176. [Google Scholar] [CrossRef]
- Cennamo, N.; D’Agostino, G.; Perri, C.; Arcadio, F.; Chiaretti, G.; Parisio, E.M.; Camarlinghi, G.; Vettori, C.; di Marzo, F.; Cennamo, R. Proof of concept for a quick and highly sensitive on-site detection of SARS-CoV-2 by plasmonic optical fibers and molecularly imprinted polymers. Sensors 2021, 21, 1681. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Khan, S.A.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Giri, B.; Pandey, S.; Shrestha, R.; Pokharel, K.; Ligler, F.S.; Neupane, B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2020, 413, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Saviñon-Flores, F.; Méndez, E.; López-Castaños, M.; Carabarin-Lima, A.; López-Castaños, K.; González-Fuentes, M.; Méndez-Albores, A. A Review on SERS-Based Detection of Human Virus Infections: Influenza and Coronavirus. Biosensors 2021, 11, 66. [Google Scholar] [CrossRef]
- Ehsan, M.; Khan, S.; Rehman, A. Screen-Printed Graphene/Carbon Electrodes on Paper Substrates as Impedance Sensors for Detection of Coronavirus in Nasopharyngeal Fluid Samples. Diagnostics 2021, 11, 1030. [Google Scholar] [CrossRef]
- Soufi, G.J.; Iravani, S.; Varma, R.S. Molecularly imprinted polymers for the detection of viruses: Challenges and opportunities. Analyst 2021, 146, 3087–3100. [Google Scholar] [CrossRef] [PubMed]
- BelBruno, J.J. Molecularly imprinted polymers. Chem. Rev. 2018, 119, 94–119. [Google Scholar] [CrossRef]
- Turiel, E.; Martín-Esteban, A. Molecularly imprinted polymers-based microextraction techniques. TrAC Trends Anal. Chem. 2019, 118, 574–586. [Google Scholar] [CrossRef]
- Vasapollo, G.; Sole, R.D.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly imprinted polymers: Present and future prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef] [Green Version]
- Fresco-Cala, B.; Mizaikoff, B. Surrogate Imprinting Strategies: Molecular Imprints via Fragments and Dummies. ACS Appl. Polym. Mater. 2020, 2, 3714–3741. [Google Scholar] [CrossRef]
- Boroznjak, R.; Reut, J.; Tretjakov, A.; Lomaka, A.; Öpik, A.; Syritski, V. A computational approach to study functional monomer-protein molecular interactions to optimize protein molecular imprinting. J. Mol. Recognit. 2017, 30, e2635. [Google Scholar] [CrossRef]
- Busato, M.; Distefano, R.; Bates, F.; Karim, K.; Bossi, A.M.; Vilariño, J.M.L.; Piletsky, S.; Bombieri, N.; Giorgetti, A. MIRATE: MIps RATional dEsign Science Gateway. J. Integr. Bioinform. 2018, 15, 20170075. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.V.; Dennison, S.R.; Archontis, G.; Reddy, S.M.; Hayes, J.M. Toward Rational Design of Selective Molecularly Imprinted Polymers (MIPs) for Proteins: Computational and Experimental Studies of Acrylamide Based Polymers for Myoglobin. J. Phys. Chem. B 2019, 123, 5432–5443. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Qu, P.; Zhang, C.; Li, M.; Ma, W.; Xiong, P.; Liu, Q.; Zou, G.; Lavillette, D.; Yin, F.; Jin, X.; et al. A new class of broadly neutralizing antibodies that target the glycan loop of Zika virus envelope protein. Cell Discov. 2020, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Van Bay, M.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock Vina Adopts More Accurate Binding Poses but Autodock4 Forms Better Binding Affinity. J. Chem. Inf. Model. 2019, 60, 204–211. [Google Scholar] [CrossRef]
- Gálvez-Vergara, A.; Fresco-Cala, B.; Cárdenas, S. Switchable Pickering emulsions stabilized by polystyrene-modified magnetic nanoparticles. Colloids Surf. A Physicochem. Eng. Asp. 2020, 606, 125462. [Google Scholar] [CrossRef]
- Fresco-Cala, B.M.; Cárdenas, S. Facile preparation of carbon nanotube-based molecularly imprinted monolithic stirred unit. Anal. Bioanal. Chem. 2020, 412, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, E.; Mosbach, K.; Haupt, K. Influence of functional and cross-linking monomers and the amount of template on the performance of molecularly imprinted polymers in binding assays. Anal. Commun. 1999, 36, 167–170. [Google Scholar] [CrossRef]
- Cheng, W.; Fan, F.; Zhang, Y.; Pei, Z.; Wang, W.; Pei, Y. A Facile Approach for Fabrication of Core-Shell Magnetic Molecularly Imprinted Nanospheres towards Hypericin. Polymers 2017, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Gu, C.; Zheng, W.; Dai, F.; Wang, X.; Zheng, Z. Facile synthesis of novel size-controlled antibacterial hybrid spheres using silver nanoparticles loaded with poly-dopamine spheres. RSC Adv. 2015, 5, 13470–13477. [Google Scholar] [CrossRef]
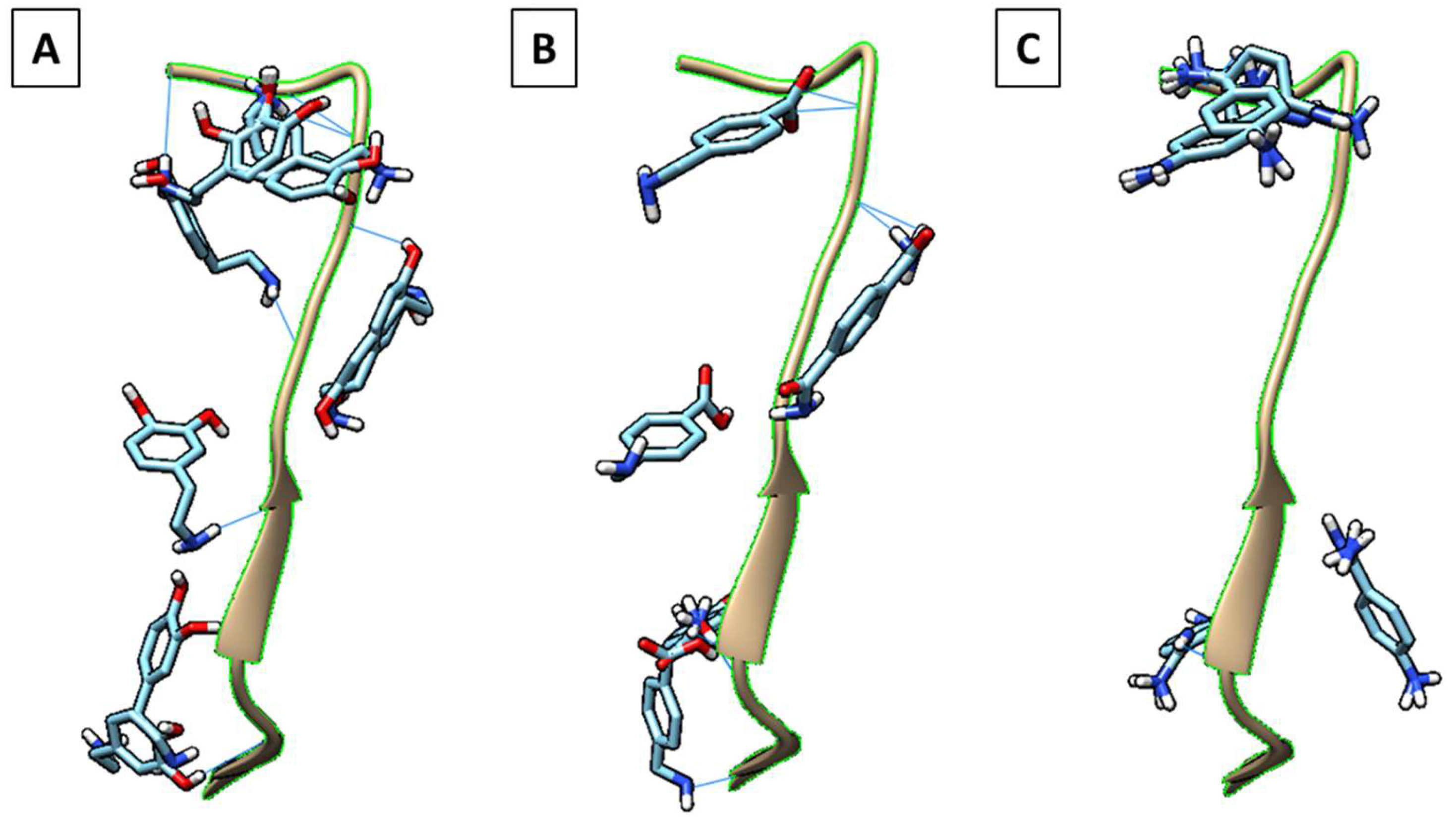
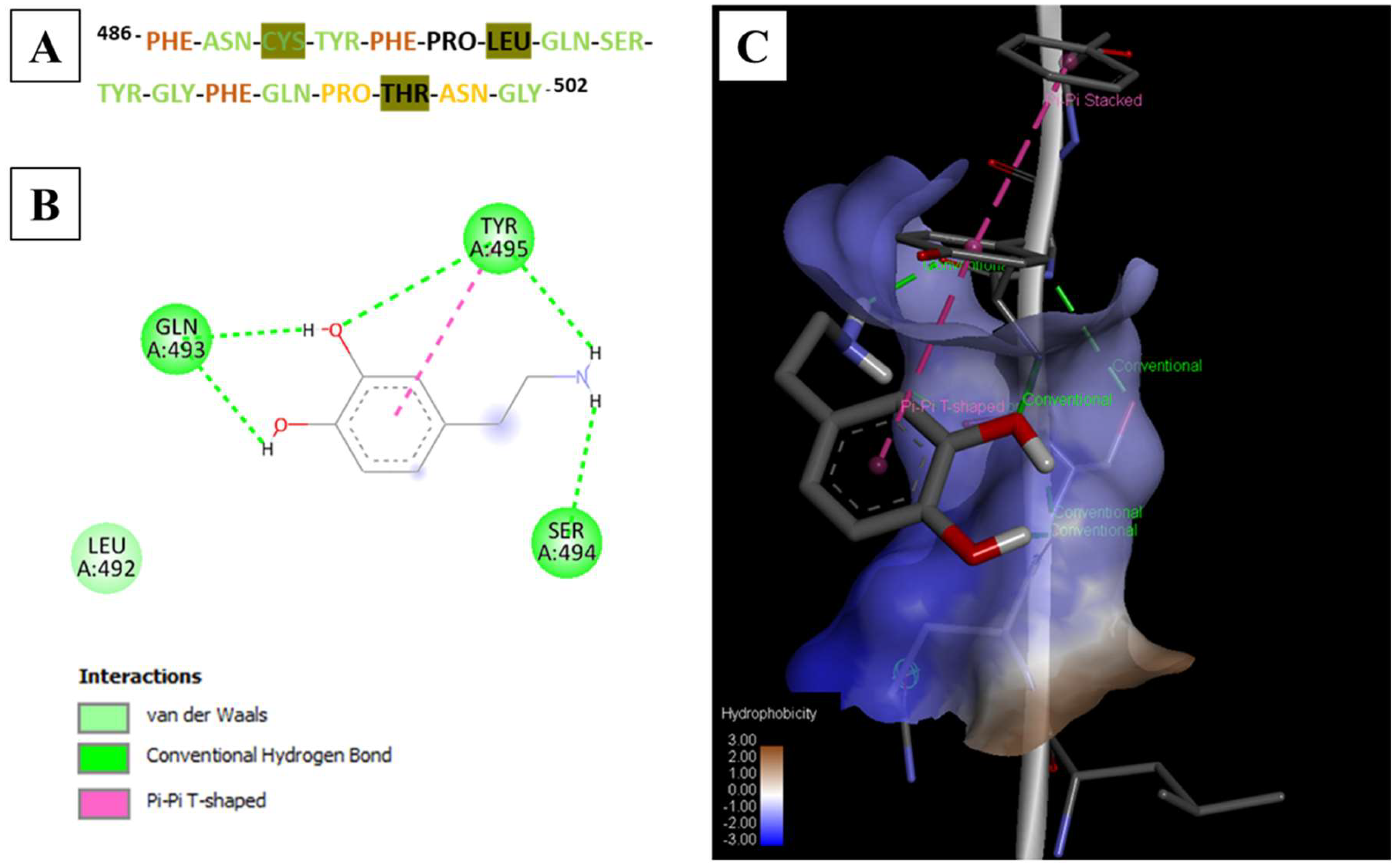

| Peptide | Sequence (One Letter Code) | Sequence (Three Letter Code) | Source Protein | Sequence Start | Sequence End |
|---|---|---|---|---|---|
| SARS-CoV-2 | FNCYFPLQSYGFQPTNG | Phe–Asn–Cys–Tyr–Phe–Pro–Leu–Gln–Ser–Tyr–Gly–Phe–Gln–Pro–Thr–Asn–Gly | PDB ID: 6M0J | 486 | 502 |
| Zika virus | MIVNDTGHETDENRA | Met–Ile–Val–Asn–Asp–Thr–Gly–His–Glu–Thr–Asp–Glu–Asn–Arg–Ala | PDB ID: 6CO8 | 151 | 165 |
| Monomers | Binding Energy (kcal/mol) |
|---|---|
| opamine | −3.4 |
| 4-(Aminomethyl)benzoic acid (MABA) | −3.4 |
| p-aminobenzamidine (PAB) | −3.4 |
| 2-Acrylamido-2-methylpropane sulfonic acid | −3.2 |
| Itaconic acid | −3.0 |
| N-(3-aminopropyl) methacrylamide | −2.9 |
| (Hydroxyethyl)methacrylate | −2.7 |
| Aniline | −2.8 |
| N-[3-(dimethylamino)-propyl]-methacrylamide | −2.8 |
| N-isopropylacrylamide | −2.8 |
| N-hydroxymethylacrylamide | −2.7 |
| N-tert-butylacrylamide | −2.7 |
| 2-(Dimethylamino)ethyl methacrylate | −2.5 |
| 2-hydroxyethyl acrylate | −2.5 |
| Glycidyl methacrylate | −2.5 |
| Methacrylic acid | −2.5 |
| Acrylamide | −2.2 |
| Acrylic acid | −2.2 |
| Methyl methacrylate | −2.3 |
| MMIPs/MNIPs | Amount of MNPs a (µL) | Amount of Peptide b (µL) | Amount of Tris Buffer (µL) | Amount of Dopamine c (µL) | Time of Polymerization |
|---|---|---|---|---|---|
| MNIP-1 | 50 | 0 | 600 | 100 | 1 |
| MNIP-2 | 2 | ||||
| MNIP-24 | 24 | ||||
| MMIP-05-1 | 50 | 37.5 | 562.5 | 100 | 1 2 24 |
| MMIP-05-2 | |||||
| MMIP-05-24 | |||||
| MMIP-1-1 | 50 | 75 | 525 | 100 | 1 |
| MMIP-1-2 | 2 | ||||
| MMIP-1-24 | 24 | ||||
| MMIP-2-1 | 50 | 150 | 450 | 100 | 1 |
| MMIP-2-2 | 2 | ||||
| MMIP-2-24 | 24 | ||||
| MMIP-3-1 | 50 | 225 | 375 | 100 | 1 |
| MMIP-3-2 | 2 | ||||
| MMIP-3-24 | 24 | ||||
| MMIP-4-1 | 50 | 300 | 300 | 100 | 1 |
| MMIP-4-2 | 2 | ||||
| MMIP-4-24 | 24 |
| Q (mg/g) ± SD (mg/g) | IF | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| MMIP-1-1/MNIP-1 | MMIP-2-2/MNIP-2 | MMIP-2-24/MNIP-24 | MMIP-PDA3/MNIP-PDA3 | MMIP-1-1 | MMIP-2-2 | MMIP-2-24 | MMIP-PDA3 | ||
| Zika | MMIP | 14.02 ± 1.02 | 18.28 ± 1.04 | 18.67 ± 4.36 | 191.55 ± 22.98 | 0.89 | 0.72 | 0.82 | 1.08 |
| MNIP | 15.69 ± 2.08 | 25.35 ± 1.93 | 22.68 ± 1.02 | 176.25 ± 9.82 | |||||
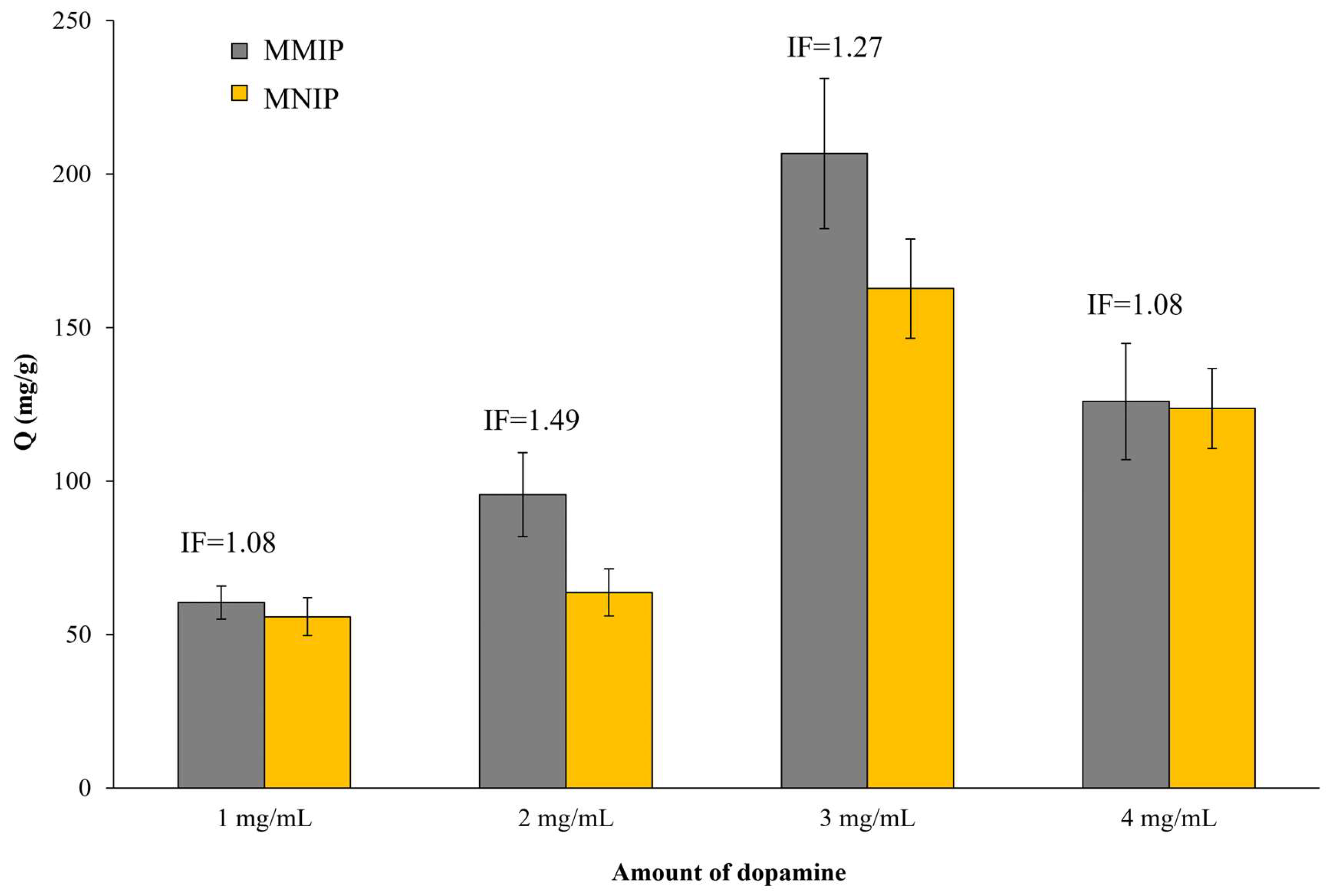
| SARS-CoV-2 | MMIP | 43.50 ± 2.76 | 74.33 ± 4.72 | 95.61 ± 9.54 | 206.73 ± 24.47 | 5.48 | 3.50 | 1.49 | 1.27 |
| MNIP | 7.94 ± 1.19 | 21.22 ± 1.69 | 63.75 ± 6.37 | 162.75 ± 16.22 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fresco-Cala, B.; Rajpal, S.; Rudolf, T.; Keitel, B.; Groß, R.; Münch, J.; Batista, A.D.; Mizaikoff, B. Development and Characterization of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers. Nanomaterials 2021, 11, 2985. https://doi.org/10.3390/nano11112985
Fresco-Cala B, Rajpal S, Rudolf T, Keitel B, Groß R, Münch J, Batista AD, Mizaikoff B. Development and Characterization of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers. Nanomaterials. 2021; 11(11):2985. https://doi.org/10.3390/nano11112985
Chicago/Turabian StyleFresco-Cala, Beatriz, Soumya Rajpal, Tamara Rudolf, Benedikt Keitel, Rüdiger Groß, Jan Münch, Alex D. Batista, and Boris Mizaikoff. 2021. "Development and Characterization of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers" Nanomaterials 11, no. 11: 2985. https://doi.org/10.3390/nano11112985
APA StyleFresco-Cala, B., Rajpal, S., Rudolf, T., Keitel, B., Groß, R., Münch, J., Batista, A. D., & Mizaikoff, B. (2021). Development and Characterization of Magnetic SARS-CoV-2 Peptide-Imprinted Polymers. Nanomaterials, 11(11), 2985. https://doi.org/10.3390/nano11112985







