A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Solutions
2.2. Apparatus and Measurements
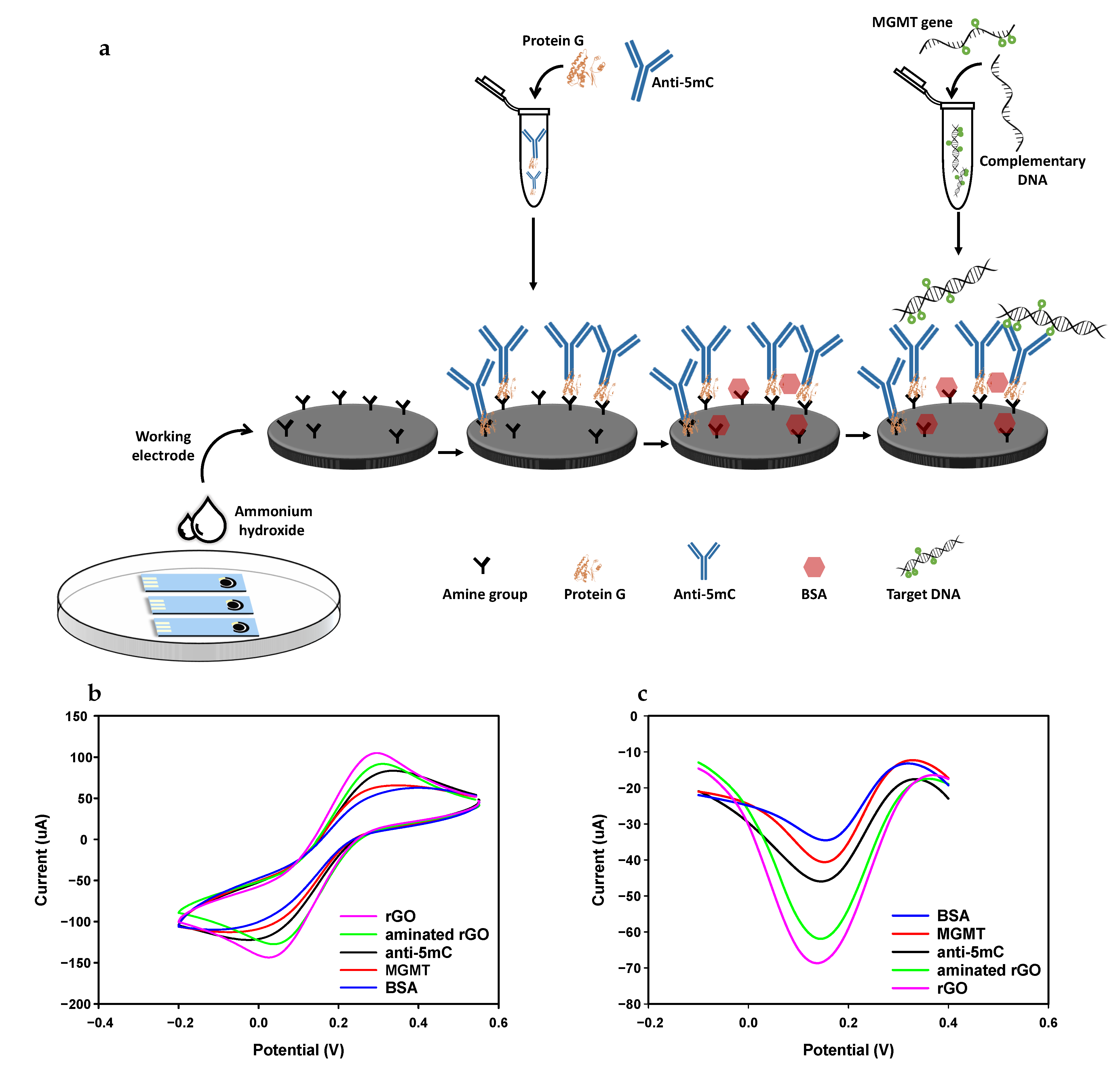
2.3. Preparation Steps
3. Results and Discussion
3.1. Characterisation of the Sensing Electrode
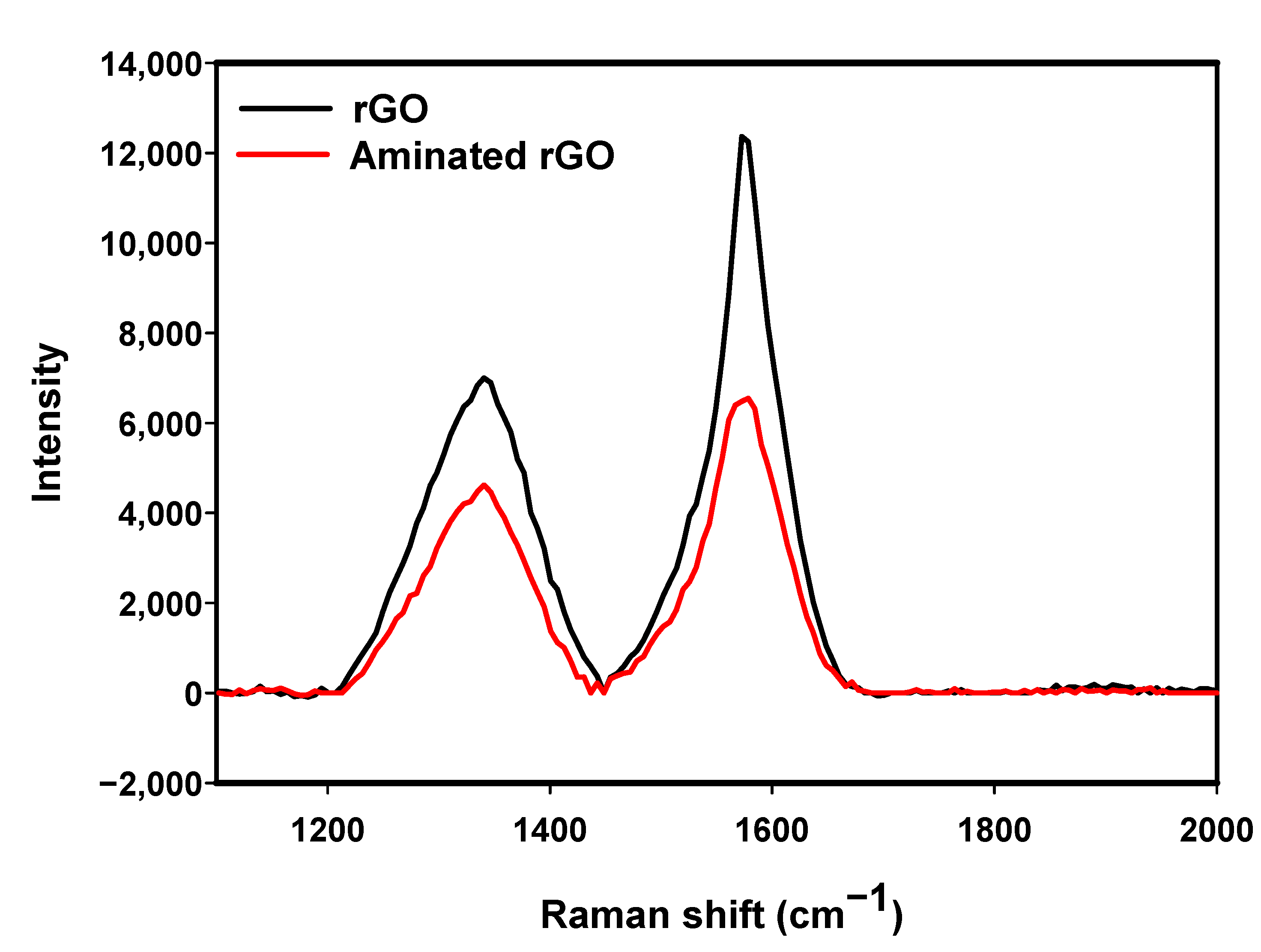
3.1.1. Raman
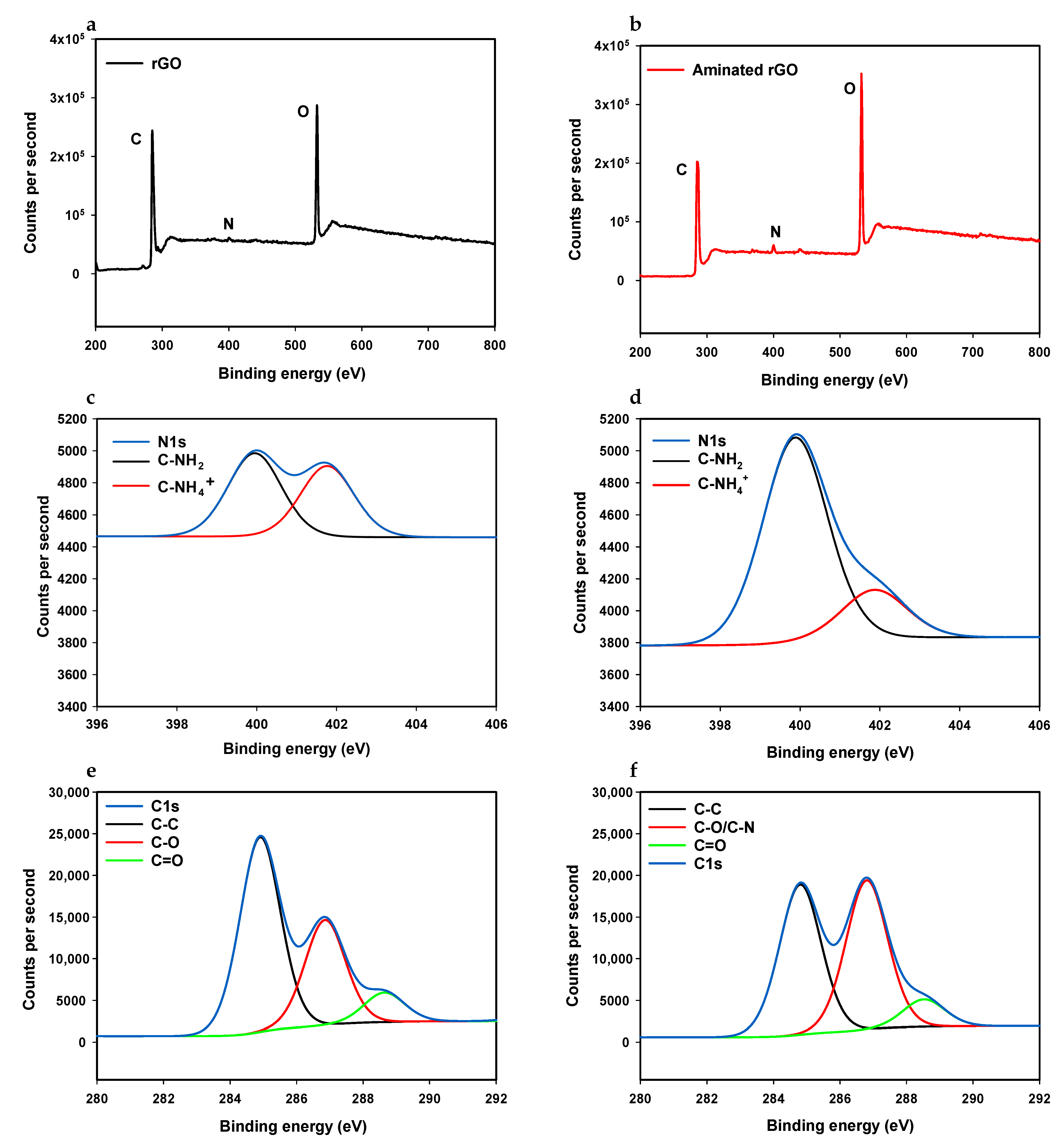
3.1.2. XPS
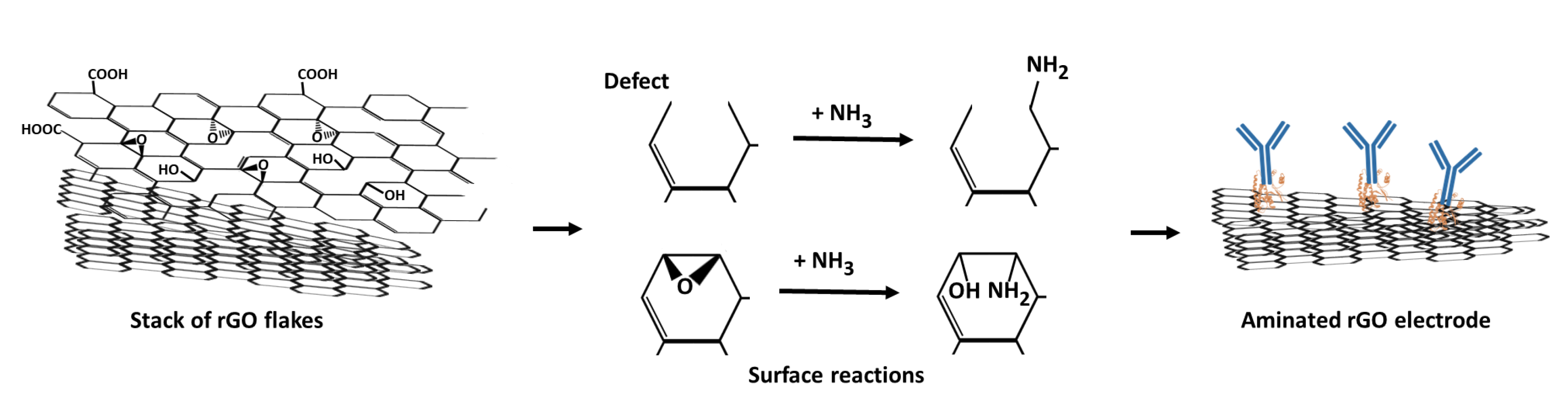
3.2. Ammonium Hydroxide Chemisorption
3.3. Electrochemical Experiments
3.3.1. Electrochemical Behaviour and Selectivity
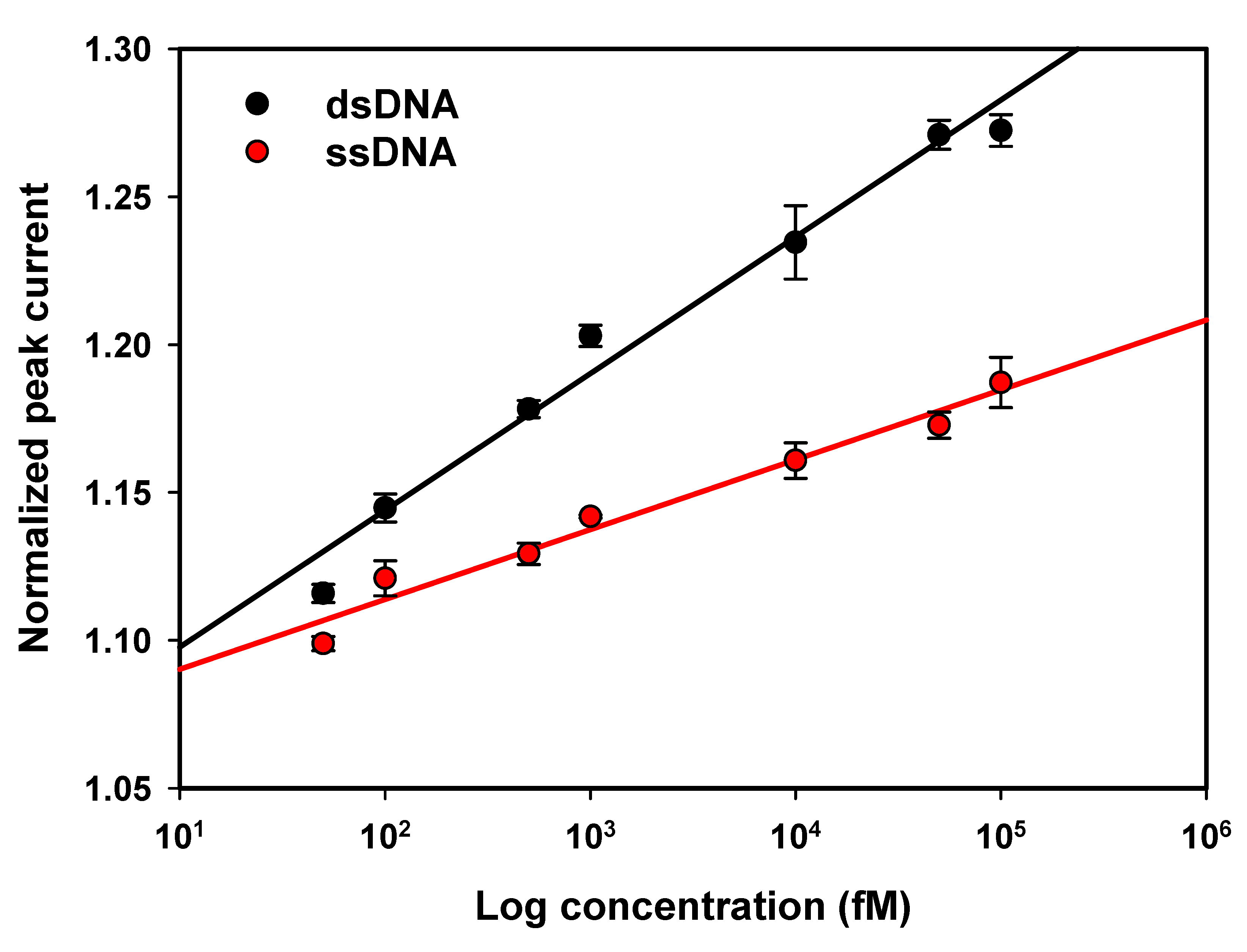
3.3.2. Biosensor Linearity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. SEM

Appendix A.2. Optimisation
| Experimental Variable | Range | Selected Value |
|---|---|---|
| Application of protein G | With-without protein G | With protein G |
| Anti-5mC incubation time, hours | 1–8 | 4 |
| BSA incubation time, hours | 5–30 | 15 |
| MGMT gene incubation time, hours | 30–120 | 60 |
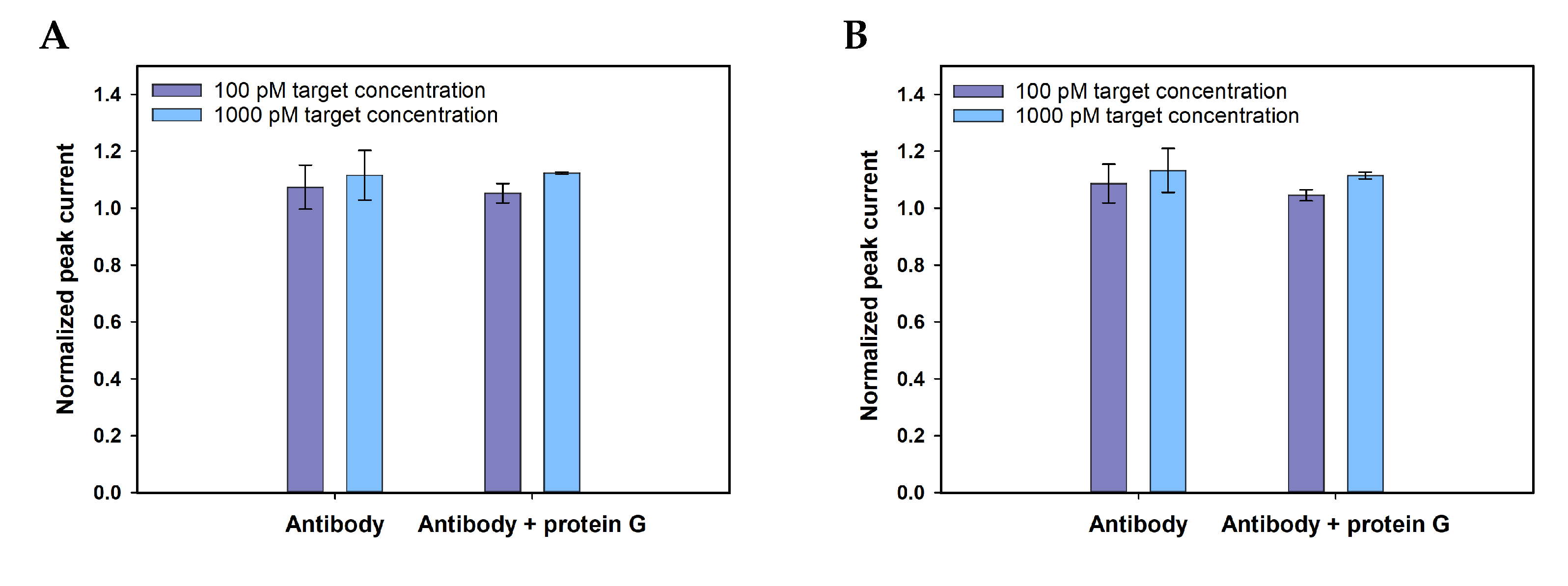

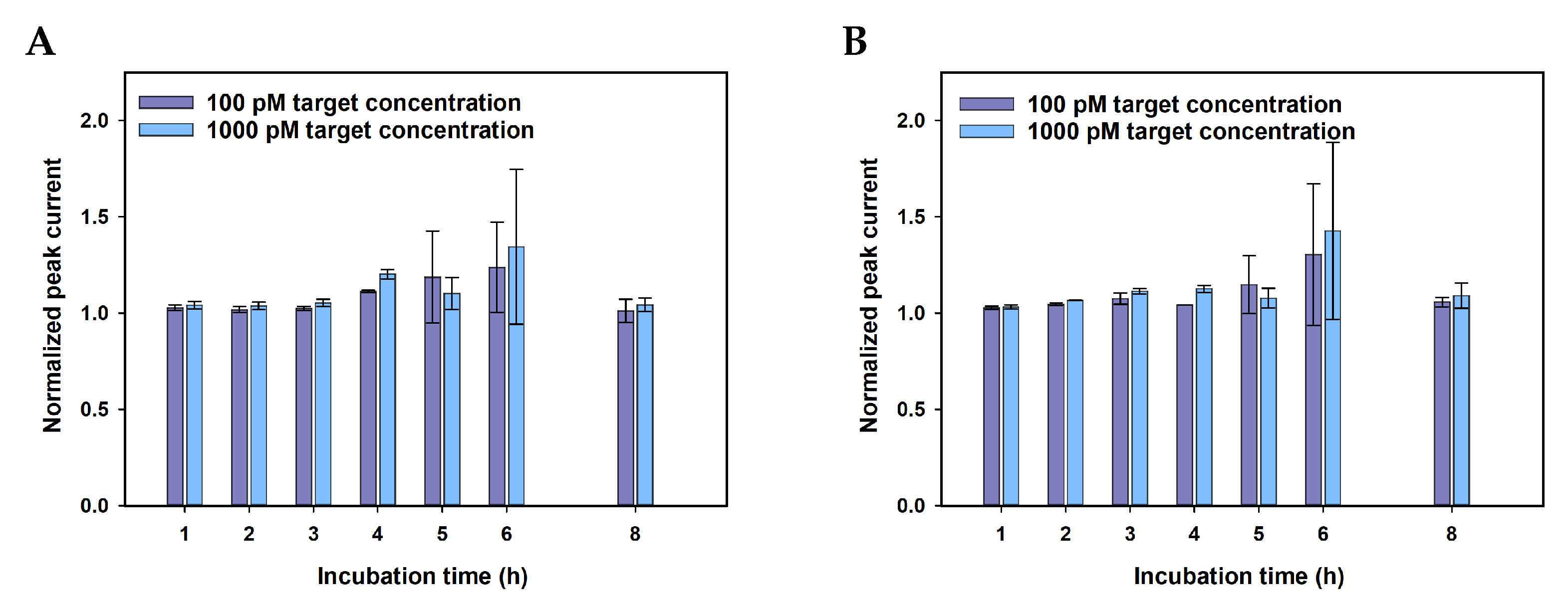

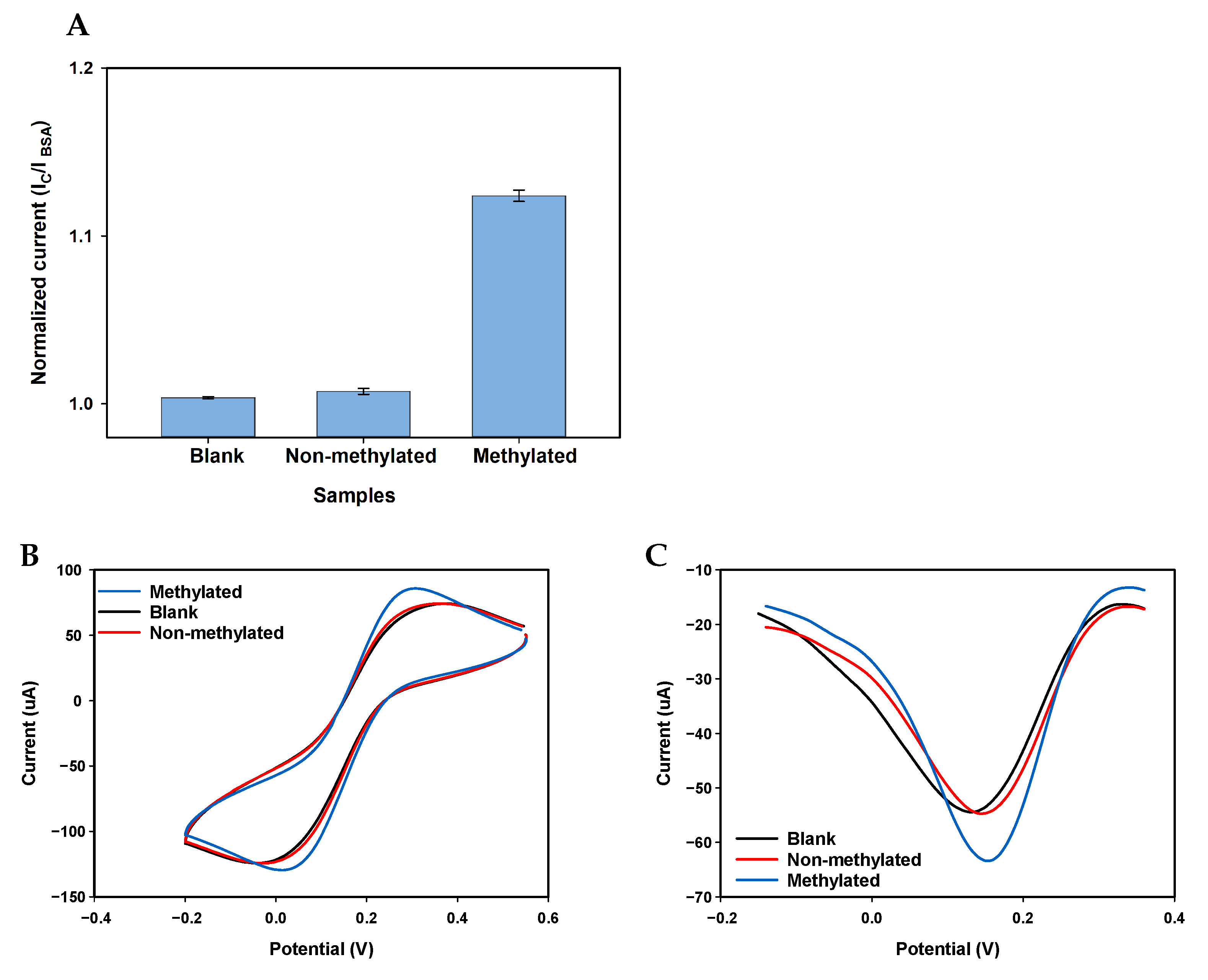
Appendix A.3. Selectivity
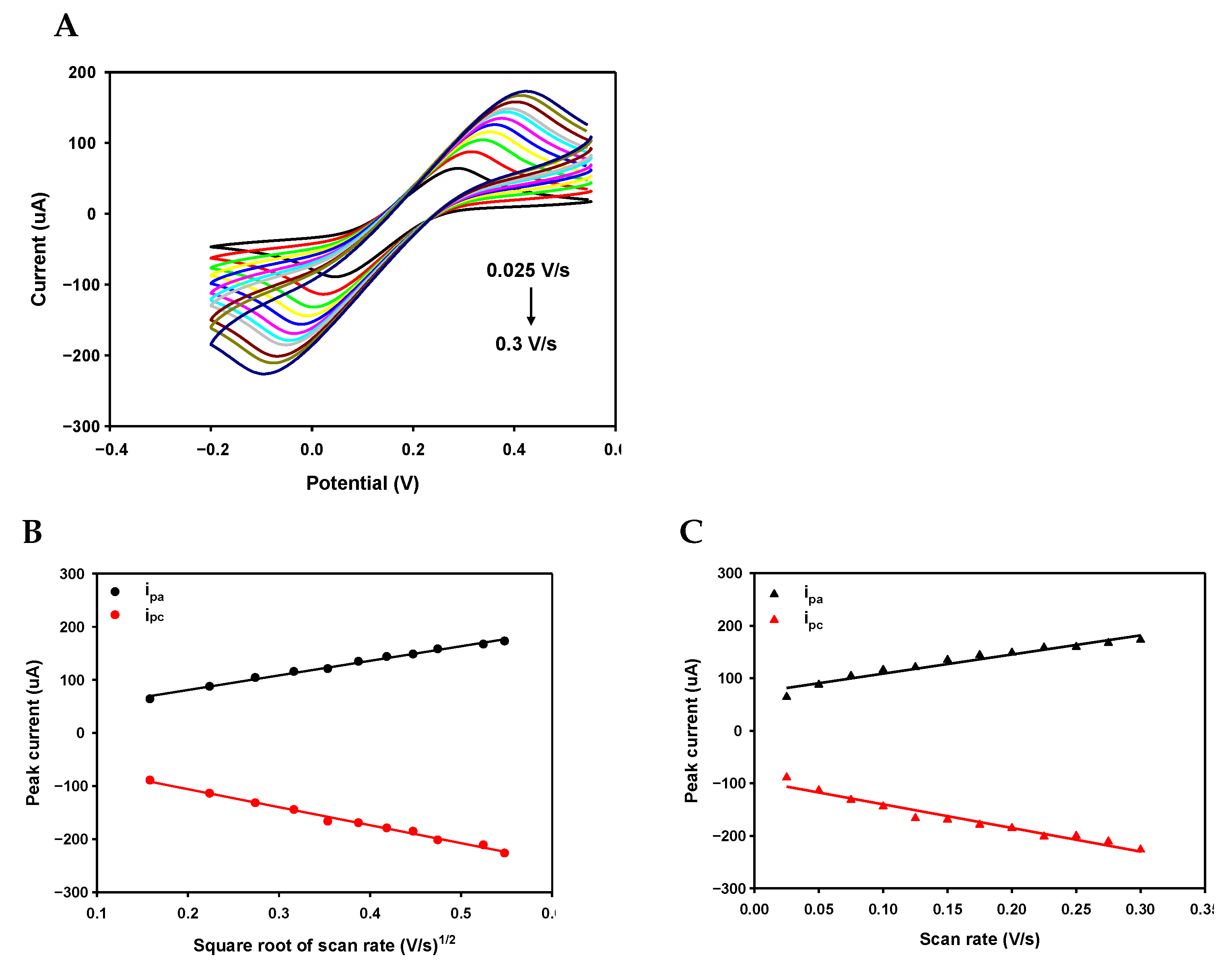
Appendix A.4. Scan Rate Experiment
Appendix A.4.1. Comparison with Other Works
| Electrode | Bioreceptor | Dynamic Range/LOD | Technique | Reference |
|---|---|---|---|---|
| Gold modified with gold nanoparticles | DNA probe, methyl binding protein, gold nanoparticle, antibody | 0.5–500 pM/0.17 pM | DPV | [54] |
| SPCE modified with polythiophene | Anti-5mC and FeO/N-trimethylchitosan/gold nanocomposite | 0.01–1000 pM/0.002 pM | DPV | [10] |
| SPCE | Biotinylated DNA probe immobilised on magnetic beads and anti-5mC | 87–2500 pM/26 pM | DPV | [46] |
| SPCE | Anti-5mC immobilised on magnetic beads and biotinylated DNA probe | 3.9–500 pM/1.2 pM | DPV | [7] |
| SPCE modified with rGO and polyvinyl alcohol | Anti-5mC immobilised and DNA probe conjugated with FeO-citric acid nanocomposites | 7×10–140.29 pM/6.31× 10 pM | DPV/EIS | [9] |
| rGO modified with ammuniom hydroxide | Anti5-mC and complementary DNA | 0.5–100 pM/0.012 pM | DPV/EIS | This work |
References
- Davis, C.D.; Uthus, E.O. DNA methylation, cancer susceptibility, and nutrient interactions. Exp. Biol. Med. 2004, 229, 988–995. [Google Scholar] [CrossRef]
- Syedmoradi, L.; Esmaeili, F.; Norton, M.L. Towards DNA methylation detection using biosensors. Analyst 2016, 141, 5922–5943. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Moriam, S.; Umer, M.; Nguyen, N.T.; Shiddiky, M.J. DNA methylation detection: Recent developments in bisulfite free electrochemical and optical approaches. Analyst 2018, 143, 4802–4818. [Google Scholar] [CrossRef]
- Kurdyukov, S.; Bullock, M. DNA methylation analysis: Choosing the right method. Biology 2016, 5, 3. [Google Scholar] [CrossRef]
- Shanmuganathan, R.; Basheer, N.B.; Amirthalingam, L.; Muthukumar, H.; Kaliaperumal, R.; Shanmugam, K. Conventional and nanotechniques for DNA methylation profiling. J. Mol. Diagn. 2013, 15, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Hossain, T.; Mahmudunnabi, G.; Masud, M.K.; Islam, M.N.; Ooi, L.; Konstantinov, K.; Al Hossain, M.S.; Martinac, B.; Alici, G.; Nguyen, N.T.; et al. Electrochemical biosensing strategies for DNA methylation analysis. Biosens. Bioelectron. 2017, 94, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Povedano, E.; Vargas, E.; Montiel, V.R.V.; Torrente-Rodríguez, R.M.; Pedrero, M.; Barderas, R.; San Segundo-Acosta, P.; Peláez-García, A.; Mendiola, M.; Hardisson, D. Electrochemical affinity biosensors for fast detection of gene-specific methylations with no need for bisulfite and amplification treatments. Sci. Rep. 2018, 8, 6418. [Google Scholar] [CrossRef]
- Chen, X.; Huang, J.; Zhang, S.; Mo, F.; Su, S.; Li, Y.; Fang, L.; Deng, J.; Huang, H.; Luo, Z.; et al. Electrochemical Biosensor for DNA Methylation Detection through Hybridization Chain-Amplified Reaction Coupled with a Tetrahedral DNA Nanostructure. ACS Appl. Mater. Interfaces 2019, 11, 3745–3752. [Google Scholar] [CrossRef] [PubMed]
- Khodaei, R.; Ahmady, A.; Khoshfetrat, S.M.; Kashanian, S.; Tavangar, S.M.; Omidfar, K. Voltammetric immunosensor for E-cadherin promoter DNA methylation using a Fe3O4-citric acid nanocomposite and a screen-printed carbon electrode modified with poly (vinyl alcohol) and reduced graphene oxide. Microchim. Acta 2019, 186, 170. [Google Scholar] [CrossRef] [PubMed]
- Daneshpour, M.; Moradi, L.S.; Izadi, P.; Omidfar, K. Femtomolar level detection of RASSF1A tumor suppressor gene methylation by electrochemical nano-genosensor based on Fe3O4/TMC/Au nanocomposite and PT-modified electrode. Biosens. Bioelectron. 2016, 77, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-based materials for biosensors: A review. Sensors 2017, 17, 2161. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Ratinac, K.R.; Ringer, S.P.; Thordarson, P.; Gooding, J.J.; Braet, F. Carbon nanomaterials in biosensors: Should you use nanotubes or graphene? Angew. Chem. Int. Ed. 2010, 49, 2114–2138. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene based electrochemical sensors and biosensors: A review. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Kuila, T.; Bose, S.; Khanra, P.; Mishra, A.K.; Kim, N.H.; Lee, J.H. Recent advances in graphene-based biosensors. Biosens. Bioelectron. 2011, 26, 4637–4648. [Google Scholar] [CrossRef] [PubMed]
- Szunerits, S.; Boukherroub, R. Graphene-based biosensors. Interface Focus 2018, 8, 20160132. [Google Scholar] [CrossRef]
- Li, B.; Pan, G.; Avent, N.D.; Lowry, R.B.; Madgett, T.E.; Waines, P.L. Graphene electrode modified with electrochemically reduced graphene oxide for label-free DNA detection. Biosens. Bioelectron. 2015, 72, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Wu, Y.; Tian, Y.; Li, G.; Liu, J.; Deng, P.; Chen, D. Facile electrochemical sensor for nanomolar rutin detection based on magnetite nanoparticles and reduced graphene oxide decorated electrode. Nanomaterials 2019, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haque, M.H.; Gopalan, V.; Yadav, S.; Islam, M.N.; Eftekhari, E.; Li, Q.; Carrascosa, L.G.; Nguyen, N.T.; Lam, A.K.; Shiddiky, M.J. Detection of regional DNA methylation using DNA-graphene affinity interactions. Biosens. Bioelectron. 2017, 87, 615–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elshafey, R.; Tavares, A.C.; Siaj, M.; Zourob, M. Electrochemical impedance immunosensor based on gold nanoparticles–protein G for the detection of cancer marker epidermal growth factor receptor in human plasma and brain tissue. Biosens. Bioelectron. 2013, 50, 143–149. [Google Scholar] [CrossRef]
- Welch, N.G.; Scoble, J.A.; Muir, B.W.; Pigram, P.J. Orientation and characterization of immobilized antibodies for improved immunoassays. Biointerphases 2017, 12, 02D301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trilling, A.K.; Beekwilder, J.; Zuilhof, H. Antibody orientation on biosensor surfaces: A minireview. Analyst 2013, 138, 1619–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Dubey, M.; Takahashi, H.; Castner, D.G.; Grainger, D.W. Immobilized antibody orientation analysis using secondary ion mass spectrometry and fluorescence imaging of affinity-generated patterns. Anal. Chem. 2010, 82, 2947–2958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Childres, I.; Jauregui, L.A.; Park, W.; Cao, H.; Chen, Y.P. Raman spectroscopy of graphene and related materials. New Dev. Photon Mater. Res. 2013, 1, 1–20. [Google Scholar]
- Zafar, Z.; Ni, Z.H.; Wu, X.; Shi, Z.X.; Nan, H.Y.; Bai, J.; Sun, L.T. Evolution of Raman spectra in nitrogen doped graphene. Carbon 2013, 61, 57–62. [Google Scholar] [CrossRef]
- Ni, Z.H.; Yu, T.; Lu, Y.H.; Wang, Y.Y.; Feng, Y.P.; Shen, Z.X. Uniaxial strain on graphene: Raman spectroscopy study and band-gap opening. ACS Nano 2008, 2, 2301–2305. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Pisana, S.; Chakraborty, B.; Piscanec, S.; Saha, S.K.; Waghmare, U.V.; Novoselov, K.S.; Krishnamurthy, H.R.; Geim, A.K.; Ferrari, A.C.; et al. Monitoring dopants by Raman scattering in an electrochemically top-gated graphene transistor. Nat. Nanotechnol. 2008, 3, 210–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Wang, P.; Yang, Y.; Luo, R.; Li, J.; Gu, X.; Zhan, Z.; Dong, Y.; Song, W.; Fan, R. Facile synthesis of nitrogen-doped reduced graphene oxide as an efficient counter electrode for dye-sensitized solar cells. J. Nanoparticle Res. 2018, 20, 110. [Google Scholar] [CrossRef]
- Baldovino, F.; Quitain, A.; Dugos, N.P.; Roces, S.A.; Koinuma, M.; Yuasa, M.; Kida, T. Synthesis and characterization of nitrogen-functionalized graphene oxide in high-temperature and high-pressure ammonia. RSC Adv. 2016, 6, 113924–113932. [Google Scholar] [CrossRef]
- Mohan, V.B.; Nieuwoudt, M.; Jayaraman, K.; Bhattacharyya, D. Quantification and analysis of Raman spectra of graphene materials. Graphene Technol. 2017, 2, 47–62. [Google Scholar] [CrossRef]
- Ederer, J.; Janoš, P.; Ecorchard, P.; Tolasz, J.; Štengl, V.; Beneš, H.; Perchacz, M.; Pop-Georgievski, O. Determination of amino groups on functionalized graphene oxide for polyurethane nanomaterials: XPS quantitation vs. functional speciation. RSC Adv. 2017, 7, 12464–12473. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.M.; Zhang, Q.; Zhao, X.C.; Zhang, B.; Kong, Q.Q.; Yang, M.G.; Yang, Q.H.; Wang, M.Z.; Yang, Y.G.; Schlögl, R.; et al. Hierarchically aminated graphene honeycombs for electrochemical capacitive energy storage. J. Mater. Chem. 2012, 22, 14076–14084. [Google Scholar] [CrossRef] [Green Version]
- Petit, C.; Seredych, M.; Bandosz, T.J. Revisiting the chemistry of graphite oxides and its effect on ammonia adsorption. J. Mater. Chem. 2009, 19, 9176–9185. [Google Scholar] [CrossRef] [Green Version]
- Al-Gaashani, R.; Najjar, A.; Zakaria, Y.; Mansour, S.; Atieh, M. XPS and structural studies of high quality graphene oxide and reduced graphene oxide prepared by different chemical oxidation methods. Ceram. Int. 2019, 45, 14439–14448. [Google Scholar] [CrossRef]
- Lai, L.; Chen, L.; Zhan, D.; Sun, L.; Liu, J.; Lim, S.H.; Poh, C.K.; Shen, Z.; Lin, J. One-step synthesis of NH2-graphene from in situ graphene-oxide reduction and its improved electrochemical properties. Carbon 2011, 49, 3250–3257. [Google Scholar] [CrossRef]
- Stobinski, L.; Lesiak, B.; Malolepszy, A.; Mazurkiewicz, M.; Mierzwa, B.; Zemek, J.; Jiricek, P.; Bieloshapka, I. Graphene oxide and reduced graphene oxide studied by the XRD, TEM and electron spectroscopy methods. J. Electron Spectrosc. Relat. Phenom. 2014, 195, 145–154. [Google Scholar] [CrossRef]
- Thakur, V.K.; Thakur, M.K. Chemical Functionalization of Carbon Nanomaterials: Chemistry and Applications; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Tang, S.; Cao, Z. Adsorption and dissociation of ammonia on graphene oxides: A first-principles study. J. Phys. Chem. C 2012, 116, 8778–8791. [Google Scholar] [CrossRef]
- Mattson, E.C.; Pande, K.; Unger, M.; Cui, S.; Lu, G.; Gajdardziska-Josifovska, M.; Weinert, M.; Chen, J.; Hirschmugl, C.J. Exploring adsorption and reactivity of NH3 on reduced graphene oxide. J. Phys. Chem. C 2013, 117, 10698–10707. [Google Scholar] [CrossRef]
- Rivera, L.; Betancur, A.; Zarate, D.; Torres, D.T.; Hoyos, L.; Garcia, A. Reduction and simultaneous doping of graphene oxide to repel LDL in treatment of atherosclerosis disease. arXiv 2019, arXiv:1902.01850. [Google Scholar]
- Tang, S.; Cao, Z. Adsorption of nitrogen oxides on graphene and graphene oxides: Insights from density functional calculations. J. Chem. Phys. 2011, 134, 044710. [Google Scholar] [CrossRef] [PubMed]
- Benvidi, A.; Firouzabadi, A.D.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Tezerjani, M.D. Ultrasensitive DNA sensor based on gold nanoparticles/reduced graphene oxide/glassy carbon electrode. Anal. Biochem. 2015, 484, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.J.; Niu, D.J.; Sun, J.Y.; Zhu, J.J. An electrochemical amperometric immunobiosensor for label-free detection of α-fetoprotein based on amine-functionalized graphene and gold nanoparticles modified carbon ionic liquid electrode. J. Electroanal. Chem. 2011, 656, 72–77. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, Z. Fabrication of an ultrasensitive electrochemical immunosensor for CEA based on conducting long-chain polythiols. Biosens. Bioelectron. 2013, 46, 1–7. [Google Scholar] [CrossRef]
- Singh, A.; Sinsinbar, G.; Choudhary, M.; Kumar, V.; Pasricha, R.; Verma, H.; Singh, S.P.; Arora, K. Graphene oxide-chitosan nanocomposite based electrochemical DNA biosensor for detection of typhoid. Sens. Actuators B Chem. 2013, 185, 675–684. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef]
- Povedano, E.; Valverde, A.; Montiel, V.R.V.; Pedrero, M.; Yáñez-Sedeño, P.; Barderas, R.; San Segundo-Acosta, P.; Peláez-García, A.; Mendiola, M.; Hardisson, D.; et al. Rapid Electrochemical Assessment of Tumor Suppressor Gene Methylations in Raw Human Serum and Tumor Cells and Tissues Using Immunomagnetic Beads and Selective DNA Hybridization. Angew. Chem. Int. Ed. 2018, 57, 8194–8198. [Google Scholar] [CrossRef]
- Trotter, M.; Borst, N.; Thewes, R.; von Stetten, F. Electrochemical DNA sensing–Principles, commercial systems, and applications. Biosens. Bioelectron. 2020, 154, 112069. [Google Scholar] [CrossRef]
- Gorodetsky, A.A.; Green, O.; Yavin, E.; Barton, J.K. Coupling into the base pair stack is necessary for DNA-mediated electrochemistry. Bioconjugate Chem. 2007, 18, 1434–1441. [Google Scholar] [CrossRef] [Green Version]
- Inouye, M.; Ikeda, R.; Takase, M.; Tsuri, T.; Chiba, J. Single-nucleotide polymorphism detection with “wire-like” DNA probes that display quasi “on–off” digital action. Proc. Natl. Acad. Sci. USA 2005, 102, 11606–11610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zhang, X.; Zhao, K. Methylation-specific electrochemical biosensing strategy for highly sensitive and quantitative analysis of promoter methylation of tumor-suppressor gene in real sample. J. Electroanal. Chem. 2016, 773, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Suijkerbuijk, K.P.; Pan, X.; Van Der Wall, E.; Van Diest, P.J.; Vooijs, M. Comparison of different promoter methylation assays in breast cancer. Anal. Cell. Pathol. 2010, 33, 133–141. [Google Scholar] [CrossRef]
- Sethi, J.; Van Bulck, M.; Suhail, A.; Safarzadeh, M.; Perez-Castillo, A.; Pan, G. A label-free biosensor based on graphene and reduced graphene oxide dual-layer for electrochemical determination of beta-amyloid biomarkers. Microchim. Acta 2020, 187, 1–10. [Google Scholar]
- Mabbott, G.A. An introduction to cyclic voltammetry. J. Chem. Educ. 1983, 60, 697. [Google Scholar] [CrossRef]
- Xu, Z.; Yin, H.; Huo, L.; Zhou, Y.; Ai, S. Electrochemical immunosensor for DNA methyltransferase activity assay based on methyl CpG-binding protein and dual gold nanoparticle conjugate-based signal amplification. Sens. Actuators B Chem. 2014, 192, 143–149. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safarzadeh, M.; Suhail, A.; Sethi, J.; Sattar, A.; Jenkins, D.; Pan, G. A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation. Nanomaterials 2021, 11, 985. https://doi.org/10.3390/nano11040985
Safarzadeh M, Suhail A, Sethi J, Sattar A, Jenkins D, Pan G. A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation. Nanomaterials. 2021; 11(4):985. https://doi.org/10.3390/nano11040985
Chicago/Turabian StyleSafarzadeh, Mina, Ahmed Suhail, Jagriti Sethi, Anas Sattar, David Jenkins, and Genhua Pan. 2021. "A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation" Nanomaterials 11, no. 4: 985. https://doi.org/10.3390/nano11040985
APA StyleSafarzadeh, M., Suhail, A., Sethi, J., Sattar, A., Jenkins, D., & Pan, G. (2021). A Label-Free DNA-Immunosensor Based on Aminated rGO Electrode for the Quantification of DNA Methylation. Nanomaterials, 11(4), 985. https://doi.org/10.3390/nano11040985








