Advances in Gold Nanoparticles-Based Colorimetric Aptasensors for the Detection of Antibiotics: An Overview of the Past Decade
Abstract
:1. Background and Introduction
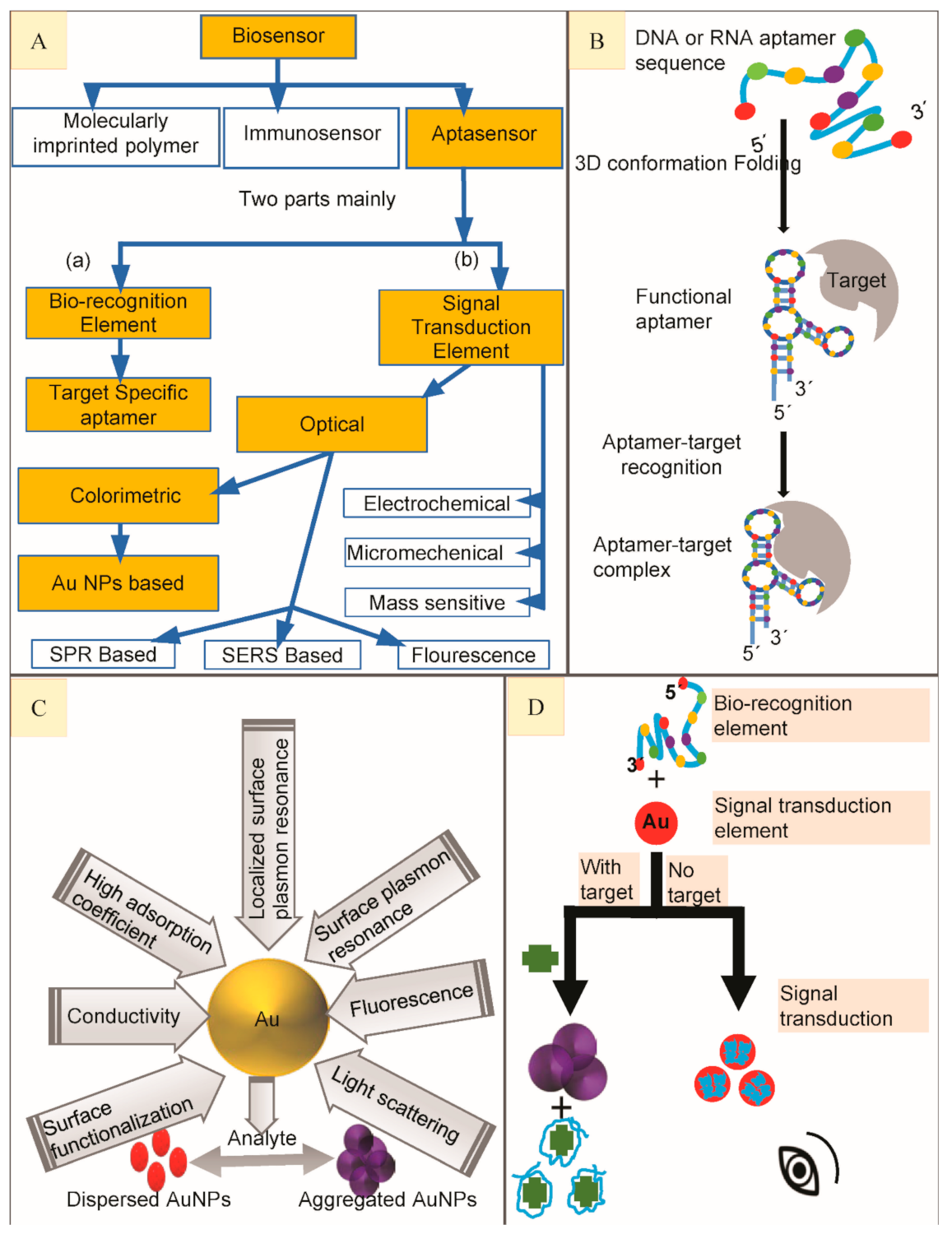
2. Biosensors
3. Aptasensors and Aptamers
4. Important Parameters to Measure the Efficiency of Aptasensors
5. Classification of Aptasensors
6. Gold Nanoparticles (AuNPs) Based Colorimetric Aptasensors
6.1. β-Lactams and Their Residue Detection
6.2. Aminoglycosides and Their Residue Detection
6.2.1. Kanamycin (KAN)
6.2.2. Tobramycin (TOB)
6.2.3. Streptomycin (STR)
6.2.4. Neomycin B (NB)
6.3. Anthracyclines and Their Residue Detection
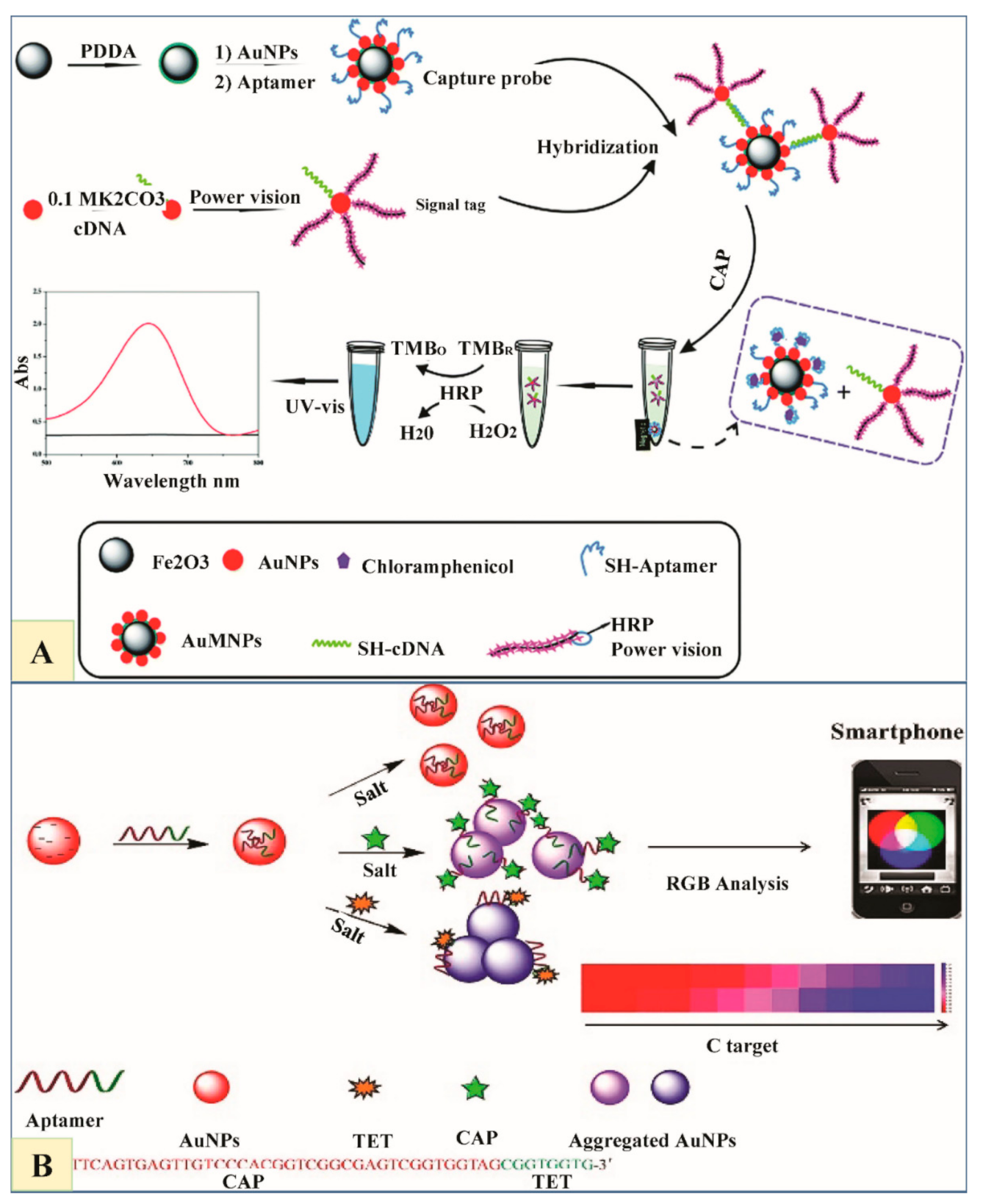
6.4. Chloramphenicol (CAP) and Their Residue Detection
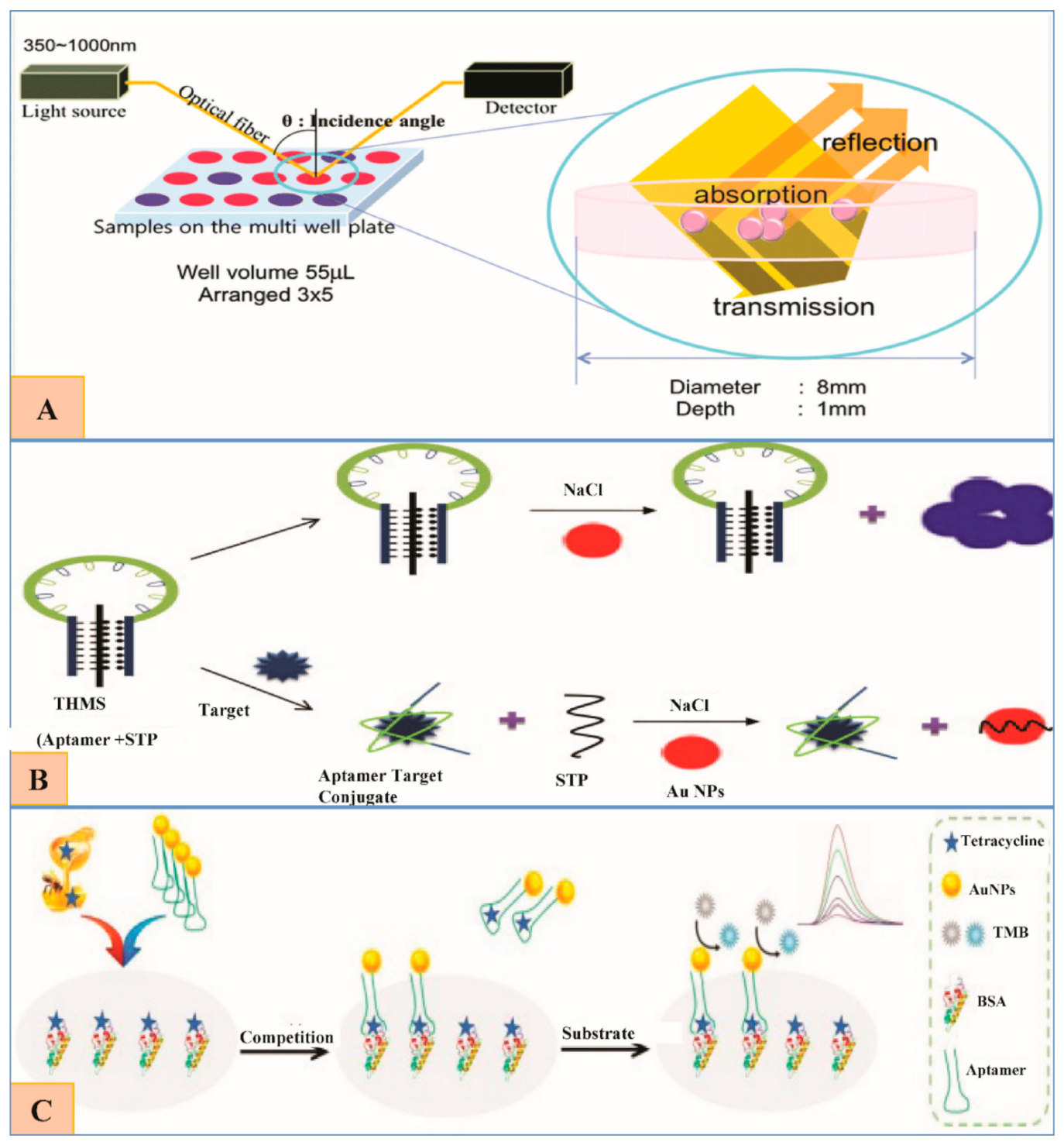
6.5. Fluoroquinolones (FQs) and Their Residue Detection
6.6. Tetracyclines and Their Residue Detection
6.6.1. Oxytetracyclines (OTC).
6.6.2. Tetracycline (TET)
6.7. Sulfonamides (SAs) and Their Residue Detection
Sulfadimethoxine (SDM)
7. A Generalized Overview, Future Perspective, Challenges and Conclusions
Funding
Conflicts of Interest
Abbreviations
| AuNPs | gold nanoparticles |
| BSA | bovine serum albumin |
| BRE | bio-recognition element |
| cDNA | complementary DNA |
| CS | complementary strand |
| dsDNA | double stranded DNA |
| KD | dissociation constant |
| LOD | limit of detection |
| SELEX | systematic evolution of ligands by exponential enrichment |
| SPR | surface plasmon resonance spectroscopy |
| STE | Signal transduction element |
| SSB | ssDNA binding protein |
| ssDNA | single stranded DNA |
| THMS | triple-helix molecular switch |
References
- Abedalwafa, M.A.; Li, Y.; Ni, C.; Wang, L. Colorimetric sensor arrays for the detection and identification of antibiotics. Anal. Methods 2019, 11, 2836–2854. [Google Scholar] [CrossRef]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277. [Google Scholar]
- Center for Disease Control and Prevention, Office of Infectious Disease. Antibiotic Resistance Threats in the United States, 2013; Center for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Kazerooni, H.; Bahreyni, A.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. A colorimetric aptasensor for selective detection of oxytetracycline in milk, using gold nanoparticles and oxytetracline-short aptamer. Nanomed. J. 2019, 6, 105–111. [Google Scholar]
- Song, Y.; Duan, F.; Zhang, S.; Tian, J.-Y.; Zhang, Z.; Wang, Z.-W.; Liu, C.-S.; Xu, W.-M.; Du, M. Iron oxide @ mesoporous carbon architectures derived from an Fe(II)-based metal organic framework for highly sensitive oxytetracycline determination. J. Mater. Chem. A 2017, 5, 19378–19389. [Google Scholar] [CrossRef]
- Briscoe, S.E.; McWhinney, B.C.; Lipman, J.; Roberts, J.A.; Ungerer, J.P. A method for determining the free (unbound) concentration of ten beta-lactam antibiotics in human plasma using high performance liquid chromatography with ultraviolet detection. J. Chromatogr. B 2012, 907, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.-H.; Xue, Y.-G.; Liu, H.-J.; Dai, L.-L.; Yan, H.; Li, N. Development of determination method of fluoroquinolone antibiotics in sludge based on solid phase extraction and HPLC-fluorescence detection analysis. Huan Jing Ke Xue 2016, 37, 1553–1561. [Google Scholar] [PubMed]
- Meersche, T.V.D.; Van Pamel, E.; Van Poucke, C.; Herman, L.; Heyndrickx, M.; Rasschaert, G.; Daeseleire, E. Development, validation and application of an ultra high performance liquid chromatographic-tandem mass spectrometric method for the simultaneous detection and quantification of five different classes of veterinary antibiotics in swine manure. J. Chromatogr. A 2016, 1429, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-J.; Wu, H.-L.; Fu, H.-Y.; Zhao, J.; Li, Y.-N.; Li, S.-F.; Kang, C.; Yu, R.-Q. Chromatographic background drift correction coupled with parallel factor analysis to resolve coelution problems in three-dimensional chromatographic data: Quantification of eleven antibiotics in tap water samples by high-performance liquid chromatography coupled with a diode array detector. J. Chromatogr. A 2013, 1302, 72–80. [Google Scholar] [CrossRef]
- Berendsen, B.J.; Wegh, R.S.; Memelink, J.; Zuidema, T.; Stolker, L.A. The analysis of animal faeces as a tool to monitor antibiotic usage. Talanta 2015, 132, 258–268. [Google Scholar] [CrossRef]
- Martínez-Carballo, E.; González-Barreiro, C.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Haller, M.Y.; Müller, S.R.; McArdell, C.S.; Alder, A.C.; Suter, M.J.-F. Quantification of veterinary antibiotics (sulfonamides and trimethoprim) in animal manure by liquid chromatography–mass spectrometry. J. Chromatogr. A 2002, 952, 111–120. [Google Scholar] [CrossRef]
- Gbylik-Sikorska, M.; Posyniak, A.; Sniegocki, T.; Zmudzki, J. Liquid chromatography–tandem mass spectrometry multiclass method for the determination of antibiotics residues in water samples from water supply systems in food-producing animal farms. Chemosphere 2015, 119, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Wang, R.; Xing, B.; Chi, H.; Wu, D.; Wei, Q. Label-free photoelectrochemical aptasensor for tetracycline detection based on cerium doped CdS sensitized BiYWO6. Biosens. Bioelectron. 2018, 106, 7–13. [Google Scholar] [CrossRef]
- Yang, S.; Zhu, X.; Wang, J.; Jin, X.; Liu, Y.; Qian, F.; Zhang, S.; Chen, J. Combustion of hazardous biological waste derived from the fermentation of antibiotics using TG–FTIR and Py–GC/MS techniques. Bioresour. Technol. 2015, 193, 156–163. [Google Scholar] [CrossRef]
- Pauter, K.; Szultka-Młyńska, M.; Buszewski, B. Determination and identification of antibiotic drugs and bacterial strains in biological samples. Molecules 2020, 25, 2556. [Google Scholar] [CrossRef]
- Mungroo, N.A.; Neethirajan, S. Biosensors for the detection of antibiotics in poultry industry—A Review. Biosensors 2014, 4, 472–493. [Google Scholar] [CrossRef] [Green Version]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Yang, Q.; Wu, W. Progress on Structured Biosensors for Monitoring Aflatoxin B1 from Biofilms: A Review. Front. Microbiol. 2020, 11, 408. [Google Scholar] [CrossRef]
- Wang, R.E.; Zhang, Y.; Cai, J.; Cai, W.; Gao, T. Aptamer-based fluorescent biosensors. Curr. Med. Chem. 2011, 18, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhou, H.; Jiang, H.; Ou, H.; Li, X.; Zhang, L. Development of a fraction collection approach in capillary electrophoresis SELEX for aptamer selection. Analyst 2015, 140, 2664–2670. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brändle, G.M.; Ewers, J.; Mayer, G. Identification and characterization of nucleobase-modified aptamers by click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, Z.; Yin, Y.; Jia, F.; Wu, Q.; Tian, P.; Wang, D. Development and evaluation of a novel in situ target-capture approach for aptamer selection of human noroviruses. Talanta 2019, 193, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Sabri, M.Z.; Hamid, A.A.A.; Hitam, S.M.S.; Rahim, M.Z.A. In-silico selection of aptamer: A review on the revolutionary approach to understand the aptamer design and interaction through computational chemistry. Mater. Today Proc. 2019, 19, 1572–1581. [Google Scholar] [CrossRef]
- Ahirwar, R.; Nahar, S.; Aggarwal, S.; Ramachandran, S.; Maiti, S.; Nahar, P. In silico selection of an aptamer to estrogen receptor alpha using computational docking employing estrogen response elements as aptamer-alike molecules. Sci. Rep. 2016, 6, 21285. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Zhu, Z.; An, Y.; Zhang, W.; Zhang, H.; Liu, D.; Yu, C.; Duan, W.; Yang, C.J. Selection of DNA Aptamers against Epithelial Cell Adhesion Molecule for Cancer Cell Imaging and Circulating Tumor Cell Capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef]
- Wolter, O.; Mayer, G. Aptamers as valuable molecular tools in neurosciences. J. Neurosci. 2017, 37, 2517–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomilin, F.N.; Moryachkov, R.; Shchugoreva, I.; Zabluda, V.N.; Peters, G.; Platunov, M.; Spiridonova, V.; Melnichuk, A.; Atrokhova, A.; Zamay, S.S.; et al. Four steps for revealing and adjusting the 3D structure of aptamers in solution by small-angle X-ray scattering and computer simulation. Anal. Bioanal. Chem. 2019, 411, 6723–6732. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Tian, Y.; Zhang, M.; Liu, J.; Yuan, Y.; Tan, J.; Ma, A. Selection, identification, and application of aptamers against Agaricus bisporus lectin to establish an aptamer-AuNPs colorimetric method for detection of ABL. J. Food Qual. 2020, 2020. [Google Scholar] [CrossRef]
- Li, L.; Xu, S.; Yan, H.; Li, X.; Yazd, H.S.; Li, X.; Huang, T.; Cui, C.; Jiang, J.; Tan, W. Nucleic acid aptamers for molecular diagnostics and therapeutics: Advances and perspectives. Angew. Chem. Int. Ed. 2020, 60, 2221–2231. [Google Scholar] [CrossRef] [PubMed]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple methods and rational design for enhancing aptamer sensitivity and specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, F.J.; Ozalp, V.C. Graphene and other nanomaterial-based electrochemical aptasensors. Biosensors 2012, 2, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, D.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar]
- Eggins, B.R. Chemical Sensors and Biosensors; John Wiley & Sons: Hoboken, NJ, USA, 2008; Volume 28. [Google Scholar]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Lim, Y.C.; Kouzani, A.Z.; Duan, W. Aptasensors: A Review. J. Biomed. Nanotechnol. 2010, 6, 93–105. [Google Scholar] [CrossRef]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef]
- Robati, R.Y.; Arab, A.; Ramezani, M.; Langroodi, F.A.; Abnous, K.; Taghdisi, S.M. Aptasensors for quantitative detection of kanamycin. Biosens. Bioelectron. 2016, 82, 162–172. [Google Scholar] [CrossRef]
- Zahra, Q.U.A.; Khan, Q.A.; Luo, Z. Advances in optical aptasensors for early detection and diagnosis of various cancer types. Front. Oncol. 2021, 11, 19. [Google Scholar] [CrossRef]
- Holzinger, M.; Le Goff, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Ragavan, K.; Thakur, M.; Raghavarao, K. Recent advances in nanoparticle based aptasensors for food contaminants. Biosens. Bioelectron. 2015, 74, 612–627. [Google Scholar] [CrossRef]
- Mohammadpour, A.H.; Tavassoli, A.; Khakzad, M.R.; Zibaee, E.; Afshar, M.; Hashemzaei, M.; Karimi, G. Effect of gold nanoparticles on postoperative peritoneal adhesions in rats. Nanomed. J. 2015, 2, 211–216. [Google Scholar]
- Babaei, M.; Ganjalikhani, M. A systematic review of gold nanoparticles as novel cancer therapeutics. Nanomed. J. 2014, 1, 211–219. [Google Scholar]
- Chen, H.; Zhou, K.; Zhao, G. Gold nanoparticles: From synthesis, properties to their potential application as colorimetric sensors in food safety screening. Trends Food Sci. Technol. 2018, 78, 83–94. [Google Scholar] [CrossRef]
- Kim, C.-H.; Lee, L.-P.; Min, J.-R.; Lim, M.-W.; Jeong, S.-H. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk. Biosens. Bioelectron. 2014, 51, 426–430. [Google Scholar] [CrossRef]
- Xu, C.; Ying, Y.; Ping, J. Colorimetric aggregation assay for kanamycin using gold nanoparticles modified with hairpin DNA probes and hybridization chain reaction-assisted amplification. Microchim. Acta 2019, 186, 448. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 249, 98–103. [Google Scholar] [CrossRef]
- Seo, H.B.; Kwon, Y.S.; Lee, J.E.; Cullen, D.; Noh, H.M.; Gu, M.B. A novel reflectance-based aptasensor using gold nanoparticles for the detection of oxytetracycline. Analyst 2015, 140, 6671–6675. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-Y.; Liu, B.-W.; Huang, P.; Wu, F.-Y. A novel colorimetric aptasensor for detection of chloramphenicol based on lanthanum ion–assisted gold nanoparticle aggregation and smartphone imaging. Anal. Bioanal. Chem. 2019, 411, 7511–7518. [Google Scholar] [CrossRef]
- Huang, P.-C.; Gao, N.; Li, J.-F.; Wu, F.-Y. Colorimetric detection of methionine based on anti-aggregation of gold nanoparticles in the presence of melamine. Sens. Actuators B Chem. 2018, 255, 2779–2784. [Google Scholar] [CrossRef]
- Abdullah, S.M.; Rachid, S. On column binding a real-time biosensor for β-lactam antibiotics quantification. Molecules 2020, 25, 1248. [Google Scholar] [CrossRef] [Green Version]
- Zheng, K.; Zhang, J.; Wang, Y.; Wang, Z.H.; Zhang, S.X.; Wu, C.M.; Shen, J.Z. Development of a rapid multi-residue assay for detecting β-lactams using penicillin binding protein 2x. Biomed. Environ. Sci. 2013, 26, 100–109. [Google Scholar]
- Lamar, J.; Petz, M. Development of a receptor-based microplate assay for the detection of beta-lactam antibiotics in different food matrices. Anal. Chim. Acta 2007, 586, 296–303. [Google Scholar] [CrossRef]
- Gustavsson, E.; Degelaen, J.; Bjurling, P.; Sternesjo, A. Determination of β-lactams in milk using a surface plasmon resonance-based biosensor. J. Agric. Food Chem. 2004, 52, 2791–2796. [Google Scholar] [CrossRef]
- Chan, P.-H.; Liu, H.-B.; Chen, Y.W.; Chan, K.-C.; Tsang, C.-W.; Leung, Y.-C.; Wong, K.-Y. Rational design of a novel fluorescent biosensor for β-lactam antibiotics from a class a β-lactamase. J. Am. Chem. Soc. 2004, 126, 4074–4075. [Google Scholar] [CrossRef] [PubMed]
- Merola, G.; Martini, E.; Tomassetti, M.; Campanella, L. New immunosensor for β-lactam antibiotics determination in river waste waters. Sens. Actuators B Chem. 2014, 199, 301–313. [Google Scholar] [CrossRef]
- Song, K.-M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence–colorimetric methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef] [PubMed]
- Shayesteh, O.H.; Ghavami, R. Two colorimetric ampicillin sensing schemes based on the interaction of aptamers with gold nanoparticles. Microchim. Acta 2019, 186, 485. [Google Scholar] [CrossRef]
- Sharma, T.K.; Ramanathan, R.; Weerathunge, P.; Mohammadtaheri, M.; Daima, H.K.; Shukla, R.; Bansal, V. Aptamer-mediated ‘turn-off/turn-on’ nanozyme activity of gold nanoparticles for kanamycin detection. Chem. Commun. 2014, 50, 15856–15859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.-M.; Cho, M.; Jo, H.; Min, K.; Jeon, S.H.; Kim, T.; Han, M.S.; Ku, J.K.; Ban, C. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal. Biochem. 2011, 415, 175–181. [Google Scholar] [CrossRef]
- Niu, S.; Lv, Z.; Liu, J.; Bai, W.; Yang, S.; Chen, A. Colorimetric aptasensor using unmodified gold nanoparticles for homogeneous multiplex detection. PLoS ONE 2014, 9, e109263. [Google Scholar] [CrossRef]
- Ha, N.-R.; Jung, I.-P.; Kim, S.-H.; Kim, A.-R.; Yoon, M.-Y. Paper chip-based colorimetric sensing assay for ultra-sensitive detection of residual kanamycin. Process. Biochem. 2017, 62, 161–168. [Google Scholar] [CrossRef]
- Liu, J.; Zeng, J.; Tian, Y.; Zhou, N. An aptamer and functionalized nanoparticle-based strip biosensor for on-site detection of kanamycin in food samples. Analyst 2017, 143, 182–189. [Google Scholar] [CrossRef]
- Zhou, N.; Zhang, J.; Tian, Y. Aptamer-based spectrophotometric detection of kanamycin in milk. Anal. Methods 2014, 6, 1569. [Google Scholar] [CrossRef]
- Abedalwafa, M.A.; Tang, Z.; Qiao, Y.; Mei, Q.; Yang, G.; Li, Y.; Wang, L. An aptasensor strip-based colorimetric determination method for kanamycin using cellulose acetate nanofibers decorated DNA–gold nanoparticle bioconjugates. Microchim. Acta 2020, 187, 1–9. [Google Scholar] [CrossRef]
- Shayesteh, O.H.; Khosroshahi, A.G. A polyA aptamer-based label-free colorimetric biosensor for the detection of kanamycin in human serum. Anal. Methods 2020, 12, 1858–1867. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Nie, J.; Zhao, S.; Tian, Y.; Zhou, N. Gold nanoparticle based photometric determination of tobramycin by using new specific DNA aptamers. Microchim. Acta 2018, 185, 4. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, J.; Zhang, J.; Li, C.; Tian, Y.; Wang, J. Selection and identification of streptomycin-specific single-stranded DNA aptamers and the application in the detection of streptomycin in honey. Talanta 2013, 108, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Xie, Y.; Sun, Y.; Bi, K.; Cui, Z.; Zhao, L.; Fan, W. An aptamer-based colorimetric sensor for streptomycin and its application in food inspection. Chem. Res. Chin. Univ. 2017, 33, 714–720. [Google Scholar] [CrossRef]
- Luan, Q.; Miao, Y.; Gan, N.; Cao, Y.; Li, T.; Chen, Y. A POCT colorimetric aptasensor for streptomycin detection using porous silica beads- enzyme linked polymer aptamer probes and exonuclease-assisted target recycling for signal amplification. Sensors Actuators B Chem. 2017, 251, 349–358. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Tao, H.; Chen, H.; Yang, W.; Qiu, S. Colorimetric detection of streptomycin in milk based on peroxidase-mimicking catalytic activity of gold nanoparticles. RSC Adv. 2017, 7, 38471–38478. [Google Scholar] [CrossRef] [Green Version]
- Emrani, A.S.; Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Colorimetric and fluorescence quenching aptasensors for detection of streptomycin in blood serum and milk based on double-stranded DNA and gold nanoparticles. Food Chem. 2016, 190, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Khavani, M.; Izadyar, M.; Housaindokht, M.R. Theoretical design and experimental study on the gold nanoparticles based colorimetric aptasensors for detection of neomycin B. Sens. Actuators B Chem. 2019, 300, 126947. [Google Scholar] [CrossRef]
- He, L.; Zhi, W.; Wu, Y.; Zhan, S.; Wang, F.; Xing, H.; Zhou, P. A highly sensitive resonance scattering based sensor using unmodified gold nanoparticles for daunomycin detection in aqueous solution. Anal. Methods 2012, 4, 2266–2271. [Google Scholar] [CrossRef]
- Abnous, K.; Danesh, N.M.; Ramezani, M.; Emrani, A.S.; Taghdisi, S.M. A novel colorimetric sandwich aptasensor based on an indirect competitive enzyme-free method for ultrasensitive detection of chloramphenicol. Biosens. Bioelectron. 2016, 78, 80–86. [Google Scholar] [CrossRef]
- Miao, Y.; Gan, N.; Ren, H.-X.; Li, T.; Cao, Y.; Hu, F.; Yan, Z.; Chen, Y. A triple-amplification colorimetric assay for antibiotics based on magnetic aptamer–enzyme co-immobilized platinum nanoprobes and exonuclease-assisted target recycling. Analyst 2015, 140, 7663–7671. [Google Scholar] [CrossRef]
- Gao, H.; Gan, N.; Pan, D.; Chen, Y.; Li, T.; Cao, Y.; Fu, T. A sensitive colorimetric aptasensor for chloramphenicol detection in fish and pork based on the amplification of a nano-peroxidase-polymer. Anal. Methods 2015, 7, 6528–6536. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, H.; Lai, G.; Liu, S.; Li, B.; Yu, A. Sensitive and rapid aptasensing of chloramphenicol by colorimetric signal transduction with a DNAzyme-functionalized gold nanoprobe. Food Chem. 2019, 270, 287–292. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Huang, P.; Wu, F.-Y. A label-free colorimetric aptasensor based on controllable aggregation of AuNPs for the detection of multiplex antibiotics. Food Chem. 2020, 304, 125377. [Google Scholar] [CrossRef]
- Javidi, M.; Housaindokht, M.R.; Verdian, A.; Razavizadeh, B.M. Detection of chloramphenicol using a novel apta-sensing platform based on aptamer terminal-lock in milk samples. Anal. Chim. Acta 2018, 1039, 116–123. [Google Scholar] [CrossRef]
- Lavaee, P.; Danesh, N.M.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Colorimetric aptamer based assay for the determination of fluoroquinolones by triggering the reduction-catalyzing activity of gold nanoparticles. Microchim. Acta 2017, 184, 2039–2045. [Google Scholar] [CrossRef]
- Kim, Y.S.; Kim, J.H.; Kim, I.A.; Lee, S.J.; Jurng, J.; Gu, M.B. A novel colorimetric aptasensor using gold nanoparticle for a highly sensitive and specific detection of oxytetracycline. Biosens. Bioelectron. 2010, 26, 1644–1649. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, C.; Sun, Y.; Shao, Y.; Cai, Y.; Zhang, Y.; Miao, J.; Miao, P. A colorimetric aptasensor for the antibiotics oxytetracycline and kanamycin based on the use of magnetic beads and gold nanoparticles. Microchim. Acta 2018, 185, 548. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Luo, Y.; Zhi, W.; Wu, Y.; Zhou, P. A Colorimetric Aptamer Biosensor Based on Gold Nanoparticles for the Ultrasensitive and Specific Detection of Tetracycline in Milk. Aust. J. Chem. 2013, 66, 485–490. [Google Scholar] [CrossRef] [Green Version]
- Ramezani, M.; Danesh, N.M.; Lavaee, P.; Abnous, K.; Taghdisi, S.M. A novel colorimetric triple-helix molecular switch aptasensor for ultrasensitive detection of tetracycline. Biosens. Bioelectron. 2015, 70, 181–187. [Google Scholar] [CrossRef]
- Luo, Y.; Xu, J.; Li, Y.; Gao, H.; Guo, J.; Shen, F.; Sun, C. A novel colorimetric aptasensor using cysteamine-stabilized gold nanoparticles as probe for rapid and specific detection of tetracycline in raw milk. Food Control. 2015, 54, 7–15. [Google Scholar] [CrossRef]
- Sheng, Y.-M.; Liang, J.; Xie, J. Indirect competitive determination of tetracycline residue in honey using an ultrasensitive gold-nanoparticle-linked aptamer assay. Molecules 2020, 25, 2144. [Google Scholar] [CrossRef]
- Chen, A.; Jiang, X.; Zhang, W.; Chen, G.; Zhao, Y.; Tunio, T.M.; Liu, J.; Lv, Z.; Li, C.; Yang, S. High sensitive rapid visual detection of sulfadimethoxine by label-freeaptasensor. Biosens. Bioelectron. 2013, 42, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Huang, Y.; Fang, Z.; Bai, W.; Zhu, C.; Zhang, C.; Yan, M.; Chen, A. Aptamer based photometric assay for the antibiotic sulfadimethoxine based on the inhibition and reactivation of the peroxidase-like activity of gold nanoparticles. Microchim. Acta 2016, 184, 59–63. [Google Scholar] [CrossRef]
- Chen, X.-X.; Lin, Z.-Z.; Hong, C.-Y.; Yao, Q.-H.; Huang, Z.-Y. A dichromatic label-free aptasensor for sulfadimethoxine detection in fish and water based on AuNPs color and fluorescent dyeing of double-stranded DNA with SYBR green I. Food Chem. 2020, 309, 125712. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.-G.; Lai, R.Y. A reagentless and reusable electrochemical aptamer-based sensor for rapid detection of ampicillin in complex samples. Talanta 2018, 176, 619–624. [Google Scholar] [CrossRef]
- Peng, X.; Tan, J.; Tang, C.; Yu, Y.; Wang, Z. Multiresidue determination of fluoroquinolone, sulfonamide, trimethoprim, and chloramphenicol antibiotics in urban waters in China. Environ. Toxicol. Chem. 2008, 27, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Popelka, P.; Nagy, J.; Marcincak, S. A comparison of BSDA and PREMI test sensitivity to penicillin standards in poultry meat and after the administration of Amuril plv. sol. Folia Vet. 2003, 47, 139–141. [Google Scholar]
- Jadhav, R.W.; Al Kobaisi, M.; Jones, L.A.; Vinu, A.; Bhosale, S.V. The supramolecular self-assembly of aminoglycoside antibiotics and their applications. ChemistryOpen 2019, 8, 1154–1166. [Google Scholar] [CrossRef]
- González-Fernández, E.; De-Los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Impedimetric aptasensor for tobramycin detection in human serum. Biosens. Bioelectron. 2011, 26, 2354–2360. [Google Scholar] [CrossRef] [PubMed]
- Haasnoot, W.; Cazemier, G.; Koets, M.; Van Amerongen, A. Single biosensor immunoassay for the detection of five aminoglycosides in reconstituted skimmed milk. Anal. Chim. Acta 2003, 488, 53–60. [Google Scholar] [CrossRef]
- Chen, S.-H.; Liang, Y.-C.; Chou, Y.-W. Analysis of kanamycin A in human plasma and in oral dosage form by derivatization with 1-naphthyl isothiocyanate and high-performance liquid chromatography. J. Sep. Sci. 2006, 29, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Granja, R.H.; Niño, A.M.M.; Zucchetti, R.A.; Niño, R.E.M.; Patel, R.; Salerno, A.G. Determination of streptomycin residues in honey by liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta 2009, 637, 64–67. [Google Scholar] [CrossRef]
- Yadavalli, R.K.; Singh, R.S.; Anand, S.K. Diffusion test for the detection of streptomycin residues in milk. J. Dairy Res. 1985, 52, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Lian, W.; Liu, S.; Yu, J.; Li, J.; Cui, M.; Xu, W.; Huang, J. Electrochemical sensor using neomycin-imprinted film as recognition element based on chitosan-silver nanoparticles/graphene-multiwalled carbon nanotubes composites modified electrode. Biosens. Bioelectron. 2013, 44, 70–76. [Google Scholar] [CrossRef]
- Wang, S.; Xu, B.; Zhang, Y.; He, J. Development of enzyme-linked immunosorbent assay (ELISA) for the detection of neomycin residues in pig muscle, chicken muscle, egg, fish, milk and kidney. Meat Sci. 2009, 82, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Wochner, A.; Menger, M.; Orgel, D.; Cech, B.; Rimmele, M.; Erdmann, V.A.; Glökler, J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Anal. Biochem. 2008, 373, 34–42. [Google Scholar] [CrossRef]
- Ferguson, J.; Baxter, A.; Young, P.; Kennedy, G.; Elliott, C.; Weigel, S.; Gatermann, R.; Ashwin, H.; Stead, S.; Sharman, M. Detection of chloramphenicol and chloramphenicol glucuronide residues in poultry muscle, honey, prawn and milk using a surface plasmon resonance biosensor and Qflex® kit chloramphenicol. Anal. Chim. Acta 2005, 529, 109–113. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, X.; Yin, H.; Meng, Q.; Zhao, Y.; Liu, L.; Wu, Z.; Xu, H. Quantitative and sensitive detection of chloramphenicol by surface-enhanced raman scattering. Sensors 2017, 17, 2962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, Y.; Gan, N.; Li, T.; Zhang, H.; Cao, Y.; Jiang, Q. A colorimetric aptasensor for chloramphenicol in fish based on double-stranded DNA antibody labeled enzyme-linked polymer nanotracers for signal amplification. Sens. Actuators B Chem. 2015, 220, 679–687. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Deng, Y.; Zhao, M.-Z.; Zhou, Y.-L.; Zhang, X.-X. Highly-sensitive detection of eight typical fluoroquinolone antibiotics by capillary electrophoresis-mass spectroscopy coupled with immunoaffinity extraction. RSC Adv. 2018, 8, 4063–4071. [Google Scholar] [CrossRef] [Green Version]
- Scherer, R.; Pereira, J.; Firme, J.; Lemos, M.; Lemos, M. Determination of ciprofloxacin in pharmaceutical formulations using HPLC method with UV detection. Indian J. Pharm. Sci. 2015, 76, 541–544. [Google Scholar]
- Speltini, A.; Sturini, M.; Maraschi, F.; Profumo, A. Fluoroquinolone antibiotics in environmental waters: Sample preparation and determination. J. Sep. Sci. 2010, 33, 1115–1131. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, Y.; Ding, S.; He, F.; Beier, R.C.; Li, J.; Jiang, H.; Feng, C.; Wan, Y.; Zhang, S.; et al. Development of a monoclonal Antibody-Based Broad-Specificity ELISA for fluoroquinolone antibiotics in foods and molecular modeling studies of cross-reactive compounds. Anal. Chem. 2007, 79, 4471–4483. [Google Scholar] [CrossRef]
- Matsunaga, T.; Kondo, T.; Osasa, T.; Kotsugai, A.; Shitanda, I.; Hoshi, Y.; Itagaki, M.; Aikawa, T.; Tojo, T.; Yuasa, M. Sensitive electrochemical detection of ciprofloxacin at screen-printed diamond electrodes. Carbon 2020, 159, 247–254. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.; Lu, J.; Xing, J.; Li, J.; Liu, T.; Xue, Q. Highly efficient detection of ciprofloxacin in water using a nitrogen-doped carbon electrode fabricated through plasma modification. N. J. Chem. 2019, 43, 15169–15176. [Google Scholar] [CrossRef]
- Johnson, A.C.; Keller, V.; Dumont, E.; Sumpter, J.P. Assessing the concentrations and risks of toxicity from the antibiotics ciprofloxacin, sulfamethoxazole, trimethoprim and erythromycin in European rivers. Sci. Total. Environ. 2015, 511, 747–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granados-Chinchilla, F.; Rodríguez, C. Tetracyclines in food and feedingstuffs: From regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017, 2017, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Jalalian, S.H.; Taghdisi, S.M.; Danesh, N.M.; Bakhtiari, H.; Lavaee, P.; Ramezani, M.; Abnous, K. Sensitive and fast detection of tetracycline using an aptasensor. Anal. Methods 2015, 7, 2523–2528. [Google Scholar] [CrossRef]
- Khosrokhavar, R.; Hosseini, M.-J.; Amini, M.; Pirali-Hamedani, M.; Ghazi-Khansari, M.; Bakhtiarian, A. Validation of an analytical methodology for determination of oxytetracycline residue in milk by HPLC with UV detection. Toxicol. Mech. Methods 2008, 18, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, Q.; Zhang, D.; Gan, N.; Li, Q.; Cuan, J. Detection and removal of antibiotic tetracycline in water with a highly stable luminescent MOF. Sens. Actuators B Chem. 2018, 262, 137–143. [Google Scholar] [CrossRef]
- He, L.; Luo, Y.; Zhi, W.; Zhou, P. Colorimetric sensing of tetracyclines in milk based on the assembly of cationic conjugated polymer-aggregated gold nanoparticles. Food Anal. Methods 2013, 6, 1704–1711. [Google Scholar] [CrossRef]
- Jamshaid, T.; Tenório-Neto, E.T.; Baraket, A.; Lebaz, N.; Elaissari, A.; Sanchís, A.; Salvador, J.-P.; Marco, M.-P.; Bausells, J.; Errachid, A.; et al. Development of novel magneto-biosensor for sulfapyridine detection. Biosensors 2020, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Dai, T.; Duan, J.; Li, X.; Xu, X.; Shi, H.; Kang, W. Determination of sulfonamide residues in food by capillary zone electrophoresis with on-line chemiluminescence detection based on an Ag(III) Complex. Int. J. Mol. Sci. 2017, 18, 1286. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Wang, G. Hazard of sulfonamides and detection technology research progress. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Singapore, 2017. [Google Scholar]
- Mohammad-Razdari, A.; Ghasemi-Varnamkhasti, M.; Izadi, Z.; Rostami, S.; Ensafi, A.A.; Siadat, M.; Losson, E. Detection of sulfadimethoxine in meat samples using a novel electrochemical biosensor as a rapid analysis method. J. Food Compos. Anal. 2019, 82, 103252. [Google Scholar] [CrossRef]
- Du, J.; Jiang, Q.; Lu, X.; Chen, L.; Zhang, Y.; Xiong, X. Detection of sulfadimethoxine using optical images of liquid crystals. Analyst 2019, 144, 1761–1767. [Google Scholar] [CrossRef] [PubMed]



| Antibiotic Class | Antibiotic | Molecular Structures |
|---|---|---|
| β-Lactams (BLCs) | Ampicillin (AMP) |  |
| Aminoglycosides (AMGs) | Kanamycin (KAN) |  |
| Tobramycin (TOB) |  | |
| Streptomycin (STR) | 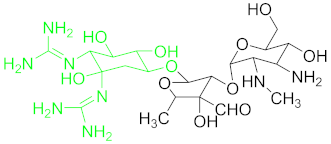 | |
| Neomycin B (NB) | 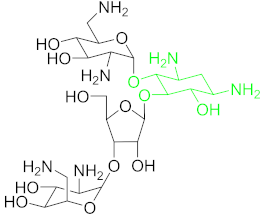 | |
| Anthracyclines (ACs) | Daunomycin (DNR) | 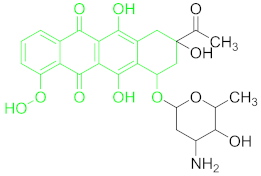 |
| Chloramphenicol | Chloramphenicol (CAP) | 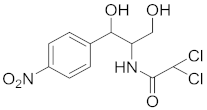 |
| Fluoroquinolones (FQs) | Ciprofloxacin (CIP) | 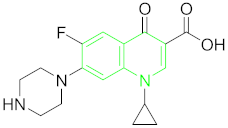 |
| Tetracyclines (TCs) | Oxytetracyclines (OTC) | 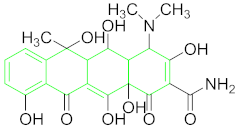 |
| Tetracycline (TET) | 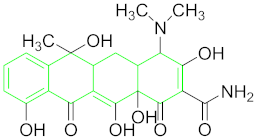 | |
| Sulfonamides (SAs) | Sulfadimethoxine (SDM) | 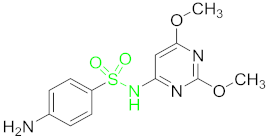 |
| Target | 5′ Linker and Spacer | Aptamer Sequences 5′→3′ | 3′ Linker and Spacer | KD (nM) | LOD (nM) | Color Change | AuNP Particle Size (nm) | AuNP Preparation Method | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| AMP | FAM | AMP17: GCG GGC GGT TGT ATA GCG G AMP18: TTA GTT GGG GTT CAG TTG G AMP4: CAC GGC ATG GTG GGC GTC GTG | biotin | 13.4 9.8 9.4 | 14.3 (dw) 28.6 (m) | Red-purple | 13 | classical citrate reduction method | [59] |
| AMP | (SH)–(CH2)6 | -GCGGGCGGTTGTATAGCGG GCGG GCGGTTGTATAGCGG- | (T)15–(A)12 | 0.10 0.49 | 10 50 | Red-purple Red-purple | 10 | Smith method | [60] |
| KAN | - | TGG GGG TTG AGG CTA AGC CGA | - | 8.38 | 1.49 | Ruby red to purplish-blue or colorless | - | classical citrate reduction method | [61] |
| KAN TOB | - | TGGGGGTTGAGGCTAAGCCGA | - | KAN = 78.8 KAN B = 84.5 TOB = 103 | 25 | Red-purple | 13 | classical citrate reduction | [62] |
| KAN SDM | - | SDM; GAGGGCAACGAGTGTTTATAGA KAN: TGGGGGTTGAGGCTAAGCCGA- | - | - | - | Red-purple/blue | 13 | Classical citrate reduction | [63] |
| KAN | - | (KBA) 3-1; CGG AAG CGC GCC ACC CCA TCG GCG GGG GCG AAG CTT GCG Ky2; TGG GGG TTG AGG CTA AGC CGA | - | - | 3.35 26.8 | wine red to purple blue | 14, 21, 27 | citrate reduction of HAuCl4 | [64] |
| KAN | Apt; HS–(CH2)6 DNA1; SH–(CH2)6 DNA2; biotin | Apt; TGG GGG TTG AGG CTA AGC CGA DNA 1: TCA GTC GGC TTA GCC GTC CAA CGT CAG ATC C DNA2: CCG ATG GAT CTG ACG T | apt: biotin | - | 0.0778 | Red- colorless or faint red | 13 | citrate reduction of HAuCl4 | [65] |
| KAN | ssDNA1; HS–(CH2)6- | KSA; TGGGGGTTGAGGCTAAGCCGA ssDNA1; CGGTCGGCTTA ssDNA2; AACCCCCATCT | ssDNA2; (CH2)3–SH | - | 01 | Red-purple | 13 | citrate reduction of HAuCl4 | [66] |
| KAN | KMC apt; NH2 | KMC apt; TGGGGGTTGAGGCTAAGCCGA cDNA; TCGG CTTAGCCTCAACCCCCA | - | - | 2.5 | Pink to white | 15–20 | citrate reduction method | [67] |
| KAN | - | TGG GGG TTG AGG CTA AGC CGA | (T)15–(A)12 | - | 0.05 | Red to blue | 15 | citrate reduction method | [68] |
| TOB | - | GGG ACT TGG TTT AGG TAA TGA GTC CC | - | - | 23.3 | red-purple-blue | 13 | classical citrate reduction | [49] |
| TOB | - | - | - | - | 37.9 | red to purple | - | - | [69] |
| STR | - | I: GGG GTC TGG TGT TCT GCT TTG TTC TGT CGG GTC GT II: TGA AGG GTC GAC TCT AGA GGC AGG TGT TCC TCA GG III: AGC TTG GGT GGG GCC ACG TAG AGG TAT AGC TTG TT IV: TGT GTG TTC GGT GCT GTC GGG TTG TTT CTT GGT TT | - | I: 199.1 II: 221.3 III: 272.0 IV: 340.6 | 200 | red to purple | 13 | citrate reduction of HAuCl4 | [70] |
| STR | I: FAM II: FAM III: FAM | I: CCC GTT TAA AGT AGT TGA GAG TAT TCC GTT TCT TTG TGT C II: GTG CGT TAT AAA CTA GTT TTG ATT CAA TGT TGG GTG TGG G III: GGG CCT GTT TTG CCT TCA CGT TCT CTT CCT TGC CGT TCT G | I: biotin II: biotin III: biotin | I: 6.07 II: 8.56 III: 13.14 | 25 nm/L | red to purple | 15 | citrate reduction of HAuCl4 | [71] |
| STR | SH–(CH2)6- | TAG GGA ATT CGT CGA CGG ATC CGG GGT CTG GTG TTC TGC TTT GTT CTG TCG GGT CGT CTG CAG GTC GAC GCA TGC GCC G | - | - | 0.0017 (b) 0.001 | colorless to blue | 19.73 | Au NPs–PV | [72] |
| STR | - | TAGGGAATTCGTCGACGGATCCGGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGTCTGCAGGTCGACGCATGCGCCG | - | 19.1 | 86 | red-grayish green | 13 | citrate reduction of HAuCl4 | [73] |
| STR | - | STR apt: TAG GGA ATT CGT CGA CGG ATC CGG GGT CTG GTG TTC TGC TTT GTT CTG TCG GGT CGT CTG CAG GTC GAC GCA TGC GCC G CS; C GGC GCA TGC GTC GAC CTG CAG ACG ACC CGA CAG AAC AAA GCA GAA CAC CAG ACC CCG GAT CCG TCG ACG AAT TCC CTA | - | - | 73.1 (b) 102.4 (b) 108.7 (m) | red to blue | 15 | citrate reduction of HAuCl4 | [74] |
| NB | - | AP-W; GGACUGGGCGAGAAGUUUAGUCC Ap-M18; GGACUAAACGAGAAGUUUAGUCC Ap-M20; GGACUAAACGAGAAGCCCAGUCC | - | - | 470 27 360 | pink red to Blue | 13 | citrate reduction of HAuCl4 | [75] |
| DNR | - | GGGAATTCGAGCTCGGTACCATCTGTGTAAAAGGGGTGGGGGTGGGTACGTCTAGCTGCAGGCATGCAAGCTTGG | - | - | 17.1 | red to purple | 13 | reduction of HAuCl4 | [76] |
| CAP | AuNPs DNA; Biotin | Apt; ACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAG AuNPs binding DNA; ACTTTCCATTCCTTTTAC | Apt; Biotin AuNPs DNA; Thiol | - | 0.451 (b) 0.697 (m) 0.601 | bright red to faint red | 14 | reduction of HAuCl4 | [77] |
| CAP | apt: SH–(CH2)6 cDNA: SH–(CH2)6 | apt: ACT TCA GTG AGT TGT CCC ACG GTC GGC GAG TCG GTG GTA G cDNA: CTA CCA CCG ACT CGCG CGA CCG TGG GAC AAC TCA CTG AAG T | - | - | 0.00093 (b) (0.3 × 10−9 g/L) | colorless to light blue | 20 | slight modified Frens method | [78] |
| CAP | apt: SH–(CH2)6 cDNA: SH–(CH2)6 | apt: ACT TCA GTG AGT TGT CCC ACG GTC GGC GAG TCG GTG GTA G cDNA: TTT TCT ACC ACC GAC TCG C | - | - | 0.02 ng/mL | colorless to light blue | 20 | slight modified Frens method | [79] |
| CAP | Apt; SH–(CH2)6 cDNA: SH–(CH2)6 | Apt; ACT TCA GTG AGT TGT CCC ACG GTC GGC GAG TCG GTG GTAG cDNA: CTA CCA CCG ACT CGC CGA CCG TGG 142 GAC AAC TCA CTG AAGT | - | - | 0.046 (b) (0.015 × 10−6 g/L) | colorless to light blue | 16 | slight modified Frens method | [78] |
| CAP | Apt;Biotin DNAzyme; (SH–(CH2)6– cDNA; (SH)–(CH2)6 | Apt; ACT TCA GTG AGT TGT CCC ACG GTC GGC GAG TCG GTG GTA G DNAzyme; AAAAAA GGG TAG GGC GGG TTG GG cDNA; AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA CTA CCA CCG ACT CGC C | - | - | 0.13 pg/mL | red to blue | 13 | reduction of HAuCl4 | [80] |
| CAP | - | ACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAG | - | - | 7.65 5.88 | red to blue | 15 | Huang et al. | [51] |
| CAP TET | - | Multi-apt; ACTTCAGTGAGTTGTCCCACGGTCGGCGAGTCGGTGGTAGCGGTGGTG | - | - | CAP.7.0 TET.32.9 | (CAP) wine red to purple (TET) wine red to blue | 15 | Huang et al. | [81] |
| CAP | - | Apt; ACT TCA GTG AGT TGT CCC ACGGTC GGC GAG TCG GTG GTAG LP; CTGAAGTTCTACCAC | - | - | 0.03 | pink red to blue | 14 | Classical reduction of HAuCl4 | [82] |
| FQs CIP | CS2; Thiol | Apt; ATACCAGCTTATTCAATTGCAGGGTATCTGAGGCTGATCTACAATGTCGTGGGGCATTTATTGGCGTTGATACGTACAATCGTAATC AGTTAG CS1; TTGAATAAGCTGGTATAAACC CS2; AAACCACCTCCGAATCCCAAGCCACC GCCGCTAACTGATTACGATTGT | CS1; Thiol | - | 1.2 (CIP) 1.3 (w) 2.6 (bs) 3.2 (m) | yellow to colorless | 12 | classic citrate reduction | [83] |
| OTC | - | CGTACGGAATTCGCTAGCGGGCGGGGGTGCTGGGGGAATGGAGTGCTGCGTGCTGCGGGT CCGAGCTCCACGTG- | - | - | 25 | red to purple | 13 | citrate reduction of HAuCl4 | [84] |
| OTC | - | CGTACGGAATTCGCTAGCACGTTGACGCTGGTGCCCGGTTGTGGTGCGAGTGTTGTGTGGATCCGAGCTCCACGTG | - | - | 01 | red to purple | 13 | citrate reduction of HAuCl4 | [50] |
| OTC and KAN | - | OTC apt; CGTACGGAATTCGCTAGCGGGCGGGGGTGTGGGGGAATGGAGTGCTGCGTGCTGCGGGGT CCGAGCTCCACGTG KAN apt; TGG GGG TTG AGG CTA AGC CGA | - | - | 1 ag mL−1 | colorless to blue or yellow | 10 | citrate reduction of HAuCl4 | [85] |
| OTC | - | CGA CGC ACA GTC GCT GGT GCG TAC CTG GTT GCC GTT GTG T | - | - | 10 (w) 20 (m) | red to blue | 13 | Classical citrate reduction | [5] |
| TET | - | CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCTCCACGTG | - | - | 122 | purple to red | 13 | citrate reduction of HAuCl4 | [86] |
| TET | - | CGTACGGAATTCGCTAGCCCCCCGGCAGGCCACGGCTTGGGTTGGTCCCACTGCGCGTGGATCCGAGCT CCACGTG | - | - | 45.8 | red to purple | 15 | citrate reduction of HAuCl4 | [86] |
| TET | - | Apt; CTCTCTCGGTGGTGTCTCTC Signal Transduction Probe; GAGAGAGAGAGAGA | - | - | 266 pM | red to blue | 15 | classical citrate reduction of HAuCl4 | [87] |
| TET | - | CGT ACG GAA TTC GCT AGC CCC CCG GCA GGC CAC GGC TTG GGT TGG TCC CAC TGC GCGTGG ATC CGA GCT CCA CGT G | - | - | 0.039 µg/mL | red to blue | 13 | - | [88] |
| TET | SH–(CH2)6 | CGT ACG GAA TTC GCT AGC CCC CCG GCA GGC CAC GGC TTG GGT TGG TCC CAC TGC GCG TGG ATC CGA GCT CCA CGT G | - | - | 0.002 ng/mL, | colorless to blue | 18 | classical citrate reduction of HAuCl4 | [89] |
| SDM | - | SDM apt; GAGGGCAACGAGTGTTTATAGA KAN apt; TGGGGGTTGAGGCTAAGCCGA | - | - | 50 ng/m | red to blue | 13 | Mayer’s method | [90] |
| SDM | - | GAGGGCAACGAGTGTTTATAGA | - | - | 10 ng/mL | red to purplish-blue | 13 | citrate reduction method | [91] |
| SDM | - | SDM apt; GGC AAC GAG TGT TTA cDNA; TAA ACA CTC GTT GCC | - | - | 3.41 ng mL−1 (w) 4.41 ng g−1 (f) | red to blue/purple | 13 | citrate reduction method | [92] |
| Antibiotic Group/Class | Advantages | Disadvantages |
|---|---|---|
| β-Lactams/(AMP) |
| - |
| Aminoglycosides/ (KAN), (TOB), (STR), (NB) |
|
|
| Anthracyclines/ (DNR) |
| - |
| Chloramphenicol/ (CAP) |
|
|
| Fluoroquinolones (CIP) |
| - |
| Tetracyclines/ (OTC), (TET) |
| - |
| Sulfonamides (SDM) |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahra, Q.u.A.; Luo, Z.; Ali, R.; Khan, M.I.; Li, F.; Qiu, B. Advances in Gold Nanoparticles-Based Colorimetric Aptasensors for the Detection of Antibiotics: An Overview of the Past Decade. Nanomaterials 2021, 11, 840. https://doi.org/10.3390/nano11040840
Zahra QuA, Luo Z, Ali R, Khan MI, Li F, Qiu B. Advances in Gold Nanoparticles-Based Colorimetric Aptasensors for the Detection of Antibiotics: An Overview of the Past Decade. Nanomaterials. 2021; 11(4):840. https://doi.org/10.3390/nano11040840
Chicago/Turabian StyleZahra, Qurat ul Ain, Zhaofeng Luo, Rizwan Ali, Muhammad Imran Khan, Fenfen Li, and Bensheng Qiu. 2021. "Advances in Gold Nanoparticles-Based Colorimetric Aptasensors for the Detection of Antibiotics: An Overview of the Past Decade" Nanomaterials 11, no. 4: 840. https://doi.org/10.3390/nano11040840








