Effect of Fluorescent Labels on DNA Affinity for Gold Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Oligonucleotides and Their Derivatives
2.3. Molecular Properties of FDs
2.4. Hydrophobicity of ONs and ON-FDs
2.5. Preparation and Characterization of Citrate-Coated GNPs
2.6. Preparation of All ON/GNPs and ON-FD/GNPs
2.7. The Langmuir Isotherm
2.8. An Equilibrium Dissociation Constant
2.9. ON Surface Density
2.10. Stability of All ON/GNPs and ON-FD/GNPs
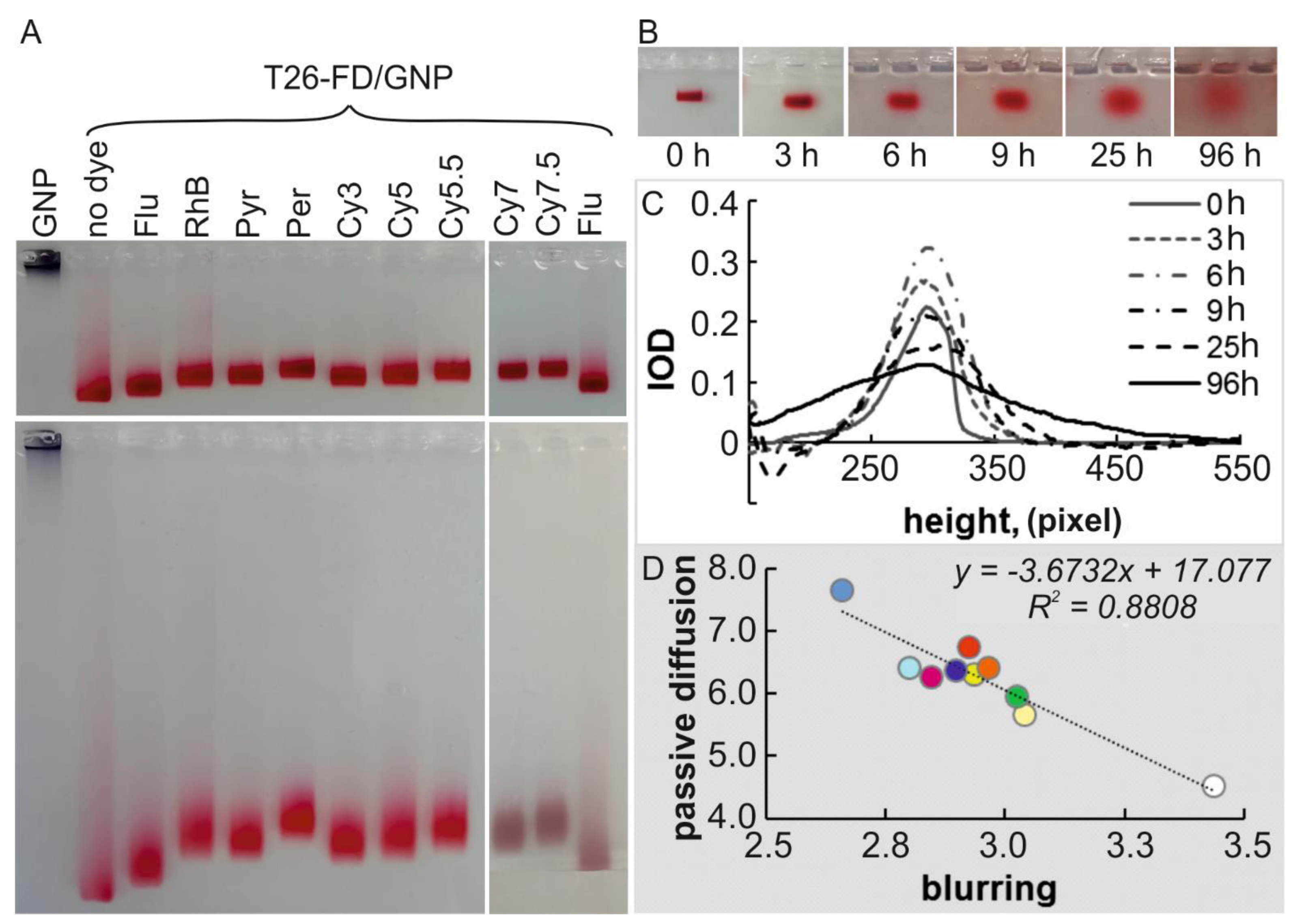
2.10.1. Blurring of Bands during Electrophoresis
2.10.2. Diffusion of Bands without Electric Field
2.11. Nonlinear Regression
2.12. Statistics
3. Results
3.1. Background Information
3.2. Adsorption of Fluorescently Labeled ONs to GNPs
3.3. Spectrophotometric and Spectrofluorometric Analysis of T26-FD/GNPs Associates
3.4. Hydrodynamic Size and Net Charge of T-26-FD/GNPs
3.5. Electrophoretic Analysis of Non-Covalent Adsorption of T26-FDs on GNPs
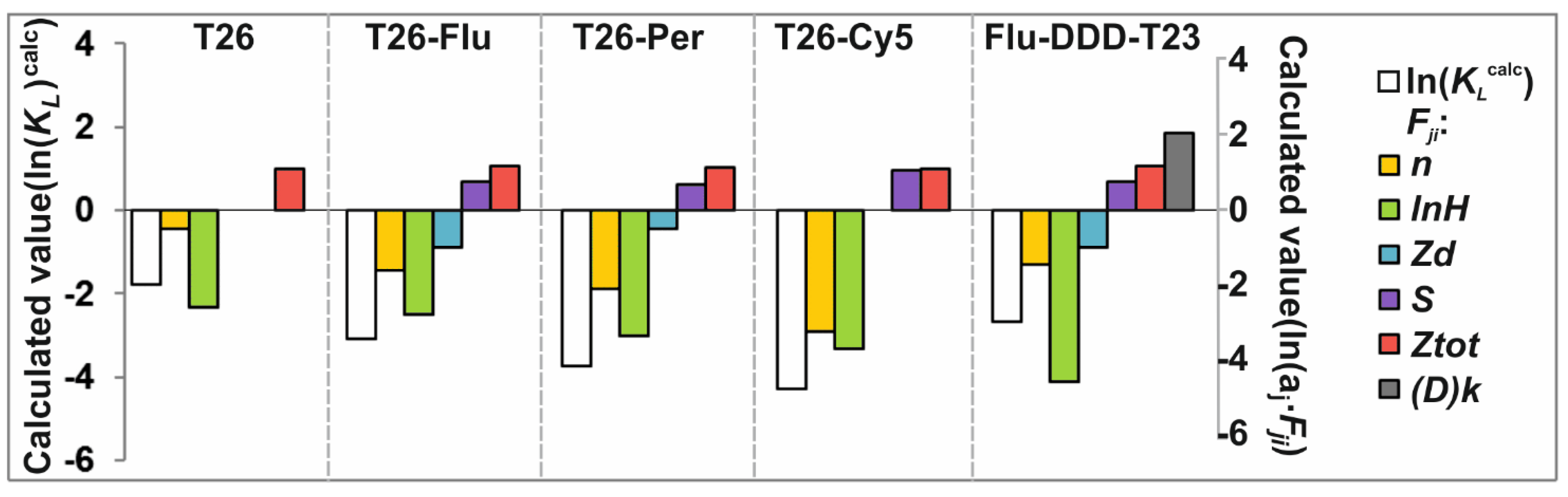
3.6. Analysis of Relationship of ON-FDs Hydrophobicity and Their Affinity for GNP
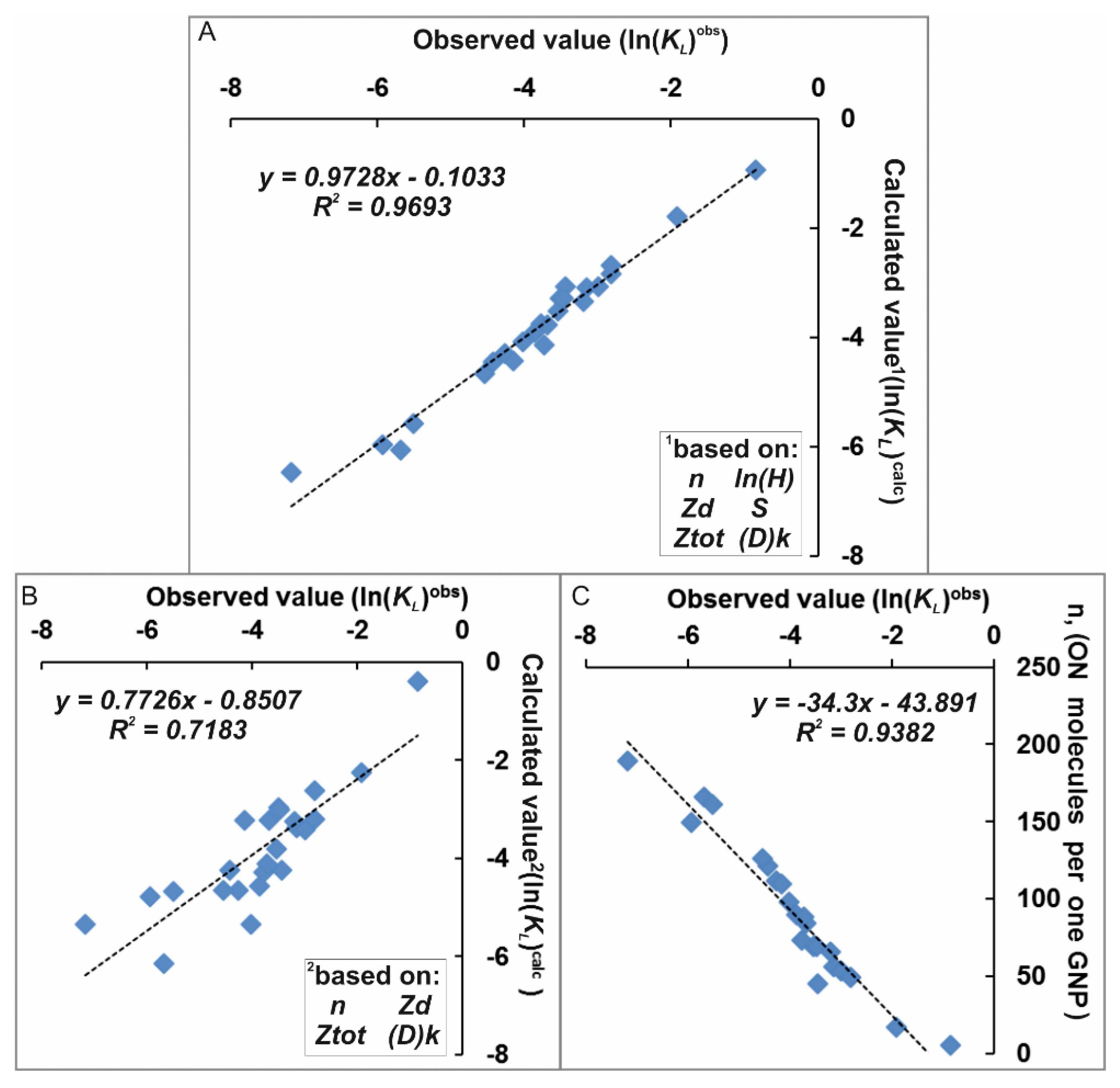
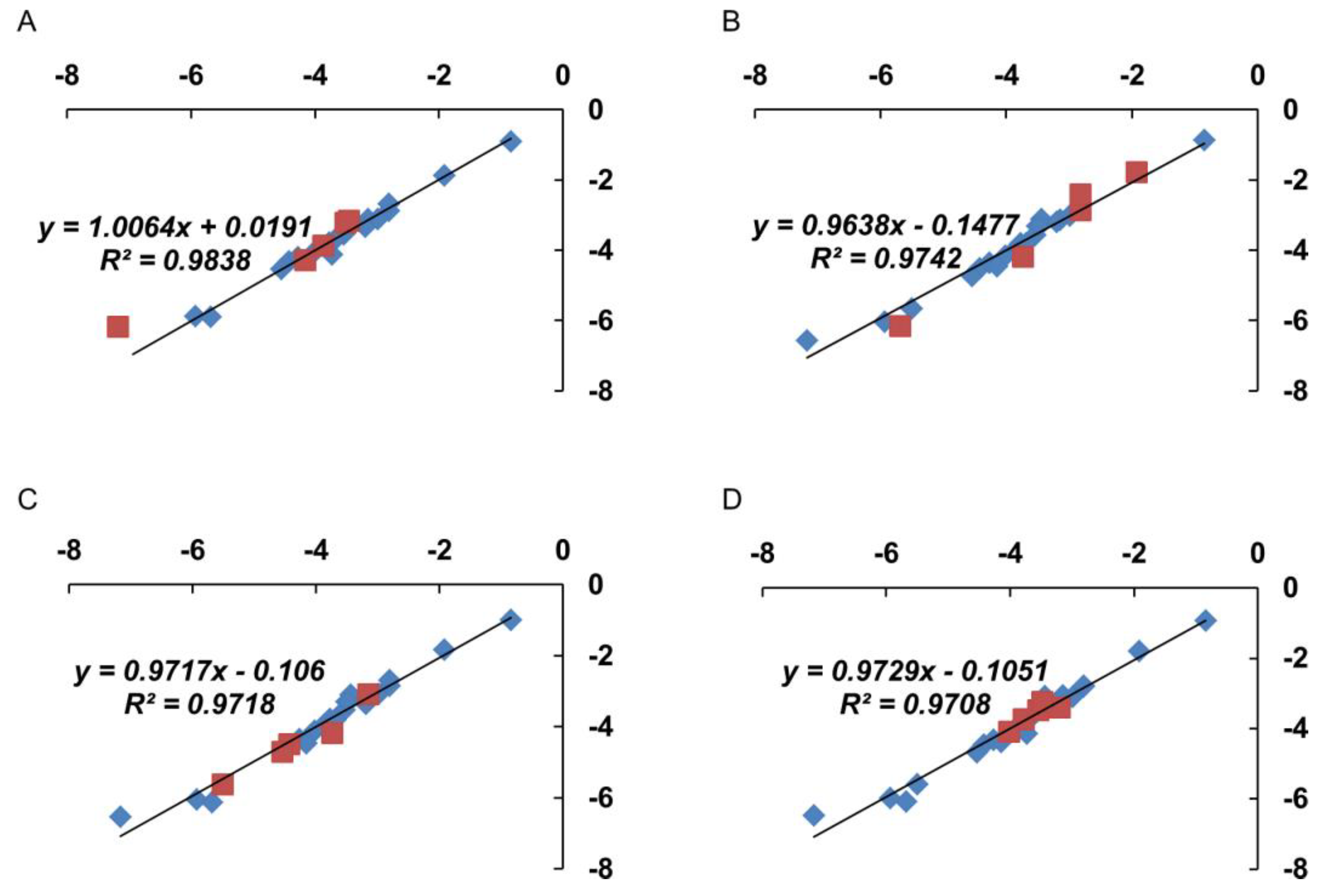
3.7. Correlation Analysis
3.8. Regression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ON | oligodeoxynucleotide |
| GNP | gold nanoparticle |
| ON/GNP | non-covalent associate–nanoconstruct obtained by non-covalent binding (synonyms: interaction, adsorption) of ON and GNP |
| ON-GNP | covalent conjugate of ON and GNP–nanoconstruct obtained by covalent binding (synonym: immobilization) of ON and GNP |
| FD | fluorescent dye (fluorescent label) |
| ON-FD | ON labeled with FD |
| ε | FD molar coefficient of absorption |
| n | surface density number of ONs attached to surface of one GNP |
| RP HPLC | reversed-phase high-performance liquid chromatography |
| obs | observed (or experimental) value |
| calc | calculated value |
| ref | reference data |
| KL | Langmuir constant of the associate |
| KD | Equilibrium Dissociation Constant of the associate |
| tr | hydrophobicity or retention time of ONs or ON-FDs |
| H | hydrophobicity coefficient of ONs or ON-FDs |
| Zd | the total charge of the FD and the linker between the FD and the ON |
| pd | passive diffusion of non-covalent associates in an agarose gel without electric field |
| bl | blurring of the main band of an associate in an agarose gel in the presence of an electric field |
| Mr | molecular weight of the ONs or ON-FDs |
| S | fluorophore area |
| dH | hydrodynamic diameter of non-covalent associate |
| ζ | associate surface potential |
| μ | normalized electrophoretic mobility of associate in the agarose gel |
| λex (λem) | fluorophore excitation (emission) wavelength |
| IOD | integrated optical density |
| (D)k | number (k) of aliphatic dodecyl residues (D) |
References
- Busatto, S.; Pham, A.; Suh, A.; Shapiro, S.; Wolfram, J. Organotropic drug delivery: Synthetic nanoparticles and extracellular vesicles. Biomed. Microdevices 2019, 21, 46. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, F.; Ding, J.; Liu, J. Tandem phosphorothioate modifications for DNA ad-sorption strength and polarity control on gold nanoparticles. ACS Appl. Mater. Interfaces 2014, 6, 14795–14800. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wan, Y.; Liu, H.; Su, Y.; Fan, C.; Wei, M.; Li, F.; Huang, Q.; Pei, H. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA–gold nanoparticle nanoconjugates. J. Am. Chem. Soc. 2012, 134, 11876–11879. [Google Scholar]
- Chen, N.; Wei, M.; Sun, Y.; Li, F.; Pei, H.; Li, X.; Su, S.; He, Y.; Wang, L.; Shi, J.; et al. Self-assembly of poly-adenine-tailed CpG oligonucleotide-gold nanoparticle nanoconjugates with immunostimulatory activity. Small 2014, 10, 368–375. [Google Scholar] [CrossRef]
- Yao, G.; Pei, H.; Li, J.; Zhao, Y.; Zhu, D.; Zhang, Y.; Lin, Y.; Huang, Q.; Fan, C. Clicking DNA to gold nanoparticles: Poly-adenine-mediated formation of monovalent DNA-gold nanoparticle conjugates with nearly quantitative yield. NPG Asia Mater. 2015, 7, e159. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, B.; Dave, N.; Servos, M.R.; Liu, J. Instantaneous attachment of an ultrahigh density of nonthiolated DNA to gold nanoparticles and its applications. Langmuir 2012, 28, 17053–17060. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wang, L.; Li, J.; Zhao, Y.; Zhou, Z.; Shi, J.; Zuo, X.; Pan, D. Quantitative investigation of the poly-adenine DNA dissociation from the surface of gold nanoparticles. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, B.; Servos, M.R.; Liu, J. Polarity control for nonthiolated DNA adsorption onto gold nanoparticles. Langmuir 2013, 29, 6091–6098. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mandal, S.; Mathew, S.P.; Selvakannan, P.R.; Mandale, A.B.; Chaudhari, R.V.; Sastry, M. Benzene- and anthracene-mediated assembly of gold nanoparticles at the liquid-liquid interface. Langmuir 2002, 18, 6478–6483. [Google Scholar] [CrossRef]
- Curry, D.; Cameron, A.; MacDonald, B.; Nganou, C.; Scheller, H.; Marsh, J.; Beale, S.; Lu, M.; Shan, Z.; Kaliaperumal, R.; et al. Adsorption of doxorubicin on citrate-capped gold nanoparticles: Insights into engineering potent chemotherapeutic delivery systems. Nanoscale 2015, 7, 19611–19619. [Google Scholar] [CrossRef]
- Li-na, M.A.; Dian-Jun, L.I.U.; Zhen-Xin, W. Synthesis and Applications of Gold Nanoparticle Probes. Chin. J. Anal. Chem. 2010, 38, 1–7. [Google Scholar]
- Oh, J.; Park, D.H.; Joo, J.H.; Lee, J. Recent advances in chemical functionalization of nano-particles with biomolecules for analytical applications. Anal. Bioanal. Chem. 2015, 407, 8627–8645. [Google Scholar] [CrossRef] [PubMed]
- Perfilieva, O.A.; Pyshnyi, D.V.; Lomzov, A.A. Molecular Dynamics Simulation of Polarizable Gold Na-noparticles Interacting with Sodium Citrate. J. Chem. Theory Comput. 2019, 15, 1278–1292. [Google Scholar] [CrossRef] [PubMed]
- Sapsford, K.E.; Algar, W.R.; Lorenzo, B.; Gimmill, K.B.; Casey, B.J.; Oh, E.; Stewart, M.H.; Medintz, I.L. Functionalizing nanoparticles with biological molecules developing chemis-tries that facilitate nanotechnology. Chem. Rev. 2013, 113, 1904–2074. [Google Scholar] [CrossRef]
- Barnaby, S.N.; Perelman, G.A.; Kohlstedt, K.L.; Chinen, A.B.; Schatz, G.C.; Mirkin, C.A. Design Considerations for RNA Spherical Nucleic Acids (SNAs). Bioconjug. Chem. 2016, 27, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Servos, M.R.; Liu, J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef] [PubMed]
- Epanchintseva, A.; Vorobjev, P.; Pyshnyi, D.; Pyshnaya, I. Fast and Strong Adsorption of Native Oligonucleotides on Citrate-Coated Gold Nanopar-ticles. Langmuir 2018, 34, 164–172. [Google Scholar] [CrossRef]
- Epanchintseva, A.; Dolodoev, A.; Grigor’eva, A.; Chelobanov, B.; Pyshnyi, D.; Ryabchikova, E.; Pyshnaya, I. Non-covalent binding of nucleic acids with gold nanoparticles provides their stability and effective desorption in environment mimicking biological media. Nanotechnology 2018, 29, 355601. [Google Scholar] [CrossRef]
- Poletaeva, J.; Dovydenko, I.; Epanchintseva, A.; Korchagina, K.; Pyshnyi, D.; Apartsin, E.; Ryabchikova, E.; Pyshnaya, I. Non-covalent associates of siRNAs and AuNPs enveloped with lipid layer and doped with amphiphilic pep-tide for efficient siRNA delivery. Int. J. Mol. Sci. 2018, 19, 2096. [Google Scholar] [CrossRef] [PubMed]
- Epanchintseva, A.; Poletaeva, J.; Pyshnyi, D.; Ryabchikova, E.; Pyshnaya, I. Long-term stability and scale-up of noncovalently bound gold nanoparticle-siRNA suspensions. Beilstein J. Nanotechnol. 2019, 10, 2568–2578. [Google Scholar] [CrossRef]
- Vorobjev, P.; Epanchintseva, A.; Lomzov, A.; Tupikin, A.; Kabilov, M.; Pyshnaya, I.; Pyshnyi, D. DNA Binding to Gold Nanoparticles through the Prism of Molecular Selection: Sequence—Affinity Relation. Langmuir 2019, 35, 7916–7928. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lu, Y.; Mao, J.; Chang, N.; Yang, J.; Liu, Y. Multidimensional sensor for pattern recognition of proteins based on DNA-gold nanoparticles conjugates. Anal. Chem. 2015, 87, 3354–3359. [Google Scholar] [CrossRef]
- Liu, J. Adsorption of DNA onto gold nanoparticles and graphene oxide: Surface science and applications. Phys. Chem. Chem. Phys. 2012, 14, 10485–10496. [Google Scholar] [CrossRef] [PubMed]
- Pylaev, T.E.; Volkova, E.K.; Kochubey, V.I.; Bogatyrev, V.A.; Khlebtsov, N.G. DNA detection assay based on fluorescence quenching of rhodamine B by gold nano-particles: The optical mechanisms. J. Quant. Spectrosc. Radiat. Transf. 2013, 131, 34–42. [Google Scholar] [CrossRef]
- Shashkova, V.V.; Epanchintseva, A.V.; Vorobjev, P.E.; Razum, K.V.; Ryabchikova, E.I.; Pyshnyi, D.V.; Pyshnaya, I.A. Multilayer associates based on oligonucleotides and gold nanoparticles. Russ. J. Bioorg. Chem. 2017, 43, 64–70. [Google Scholar] [CrossRef]
- Giusti, W.G.; Adriano, T. Synthesis and characterization of 5’-fluorescent-dye-labeled oli-gonucleotides. PCR Methods Appl. 1993, 2, 223–227. [Google Scholar] [CrossRef]
- Astakhova, I.K.; Wengel, J. Interfacing Click Chemistry with Automated Oligonucle-otide Synthesis for the Preparation of Fluorescent DNA Probes Containing Internal Xan-thene and Cyanine Dyes. Chem. A Eur. J. 2013, 19, 3528. [Google Scholar] [CrossRef]
- Borer, P.N. Optical properties of nucleic acids, adsorption, and circular dicroism spectra. In Handbook of Biochemistry and Molecular Biology: Nucleic Acids; Fasman, T.E., Ed.; CRC Press: Cleveland, OH, USA, 1975; Volume 1, pp. 589–595. [Google Scholar]
- Glen Research: Oligonucleotide Synthesis Supplies & Supports. Available online: https://www.glenresearch.com (accessed on 31 March 2021).
- Laser Photomedicine and Biomedical Optics at the Oregon Medical Laser Center. Available online: http://www.omlc.org (accessed on 31 March 2021).
- Thermo Fisher Scientific. Available online: https://www.thermofisher.com (accessed on 31 March 2021).
- Vailaya, A.; Horva´th, C. Retention Thermodynamics in Hydrophobic In-teraction Chromatography. Ind. Eng. Chem. Res. 1996, 35, 2964–2981. [Google Scholar] [CrossRef]
- Vailaya, A. Fundamentals of Reversed Phase Chromatography: Thermodynamic and Exothermodynamic Treatment. J Liq. Chrom. Relat. Tech. 2005, 28, 965–1054. [Google Scholar] [CrossRef]
- Wei Tsai, C.; Yih Chen, W.; Chyu Ruaan, R. Retention Prediction of Peptide Diastereomers in Reversed-Phase Liquid Chromatography Assisted by Molecular Dynamics Simulation. Langmuir 2012, 28, 13601–13608. [Google Scholar] [CrossRef]
- Frens, G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Liu, X.; Atwater, M.; Wang, J.; Huo, Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf. B Biointerfaces 2007, 58, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Ryder, S.P.; Recht, M.I.; Williamson, J.R. Quantitative Analysis of Protein-RNA Interactions by Gel Mobility Shift. In RNA-Protein Interaction Protocols. Methods in Molecular Biology; Lin, R.J., Ed.; Humana Press: Totowa, NJ, USA, 2008; Volume 488, pp. 99–115. [Google Scholar] [CrossRef]
- Golyshev, V.M.; Abramova, T.V.; Pyshnyi, D.V.; Lomzov, A.A. A new approach to precise thermodynamic characterization of hybridization properties of modified oligonucleotides: Comparative studies of deoxyribo- and glycine morpholine pentaadenines. Biophys. Chem. 2018, 234, 24–33. [Google Scholar] [CrossRef]
- Amaya-González, S.; López-López, L.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Affinity of aptamers binding 33-mer gliadin peptide and gluten proteins: Influence of immobilization and labeling tags. Anal. Chim. Acta 2015, 873, 63–70. [Google Scholar] [CrossRef]
- Tuleuova, N.; Jones, C.N.; Yan, J.; Ramanculov, E.; Yokobayashi, Y.; Revzin, A. Development of an Aptamer Beacon for Detection of Interferon-Gamma. Anal. Chem. 2010, 82, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Wang, J.; Jasinski, J.B.; Achilefu, S. Fluorescence Manipulation by Gold Nanoparticles: From Complete Quenching to Extensive Enhancement. J. Nanobiotechnol. 2011, 9, 16. [Google Scholar] [CrossRef]
- Borovok, N.; Gillon, E.; Kotlyar, A. Synthesis and Assembly of Conju-gates Bearing Specific Numbers of DNA Strands per Gold Nanoparticle. Bioconjugate Chem. 2012, 23, 916–922. [Google Scholar] [CrossRef]
- Kupryushkin, M.S.; Nekrasov, M.D.; Stetsenko, D.A.; Pyshnyi, D.V. Ef-ficient Functionalization of Oligonucleotides by New Achiral Nonnucleosidic Monomers. Org. Lett. 2014, 16, 2842–2845. [Google Scholar] [CrossRef]
- Pavlova, A.S.; Dovydenko, I.S.; Kupryushkin, M.S.; Grigor’eva, A.E.; Pyshnaya, I.A.; Pyshnyi, D.V. Amphiphilic “Like-A-Brush” Oligonucleotide Conjugates with Three Dodecyl Chains: Self-Assembly Features of Novel Scaffold Compounds for Nucleic Acids Delivery. Nanomaterials 2020, 10, 1948. [Google Scholar] [CrossRef]
- Markov, O.V.; Filatov, A.V.; Kupryushkin, M.S.; Chernikov, I.V.; Patutina, O.A.; Strunov, A.A.; Chernolovskaya, E.L.; Vlassov, V.V.; Pyshnyi, D.V.; Zenkova, M.A. Transport Oligonucleotides—A Novel System for Intracellular Delivery of Antisense Therapeutics. Molecules 2020, 25, 3663. [Google Scholar] [CrossRef]
- Verma, J.; Khedkar, V.M.; Coutinho, E.C. 3D-QSAR in Drug Design—A Review. Curr. Top Med. Chem. 2010, 10, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Santa Lucia, J., Jr. How much free energy is absorbed upon breaking DNA base pairs? Comment on “DNA melting and energetics of the double helix” by Maxim Frank-Kamenetskii. Phys. Life Rev. 2018, 25, 29–33. [Google Scholar] [CrossRef]
- Santa Lucia, J., Jr.; Hicks, D. The thermodynamics of DNA structuralmotifs. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 415–440. [Google Scholar] [CrossRef]
- Lomzov, A.A.; Pyshnyi, D.V. Considering the oligonucleotide secondary structures in thermodynamic and kinetic analysis of DNA duplex formation. Biophysics 2012, 57, 19–34. [Google Scholar] [CrossRef]
- Gu, X.-B.; Nakano, S.; Sugimoto, N. Consecutive GC base pairs determine the energy barrier of DNA duplex formation under molecularly crowded conditions. Chem. Commun. 2007, 26, 2750–2752. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.-C.; Macgregor, R.B., Jr. Activation Volume of DNA Duplex Formation. Biochemistry 1997, 36, 6539–6544. [Google Scholar] [CrossRef] [PubMed]

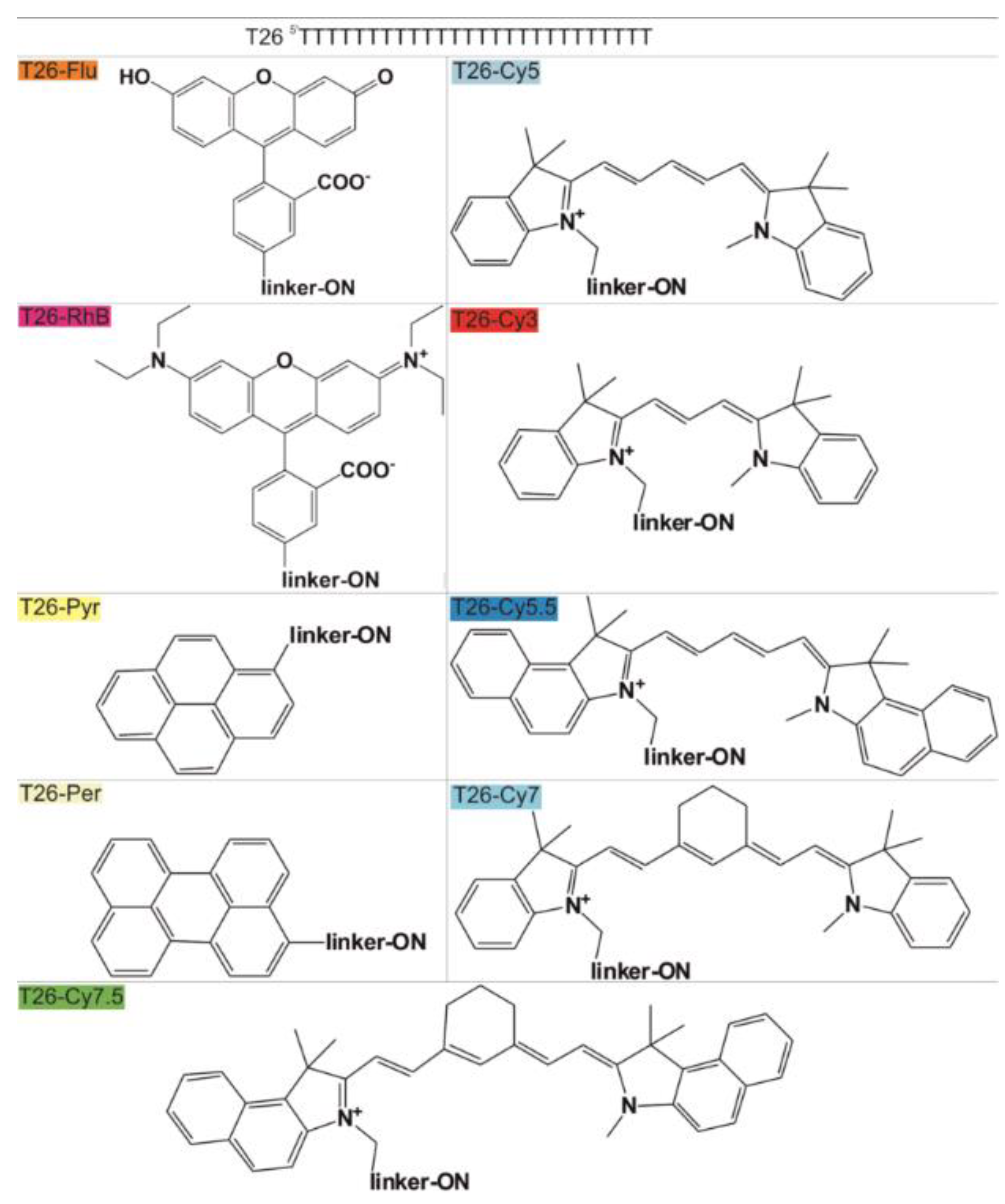
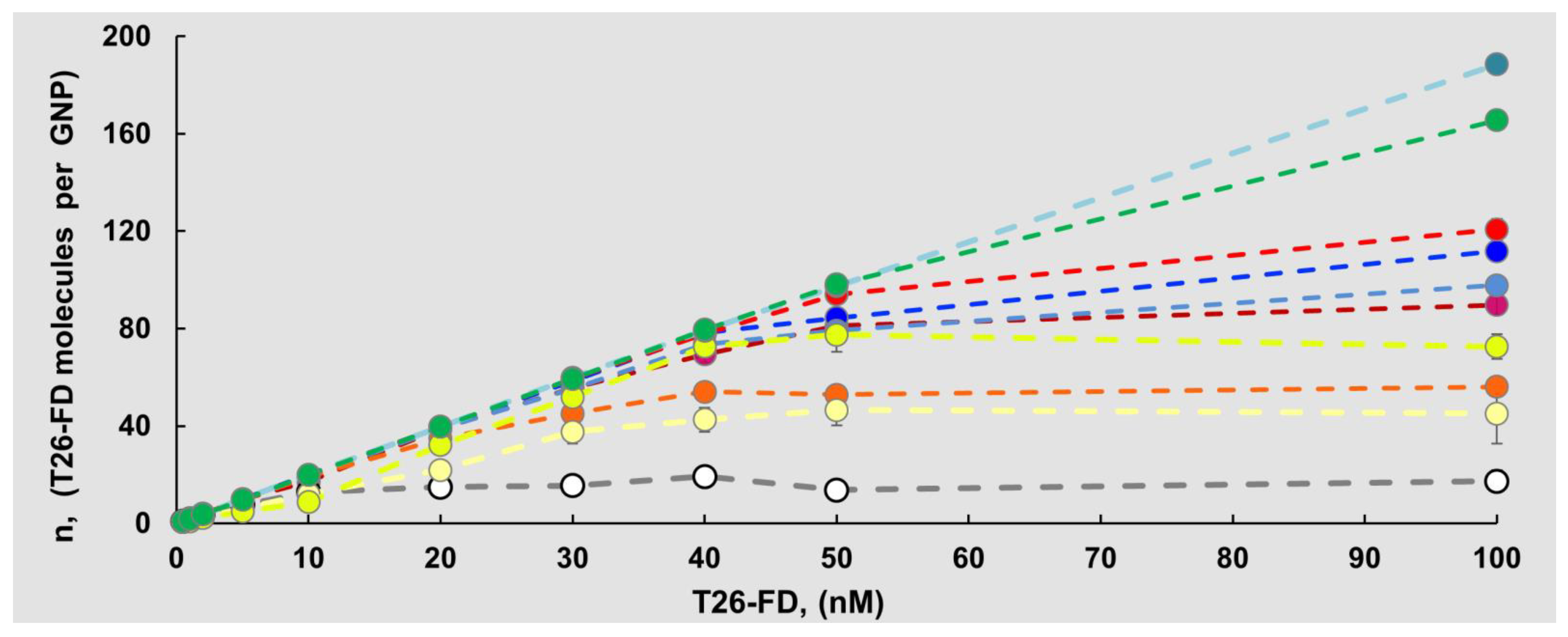


| ON-FD | KL (nM−1) | n (Molecules per Particle) | KD * (nM) |
|---|---|---|---|
| T26 (without FD) | 0.147 ± 0.0510 | 17 ± 5 | - |
| T26-Pyr | 0.032 ± 0.0110 | 45 ± 13 | 0.181 ± 0.074 |
| T26-Per | 0.023 ± 0.0120 | 73 ± 5 | - |
| T26-Flu | 0.043 ± 0.0100 | 56 ± 4 | 0.224 ± 0.021 |
| T26-RhB | 0.021 ± 0.0050 | 90 ± 4 | - |
| T26-Cy3 | 0.012 ± 0.0020 | 121 ± 4 | - |
| T26-Cy5 | 0.014 ± 0.0020 | 112 ± 4 | 0.064 ± 0.005 |
| T26-Cy5.5 | 0.018 ± 0.0030 | 98 ± 2 | - |
| T26-Cy7.5 | 0.003 ± 0.0003 | 166 ± 2 | - |
| T26-Cy7 | 0.00077 ± 0.00004 | 189± 1 | - |
| ON-FD | Sequence (5′ → 3′) | KL (nM−1) | n (Molecules per Particle) |
|---|---|---|---|
| *-Flu | GATATGATGACGTTAGTTAG-Flu | 0.0286 ± 0.0056 | 69 ± 7 |
| N*1-Flu | TTTTTTGATATGATGACGTTAGTTAG-Flu | 0.0312 ± 0.0074 | 69 ± 5 |
| N*2-Flu | TTGTTGGATATGATGACGTTAGTTAG-Flu | 0.0300 ± 0.0065 | 70 ± 4 |
| FAM-D-T25 | Flu-D-TTTTTTTTTTTTTTTTTTTTTTTTT | 0.0503 ± 0.0175 | 54 ± 2 |
| FAM-DD-T24 | Flu-DD-TTTTTTTTTTTTTTTTTTTTTTTT | 0.0410 ± 0.0143 | 66 ± 2 |
| FAM-DDD-T23 | Flu-DDD- TTTTTTTTTTTTTTTTTTTTTTT | 0.0600 ± 0.0243 | 49 ± 1 |
| A26-FITC | AAAAAAAAAAAAAAAAAAAAAAAAAA-Flu | 0.0251 ± 0.0060 | 84 ± 4 |
| A26-Pyr | AAAAAAAAAAAAAAAAAAAAAAAAAA-Pyr | 0.0240 ± 0.0072 | 89 ± 3 |
| A26-Cy5 | AAAAAAAAAAAAAAAAAAAAAAAAAA-Cy5 | 0.0106 ± 0.0025 | 126 ± 1 |
| C26-FITC | CCCCCCCCCCCCCCCCCCCCCCCCCC-Flu | 0.0157 ± 0.0036 | 110 ± 1 |
| C26-Cy5 | CCCCCCCCCCCCCCCCCCCCCCCCCC-Cy5 | 0.0041 ± 0.0005 | 161 ± 4 |
| T40 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | 0.4299 ± 0.1270 | 5 ± 3 |
| T6 | TTTTTT | 0.0026 ± 0.0004 | 149 ± 16 |
| ln(KLobs) | ln(KLcalc) | Std. Err. | n | ln(Hobs) | Zd | Mr | S | Ztot | (D)k | 5′End Dye | Dye | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T26 | −1.917 | −1.788 | 0.149 | 17 | 1.403 | 0 | 0 | 0 | −25 | 0 | 0 | 0 |
| T40 | −0.844 | −0.936 | 0.224 | 5 | 1.408 | 0 | 0 | 0 | −39 | 0 | 0 | 0 |
| T6 | −5.934 | −5.961 | 0.234 | 149 | 1.367 | 0 | 0 | 0 | −5 | 0 | 0 | 0 |
| T26-Pyr | −3.442 | −3.063 | 0.132 | 45 | 1.812 | −1 | 466 | 399 | −26 | 0 | 0 | 1 |
| T26-Per | −3.772 | −3.742 | 0.089 | 73 | 1.822 | −1 | 744 | 446 | −26 | 0 | 0 | 1 |
| T26-RdB | −3.863 | −3.908 | 0.090 | 90 | 1.892 | −1 | 695 | 736 | −26 | 0 | 0 | 1 |
| T26-Flu | −3.147 | −4.449 | 0.086 | 56 | 1.503 | −2 | 584 | 513 | −27 | 0 | 0 | 1 |
| T26-Cy3 | −4.423 | −4.291 | 0.093 | 121 | 1.902 | 0 | 647 | 642 | −25 | 0 | 0 | 1 |
| T26-Cy5 | −4.269 | −4.069 | 0.143 | 112 | 2.003 | 0 | 673 | 710 | −25 | 0 | 0 | 1 |
| T26-Cy5.5 | −4.017 | −3.089 | 0.093 | 98 | 2.177 | 0 | 773 | 818 | −25 | 0 | 0 | 1 |
| T26-Cy7 | −7.169 | −6.463 | 0.139 | 189 | 2.177 | 0 | 739 | 797 | −25 | 0 | 0 | 1 |
| T26-Cy7.5 | −5.684 | −6.056 | 0.117 | 166 | 2.378 | 0 | 826 | 903 | −25 | 0 | 0 | 1 |
| Flu-T26 | −2.813 | −2.836 | 0.096 | 49 | 1.461 | −2 | 456 | 513 | −27 | 0 | 1 | 1 |
| Flu-D-T25 | −2.990 | −3.070 | 0.091 | 54 | 1.899 | −2 | 559 | 513 | −27 | 1 | 1 | 1 |
| Flu-DD-T24 | −3.194 | −3.345 | 0.140 | 66 | 2.245 | −2 | 662 | 513 | −27 | 2 | 1 | 1 |
| Flu-DDD-T23 | −2.814 | −2.686 | 0.211 | 49 | 2.474 | −2 | 765 | 513 | −27 | 3 | 1 | 1 |
| N*-Flu | −3.540 | −3.511 | 0.103 | 69 | 1.412 | −2 | 456 | 513 | −21 | 0 | 0 | 1 |
| N*1-Flu | −3.507 | −3.281 | 0.098 | 70 | 1.400 | −2 | 456 | 513 | −27 | 0 | 0 | 1 |
| N*2-Flu | −3.474 | −3.279 | 0.096 | 69 | 1.415 | −2 | 456 | 513 | −27 | 0 | 0 | 1 |
| A26-Flu | −3.686 | −3.771 | 0.102 | 84 | 1.470 | −2 | 584 | 513 | −27 | 0 | 0 | 1 |
| A26-Pyr | −3.729 | −4.139 | 0.078 | 89 | 1.780 | −1 | 466 | 399 | −26 | 0 | 0 | 1 |
| A26-Cy5 | −4.543 | −4.656 | 0.090 | 126 | 2.004 | 0 | 673 | 710 | −25 | 0 | 0 | 1 |
| C26-Flu | −4.152 | −4.424 | 0.135 | 110 | 1.470 | −2 | 584 | 513 | −27 | 0 | 0 | 1 |
| C26-Cy5 | −5.504 | −5.576 | 0.119 | 161 | 2.012 | 0 | 673 | 710 | −25 | 0 | 0 | 1 |
| Factors, F j | Std. Err. | p-Level | |
|---|---|---|---|
| n | −0.02598 | 0.00214 | 4.1 × 10−10 |
| ln(H) | −1.66333 | 0.28448 | 1.5 × 10−5 |
| Zd | 0.44722 | 0.07271 | 8.3 × 10−6 |
| S | 0.00136 | 0.00033 | 7.2 × 10−4 |
| Ztot | −0.03948 | 0.01328 | 8.1 × 10−3 |
| (D)k | 0.61519 | 0.13007 | 1.7 × 10−4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Epanchintseva, A.V.; Gorbunova, E.A.; Ryabchikova, E.I.; Pyshnaya, I.A.; Pyshnyi, D.V. Effect of Fluorescent Labels on DNA Affinity for Gold Nanoparticles. Nanomaterials 2021, 11, 1178. https://doi.org/10.3390/nano11051178
Epanchintseva AV, Gorbunova EA, Ryabchikova EI, Pyshnaya IA, Pyshnyi DV. Effect of Fluorescent Labels on DNA Affinity for Gold Nanoparticles. Nanomaterials. 2021; 11(5):1178. https://doi.org/10.3390/nano11051178
Chicago/Turabian StyleEpanchintseva, Anna V., Ekaterina A. Gorbunova, Elena I. Ryabchikova, Inna A. Pyshnaya, and Dmitrii V. Pyshnyi. 2021. "Effect of Fluorescent Labels on DNA Affinity for Gold Nanoparticles" Nanomaterials 11, no. 5: 1178. https://doi.org/10.3390/nano11051178
APA StyleEpanchintseva, A. V., Gorbunova, E. A., Ryabchikova, E. I., Pyshnaya, I. A., & Pyshnyi, D. V. (2021). Effect of Fluorescent Labels on DNA Affinity for Gold Nanoparticles. Nanomaterials, 11(5), 1178. https://doi.org/10.3390/nano11051178







