Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes
Abstract
1. Introduction
2. Materials and Methods
2.1. HN Dispersion
2.2. HNs–DNA Mixing
2.3. ATR-FTIR Measurements
2.4. Nano-FTIR Measurements
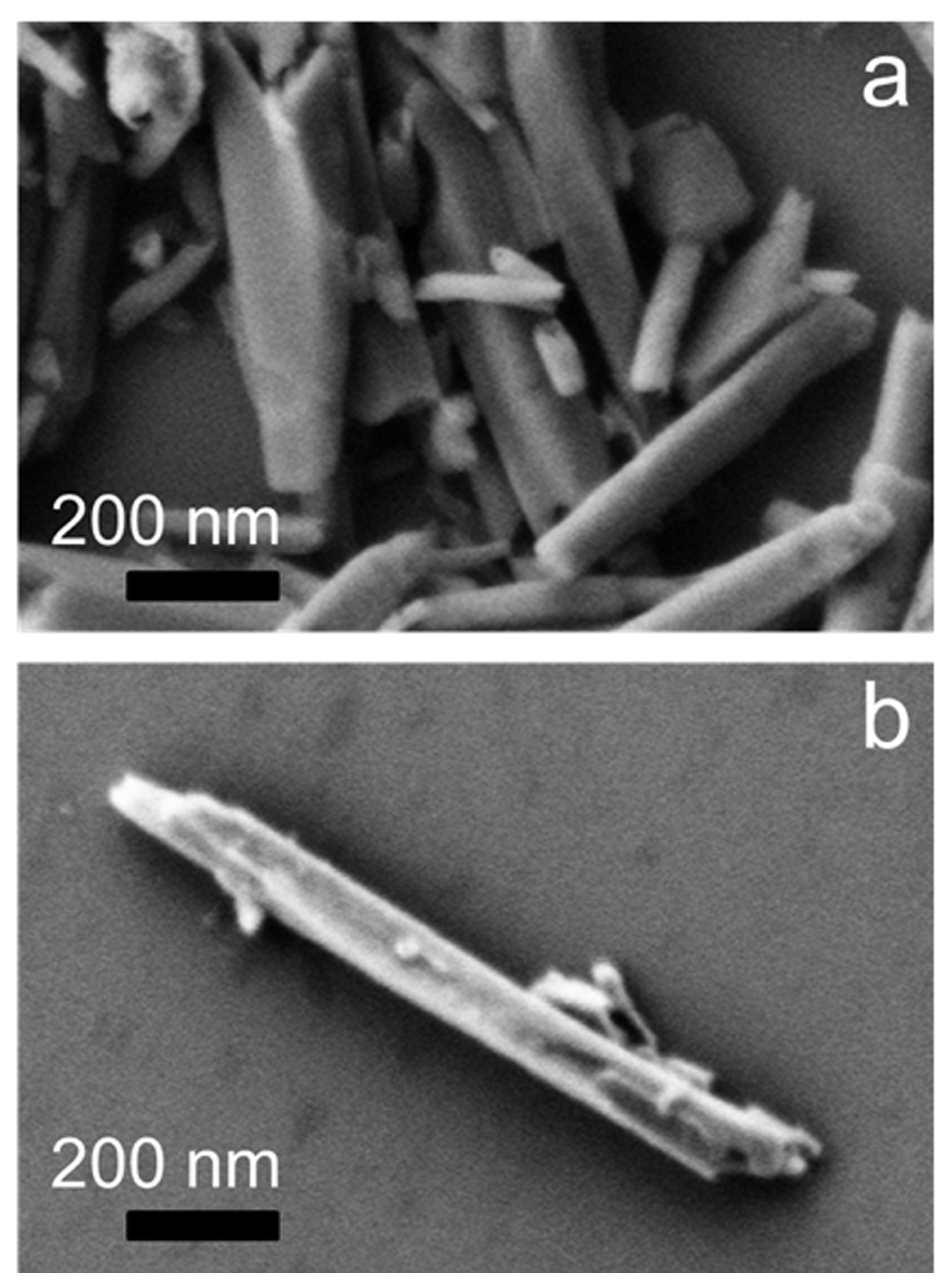
2.5. SEM Microscopy
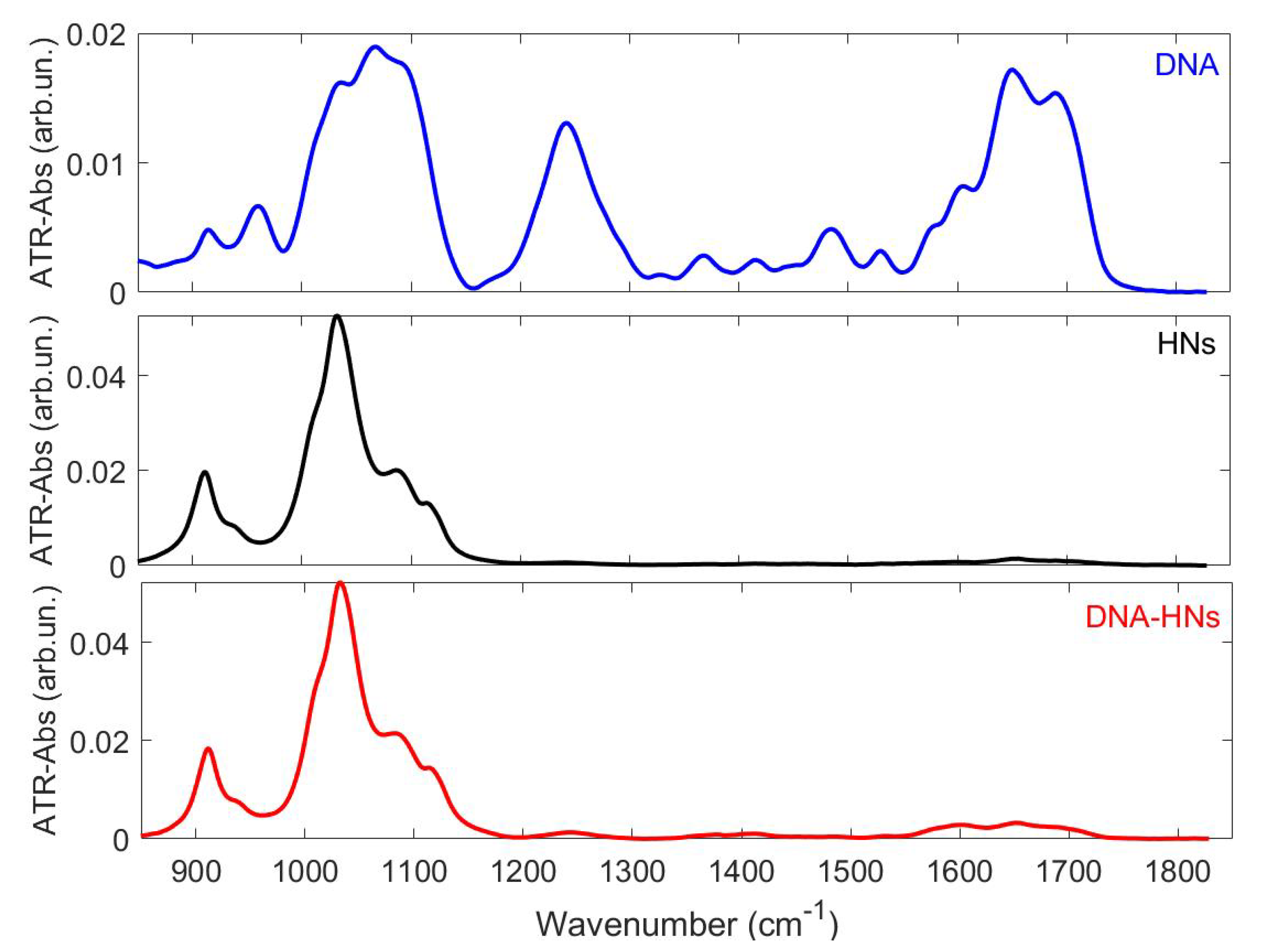
3. Results and Discussion
3.1. Far-Field FTIR Analysis
3.2. Nano-FTIR Analysis: DNA Phosphates and Nitrogen Bases Absorption
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bianco, A.; Kostaleros, K.; Prato, M. Applications of carbon nanotubes in drug delivery. Curr. Opin. Chem. Biol. 2005, 9, 674–679. [Google Scholar] [CrossRef]
- Yoshida, N. Discovery and application of the Yoshida effect: Nano-sized acicular materials enable penetration of bacterial cells by sliding friction force. Recent Patents Biotechnol. 2007, 1, 194–201. [Google Scholar] [CrossRef]
- Santos, A.C.; Pereira, I.; Reis, S.; Veiga, F.; Saleh, M.; Lvov, Y. Biomedical potential of clay nanotube formulations and their toxicity assessment. Expert Opin. Drug Deliv. 2019, 16, 1169–1182. [Google Scholar] [CrossRef]
- Setter, O.P.; Segal, E. Halloysite nanotubes at the nano-bio interface. Nanoscale 2020, 12, 23444–23460. [Google Scholar] [CrossRef]
- Lvov, Y.; DE Villiers, M.; Fakhrullin, R. The application of halloysite tubule nanoclay in drug delivery. Expert Opin. Drug Deliv. 2016, 13, 977–986. [Google Scholar] [CrossRef]
- Shamsi, M.H.; Geckeler, K.E. The first biopolymer-wrapped non-carbon nanotubes. Nanotechnology 2008, 19, 075604. [Google Scholar] [CrossRef]
- Shi, Y.F.; Tian, Z.; Zhang, Y.; Shen, H.B.; Jia, N.Q. Functionalized halloysite nanotube-based carrier for intracellular delivery of antisense oligonucleotides. Nanoscale Res. Lett. 2011, 6, 608. [Google Scholar] [CrossRef]
- Rawtani, D.; Agrawal, Y. Study the interaction of DNA with halloysite nanotube-gold nanoparticle based composite. J. Bionanosci. 2012, 6, 95–98. [Google Scholar] [CrossRef]
- Wu, H.; Shi, Y.; Huang, C.; Zhang, Y.; Wu, J.; Shen, H.; Jia, N. Multifunctional nanocarrier based on clay nanotubes for efficient intracellular siRNA delivery and gene silencing. J. Biomater. Appl. 2014, 28, 1180–1189. [Google Scholar] [CrossRef]
- Santos, A.C.; Ferreira, C.; Veiga, F.; Ribeiro, A.J.; Panchal, A.; Lvov, Y.; Agarwal, A. Halloysite clay nanotubes for life sciences applications: From drug encapsulation to bioscaffold. Adv. Colloid Interface Sci. 2018, 257, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jung, G.E.; Cho, S.J.; Geckeler, K.E.; Fuchs, H. Cellular interactions of doxorubicin-loaded DNA-modified halloysite nanotubes. Nanoscale 2013, 5, 8577–8585. [Google Scholar] [CrossRef] [PubMed]
- Massaro, M.; Iborra, C.V.; Cavallaro, G.; Colletti, C.G.; Garcia-Villen, F.; Lazzara, G.; Riela, S. Synthesis and Characterization of Nanomaterial Based on Halloysite and Hectorite Clay Minerals Covalently Bridged. Nanomaterials 2021, 11, 506. [Google Scholar] [CrossRef]
- Yuan, P.; Tan, D.; Annabi-Bergaya, F. Properties and applications of halloysite nanotubes: Recent research advances and future prospects. Appl. Clay Sci. 2015, 112, 75–93. [Google Scholar] [CrossRef]
- Buck, J.; Grossen, P.; Cullis, P.R.; Huwyler, J.; Witzigmann, D. Lipid-Based DNA Therapeutics: Hallmarks of Non-Viral Gene Delivery. ACS Nano 2019, 13, 3754–3782. [Google Scholar] [CrossRef] [PubMed]
- Massaro, M.; Barone, G.; Biddeci, G.; Cavallaro, G.; Di Blasi, F.; Lazzara, G.; Nicotra, G.; Spinella, N.; Spinelli, G.; Riela, S. Halloysite nanotubes-carbon dots hybrids multifunctional nanocarrier with positive cell target ability as a potential non-viral vector for oral gene therapy. J. Colloid Interface Sci. 2019, 552, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Vergaro, V.; Abdullayev, E.; Lvov, Y.M.; Zeitoun, A.; Cingolani, R.; Rinaldi, R.; Leporatti, S. Cytocompatibility and uptake of halloysite clay nanotubes. Biomacromolecules 2010, 11, 820–826. [Google Scholar] [CrossRef]
- Tully, J.; Yendluri, R.; Lvov, Y. Halloysite clay nanotubes for enzyme immobilization. Biomacromolecules 2016, 17, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Duce, C.; Della Porta, V.; Bramanti, E.; Campanella, B.; Spepi, A.; Tiné, M.R. Loading of halloysite nanotubes with BSA α-Lac and β-Lg: A Fourier transform infrared spectroscopic and thermogravimetric study. Nanotechnology 2016, 28, 055706. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, D.; Liu, Q.; Komarneni, S. A comparative study of synthetic tubular kaolinite nanoscrolls and natural halloysite nanotubes. Appl. Clay Sci. 2019, 168, 421–427. [Google Scholar] [CrossRef]
- Batasheva, S.; Kryuchkova, M.; Fakhrullin, R.; Cavallaro, G.; Lazzara, G.; Akhatova, F.; Nigamatzyanova, L.; Evtugyn, V.; Rozhina, E.; Fakhrullin, R. Facile Fabrication of Natural Polyelectrolyte-Nanoclay Composites: Halloysite Nanotubes, Nucleotides and DNA Study. Molecules 2020, 25, 3557. [Google Scholar] [CrossRef]
- Piccirilli, F.; Plotegher, N.; Ortore, M.; Tessari, I.; Brucale, M.; Spinozzi, F.; Beltramini, M.; Mariani, P.; Militello, V.; Lupi, S.; et al. High-Pressure-Driven Reversible Dissociation of α-Synuclein Fibrils Reveals Structural Hierarchy. Biophys. J. 2017, 113, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Vetri, V.; Piccirilli, F.; Krausser, J.; Buscarino, G.; Łapinska, U.; Vestergaard, B.; Zaccone, A.; Foderá, V. Ethanol controls the self-assembly and mesoscopic properties of human insulin amyloid spherulites. J. Phys. Chem. B 2018, 122, 3101–3112. [Google Scholar] [CrossRef]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Billè, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of ribonucleic acid (RNA) to the Fourier transform infrared (FTIR) spectrum of eukaryotic cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef]
- Lupi, S.; Nucara, A.; Perucchi, A.; Calvani, P.; Ortolani, M.; Quaroni, L.Q.; Kiskinova, M. Performance of SISSI, the infrared beamline of the ELETTRA storage ring. J. Opt. Soc. Am. B 2007, 24, 959–964. [Google Scholar] [CrossRef]
- Roy, R.; Brubach, J.B.; Calvani, P.; de Marzi, G.; Filabozzi, A.; Gerschel, A.; Giura, P.; Lupi, S.; Marcouillas, O.; Mermet, A.; et al. Infrared synchrotron radiation: From the production to the spectroscopic and microscopic applications. In Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment; Elsevier: Amsterdam, The Netherlands, 2001; Volume 467–468, pp. 426–436. [Google Scholar] [CrossRef]
- Huth, F.; Govyadinov, A.; Amarie, S.; Nuansing, W.; KEilman, F.; Hillenbrand, R. Nano-FTIR Absorption Spectroscopy of Molecular Fingerprints at 20 nm Spatial Resolution. Nano Lett. 2012, 12, 3973–3978. [Google Scholar] [CrossRef] [PubMed]
- Taubner, T.; Hillenbrand, R.; Keilmann, F. Nanoscale polymer recognition by spectral signature in scattering infrared near-field microscopy. Appl. Phys. Lett. 2004, 85, 5064–5066. [Google Scholar] [CrossRef]
- Birarda, G.; Delneri, A.; Lagatolla, C.; Parisse, P.; Cescutti, P.; Vaccari, L.; Rizzo, R. Multi-technique microscopy investigation on bacterial biofilm matrices: A study on Klebsiella pneumoniae clinical strains. Anal. Bioanal. Chem. 2019, 411, 7315–7325. [Google Scholar] [CrossRef]
- Tardani, F.; Casciardi, S.; Ruzicka, B.; Sennato, S. Salt enhanced sedimentation of halloysite nantubes for precise determination of DNA absorption isotherm. Colloids Surf. Physicochem. Eng. Asp. 2020, 605, 125400. [Google Scholar] [CrossRef]
- Tardani, F.; Sarti, S.; Sennato, S.; Leo, M.; Filetici, P.; Casciardi, S.; Schiavi, P.G.; Bordi, F. Experimental evidence of single-stranded DNA adsorption on multi-walled carbon nanotubes. J. Phys. Chem. B 2020, 124, 2305–2526. [Google Scholar] [CrossRef]
- Beall, G.W.; Sowersby, D.S.; Roberts, R.D.; Robson, M.H.; Lewis, L.K. Analysis of Oligonucleotide DNA Binding and Sedimentation Properties of Montmorillonite Clay Using Ultraviolet Light Spectroscopy. Biomacromolecules 2009, 10, 105–112. [Google Scholar] [CrossRef]
- Piccirilli, F.; Mangialardo, S.; Postorino, P.; Lupi, S.; Perucchi, A. FTIR analysis of the high pressure response of native insulin assemblies. J. Mol. Struct. 2012, 1050, 159–165. [Google Scholar] [CrossRef]
- Piccirilli, F.; Schirò, G.; Vetri, V.; Lupi, S.; Perucchi, A.; Militello, V. Decoding vibrational states of Concanavalin A amyloid fibrils. Biophys. Chem. 2015, 199, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zha, Y.; Zhu, T.; Li, Y.; Ruan, J.; Li, G. Effective solvent-free oxidation of cyclohexene to allylic products with oxygen by mesoporous etched halloysite nanotube supported Co2+. RSC Adv. 2018, 8, 14870. [Google Scholar] [CrossRef]
- Oh, T.; Choi, C.K. Spectroscopic Characterization of PEG-DNA Biocomplexes by FTIR. J. Korean Phys. Soc. 2010, 56, 1150–1155. [Google Scholar] [CrossRef]
- Dovbeshko, G.I.; Gridina, N.Y.; Kruglova, E.B.; Pashchuk, O.P. FTIR spectroscopy studies of nucleic acid damage. Talanta 2000, 53, 233–246. [Google Scholar] [CrossRef]
- Mello, M.L.S.; Vidal, B.C. Changes in the Infrared Microspectroscopic Characteristics of DNA Caused by Cationic Elements, Different Base Richness and Single-Stranded Form. PLoS ONE 2012, 7, e43169. [Google Scholar] [CrossRef]
- Tobin, M.C. The Raman spectra of calf thymus and of salmon testes deoxyribonucleic acid. Spectrochim. Acta 1969, 25A, 1855. [Google Scholar] [CrossRef]
- Han, Y.; Han, L.; Yao, Y.; Li, Y.; Liu, X. Key factors in FTIR spectroscopic analysis of DNA: The sampling 1 technique, pretreatment temperature and sample concentration. Anal. Methods 2018, 21, 2436–2443. [Google Scholar] [CrossRef]
- Kelly, J.G.; Najand, G.M.; Martin, F.L. Characterisation of DNA methylation status using spectroscopy (mid-IR versus Raman) with multivariate analysis. J. Biophotonics 2010, 4, 345–354. [Google Scholar] [CrossRef]
- Elmlund, L.; Söderberg, P.; Suriyanarayanan, S.; Nicholls, I.A. A Phage Display Screening Derived Peptide with Affinity for the Adeninyl Moiety. Biosensors 2014, 4, 137–149. [Google Scholar] [CrossRef]
- Neault, J.F.; Tajmir-Riahi, H.A. Spectroscopic Characterization of PEG-DNA Biocomplexes by FTIR. Biophys. J. 1999, 76, 2177–2182. [Google Scholar] [CrossRef]
- Jangir, D.K.; Kundu, S.; Mehrotra, R. Role of Minor Groove Width and Hydration pattern on Amsacrine interaction with DNA. PLoS ONE 2013, 8, e69933. [Google Scholar] [CrossRef]
- Hassan, A.; Macedo, L.J.A.; de Souza, J.C.P.; Lima, F.C.D.A.; Crespiho, F.N. A combined Far-FTIR, FTIR spectromicroscopy, and DFT study of the effect of DNA Binding on the [4Fe4S] Cluster site in EndoII. Sci. Rep. 2020, 10, 1931. [Google Scholar] [CrossRef]
- Tallandier, E.; Liquier, J. Infrared spectroscopy of DNA. Method Enzym. 1991, 221, 307–335. [Google Scholar]
- Banyay, M.; Sarkar, M.; Graslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2015, 45, 1980–1988. [Google Scholar] [CrossRef] [PubMed]
- Olsztynska-janus, S.; Szymborska-Malek, K.; Gasior-Glogowska, M.; Walski, T.; Komorowska, M.; Witkiewicz, W.; Pezowicz, C.; Kobielarz, M.; Szotek, S. Spectroscopic techniques in the study of human tissues and their components. Part I: IR spectroscopy. Acta Bioeng. Biomech. 2012, 14, 101–115. [Google Scholar] [PubMed]
- Neumann, T.; Gajria, S.; Tirrell, M.; Jaeger, L. Reversible structural switching of a DNA-DDAB film. J. Am. Chem. Soc. 2009, 131, 3440. [Google Scholar] [CrossRef]
- Auffinger, P.; Bielecki, L.; Westhof, E. Anion Binding to Nucleic Acids. Structure 2004, 12, 379–388. [Google Scholar] [CrossRef]


| Chemical Assignment | DNA cm | HNs cm | DNA–HNs cm | Nano DNA–HNs cm |
|---|---|---|---|---|
| Si-O (amorphous) [35], PO as [23] | 1242 | 1243 | 1243 | 1253 |
| C-N s (A,T) [36] | 1330 | - | - | 1330 |
| C-N s (C,G)) [36] | 1368 | - | 1366 | 1358 |
| Base in-plane vibration (C,G) [37,38,39] | 1415 | - | 1415 | 1421 |
| C-C C-N (G,C) [40] | 1485 | - | 1484 | 1482 |
| Base in-plane vibration (C,G) | 1530 | - | 1530 | 1520 |
| C-C C-N (C) [40], N-H (A) [41] | 1575 | - | - | 1570 |
| (A) [42] | 1602 | - | 1602 | - |
| OH (G,C,T,A) | 1650 | 1650 | 1650 | 1652 |
| C=O (T) [43,44] | 1698 | - | 1698 | - |
| C=O of nucleobases [37,39,45,46] | 1707 | - | 1710 | 1706 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccirilli, F.; Tardani, F.; D’Arco, A.; Birarda, G.; Vaccari, L.; Sennato, S.; Casciardi, S.; Lupi, S. Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes. Nanomaterials 2021, 11, 1103. https://doi.org/10.3390/nano11051103
Piccirilli F, Tardani F, D’Arco A, Birarda G, Vaccari L, Sennato S, Casciardi S, Lupi S. Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes. Nanomaterials. 2021; 11(5):1103. https://doi.org/10.3390/nano11051103
Chicago/Turabian StylePiccirilli, Federica, Franco Tardani, Annalisa D’Arco, Giovanni Birarda, Lisa Vaccari, Simona Sennato, Stefano Casciardi, and Stefano Lupi. 2021. "Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes" Nanomaterials 11, no. 5: 1103. https://doi.org/10.3390/nano11051103
APA StylePiccirilli, F., Tardani, F., D’Arco, A., Birarda, G., Vaccari, L., Sennato, S., Casciardi, S., & Lupi, S. (2021). Infrared Nanospectroscopy Reveals DNA Structural Modifications upon Immobilization onto Clay Nanotubes. Nanomaterials, 11(5), 1103. https://doi.org/10.3390/nano11051103









